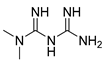
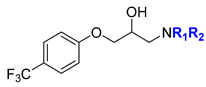
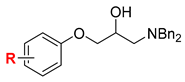
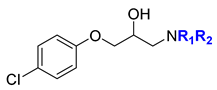
Abstract
Targeting growth differentiation factor 15 (GDF15) is a recent strategy for the treatment of obesity and type 2 diabetes mellitus (T2DM). Here, we designed, synthesized, and pharmacologically evaluated in vitro a novel series of AMPK activators to upregulate GDF15 levels. These compounds were structurally based on the (1-dibenzylamino-3-phenoxy)propan-2-ol structure of the orphan ubiquitin E3 ligase subunit protein Fbxo48 inhibitor, BC1618. This molecule showed a better potency than metformin, increasing GDF15 mRNA levels in human Huh-7 hepatic cells. Based on BC1618, structural modifications have been performed to create a collection of diversely substituted new molecules. Of the thirty-five new compounds evaluated, compound 21 showed a higher increase in GDF15 mRNA levels compared with BC1618. Metformin, BC1618, and compound 21 increased phosphorylated AMPK, but only 21 increased GDF15 protein levels. Overall, these findings indicate that 21 has a unique capacity to increase GDF15 protein levels in human hepatic cells compared with metformin and BC1618.
1. Introduction
Growth differentiation factor 15 (GDF15) (also known as nonsteroidal anti-inflammatory drug-activated gene, placental bone morphogenetic protein, placental transforming growth factor-ß, and prostate-derived factor) is a stress-induced cytokine that is involved in appetite regulation [1]. In fact, the increase in GDF15 levels promotes an anorectic effect that leads to a reduction in body weight through its binding to its central receptor glial cell-line-derived neurotrophic factor (GDNF)-like alpha-1 (glial cell-derived neurotrophic factor family receptor alpha-like (GFRAL)) [1]. Consistent with this role of GDF15, transgenic mice overexpressing Gdf15 display a lean phenotype and a reduction in food intake and are more resistant to obesity, metabolic inflammation, and glucose intolerance [2,3]. Likewise, administration of recombinant GDF15 (rGDF15) has been reported to reduce food intake and body weight in mice, but these effects are absent in Gfral-knockout mice [2,4,5]. Overall, these findings suggest that pharmacological modulation of GDF15 shows promise for the treatment of obesity and its complications, such as type 2 diabetes mellitus (T2DM). Indeed, GDF15 analogs, which try to overcome the pharmacokinetic (e.g., a short half-life of ~3 h in mice and non-human primates) and physicochemical (e.g., high aggregation propensity) limitations of native GDF15 [1,6,7], have been developed for the treatment of obesity. Another potential strategy to target the GDF15–GFRAL pathway involves the pharmacological regulation of endogenous GDF15 levels. Interestingly, metformin, the most prescribed drug for the treatment of T2DM, has been reported to increase serum GDF15 in patients [8]. Subsequently, two studies reported that the anorectic response of mice to metformin requires an intact GDF15–GFRAL pathway [9,10]. In addition, the increase in GDF15 levels caused by metformin was associated with the reduction in body weight in patients with [10] or without [9] T2DM. Although a recent study reported that the effects of metformin on body weight were independent of the GDF15–GFRAL signaling in some circumstances [11], a new study confirmed the involvement of this pathway on the effects of metformin on body weight [12]. These findings validate that targeting endogenous GDF15 may serve as a pharmacological approach to treat obesity and T2DM. Interestingly, many of the antidiabetic effects of metformin are mediated by the activation of the adenosine monophosphate-activated protein kinase (AMPK) [13], and we reported that the increase in GDF15 caused by metformin involves the activation of this kinase [14]. Moreover, we observed that the antidiabetic effect of a low dose of metformin is not observed in Gdf15-knockout mice [14]. These findings suggest that targeting AMPK may provide new possibilities to increase endogenous levels of GDF15 to treat obesity and T2DM.
In this study, we designed, synthesized, and pharmacologically evaluated in vitro a novel series of AMPK activators, aiming at increasing GDF15 expression. These compounds are structurally based on the (1-dibenzylamino-3-phenoxy)propan-2-ol skeleton of the orphan ubiquitin E3 ligase subunit protein Fbxo48 inhibitor, BC1618, which exceeds metformin potency for stimulating AMPK [15]. In this context, we carried out conservative structural modifications on the substituents in the phenoxy ring and in the amine moiety. Our findings show that one of the new compounds synthesized, 21, exhibits substantially better potency compared to metformin and BC1618 towards increased GDF15 mRNA and protein levels in human hepatic cells.
2. Results
2.1. Synthesis of GDF15 Inducers
The synthesis of BC1618 was performed as described [16]. The other products were synthetized following the same synthetic strategy (Scheme 1). In this manner, a methanol solution of the required phenol (1 equiv.) with epichlorohydrin (10 equiv.) in the presence of potassium carbonate (1.2 equiv.) was heated at reflux for 4 h, leading to the corresponding phenoxymethyloxirane intermediates. In a second synthetic step, an equimolecular mixture of the phenoxymethyloxirane derivatives and the required primary or secondary amine were heated at 70 °C for 24 h to afford the final compounds (1–35) in good yields (see Section 4 for further details).
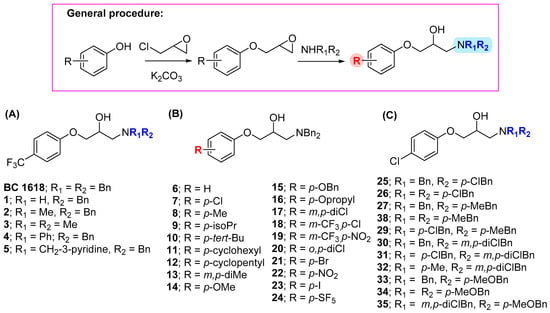
Scheme 1.
General procedure for the synthesis of compounds (1–35) with modifications in the amino substituents (in blue) and in the substituents of the phenoxy group (in red). General structures, (A) variations in the substituents in the amino group of the p-trichlorophenyl general structure, (B) variations in the substituents in the phenoxy group, and (C) variations in the substituents in the amino group of the p-chlorophenyl general structure.
All the compounds synthesized for in vitro evaluation were fully characterized through their spectroscopic data (1H and 13C), infrared (IR), and high-resolution mass spectrometry (HMRS). The purity was determined by high-performance liquid chromatography (HPLC)/mass spectrometry, and only compounds with a purity >95% were considered for biological studies (see the Section 4 and the Supplementary Materials for further details).
2.2. Assessment of the Activity of the New Compounds Increasing GDF15 mRNA Levels in Huh-7 Human Hepatic Cells and Structural–Activity Relationship of the New Compounds
To evaluate the activity of the synthesized compounds (Scheme 1A–C) in increasing GDF15 expression, we incubated Huh-7 human hepatic cells with the new compounds (30 µM) for 24 h, using metformin (5 mM) and BC1618 (30 µM) as standards for comparison purposes. Hepatic cells were used since circulating GDF15 is primarily derived from the liver [17]. As shown in Table 1, metformin caused a 2.7-fold increase in GDF15 mRNA levels compared with control cells. By contrast, a much lower concentration of BC1618 caused a higher increase in GDF15 expression (10.4-fold induction), thereby indicating that this compound shows a higher potency, which is consistent with the reported higher activation of AMPK [15].

Table 1.
Assessment of the effects of the compounds 1–35 on human GDF15 expression. GDF15 mRNA levels in the human liver cell line Huh-7 exposed to 5 mM metformin, 30 µM BC1618, or 30 µM of new compound for 24 h. Data are presented as the mean ± SEM (n = 3). * p < 0.05, ** p < 0.01, and *** p < 0.001 vs. control. p-values determined by one-way ANOVA with Tukey’s post hoc test.
To determine the impact that modifications in the different substituents could have in the biological activities (Table 1), we considered two main approximations starting from BC1618: (a) an exploration of the substituents in the amino moiety (right-hand side, RHS; highlighted in blue in the general structure in the Scheme 1) and (b) a study of the substituents at the phenoxy group (left-hand side, LHS; highlighted in red in the general structure in the Scheme 1).
In this context, the simplification of the dibenzylamino substituent of BC1618 to a benzylamino was considered, leading to compound 1, which exhibited important increases in GDF15 expression compared with BC1618 but also increased cell mortality, suggesting some toxicity that rendered it unsuitable for further work. Compared with BC1618, an N-benzyl,N-methylamine substituent in 2 or N,N-dimethylamine in 3 resulted in substantial reductions in the expression of GDF15. An N-benzyl-N-phenylamino moiety in 4 caused no significant changes in GDF15 levels compared with BC1618, whereas the presence of an electron-deficient aromatic ring such as a 3-pyridine in 5 caused a dramatic reduction in GDF15 levels.
At this point, we maintained unaltered the RHS part of the initial scaffold (N-dibenzylamino) and focused on the modifications of the substituents at the phenoxy moiety. Removal of the trifluoromethyl group, as in 6, had no effect on GDF15 levels compared with BC1618. Comprehensive exploration of the RHS involved the replacement of the trifluoromethyl group by a chlorine atom in 7 or a methyl group in 8, leading to small increases in the GDF15 mRNA levels. Other replacements, featuring bulkier p-alkyl substituents, such as those conducted in 9 (isopropyl), 10 (tert-butyl), 11 (cyclohexyl), or 12 (cyclopentyl), did not improve the levels of the biomarker. Compound 13, featuring a 3,4-dimethylphenoxy group, exhibited a remarkable increase in the GDF15 levels, although it was somehow toxic, showing an increase in the cell mortality. Compound 14, embodying a p-methoxy group, depicted similar activity as BC1618. With the aim of exploring alternative p-alkyloxy substituents, 15 with a p-benzyloxy group and 16 with a p-propyloxy unit were synthesized, leading to a significant reduction in GDF15 expression compared with BC1618. Next, we undertook the synthesis of disubstituted compounds 17–19, featuring m,p-dichloro-, p-chloro,m-trifluoromethyl-, and p-nitro,m-trifluoromethyl-phenoxy units, respectively. The three new compounds did not improve the activity of BC1618. The presence of a o,p-dichloro atoms at the phenoxy group in 20 led to an increase in GDF15 expression.
Considering the abovementioned results, in a final round, we selected compound 7, with a p-chlorophenoxy group, and we introduced substituents in the dibenzylamino moiety. Therefore, we synthesized eleven new analogs, including 25 and 26, with a p-chloro in one or two of the benzyl groups, respectively; 27 and 28, with a p-methyl in one or two of the benzyl groups, respectively; and 29, with a p-chloro and a p-methyl group in each benzyl group. All these compounds depicted a very important decrease in the GDF15 levels compared with BC1618. Particularly, 25 was toxic for the cells, and the number of cells collected was too low to analyze GDF15 expression. Increasing the number of chloro atoms in the benzyl substituents, as in 30 (m,p-dichloro), 31 (m,p-dichloro and p-chloro), and 32 (m,p-dichloro and p-methyl), resulted in a dramatical decrease in GDF15 levels. Finally, moving to electron-donating groups, such as a p-methoxy group (33 in one benzyl or 34 in both benzyl groups), or to a combination of m,p-dichloro on one ring and p-methoxy in the other ring (35) was deleterious for the biological activity.
Considering that compound 7, featuring a dibenzylamino unit at the RHS of the molecule and a p-chlorophenoxy group at the LHS, provided the highest GDF15 levels without toxicity, we finally explored the effect of replacing the p-chlorine atom by another electron-withdrawing group. Thus, we synthesized compound 21, with a p-bromophenoxy group. Interestingly, this compound exhibited a marked increase in GDF15 expression without toxicity issues. When a nitro group or an iodine atom occupied the para position of the phenoxy group in 22 and 23, respectively, a decrease in GDF15 levels was observed. Our experience in the study of the rather scarcely explored pentafluorosulfanyl substituent [18] prompted us to prepare compound 24, which did not represent a biological improvement. Considering all the above-mentioned results, compound 21 was selected for further experiments.
2.3. Evaluation of the Activity of 21 on AMPK Activation and GDF15 Protein Levels in Huh-7 Human Hepatic Cells
The effect of compound 21 on GDF15 protein levels compared with metformin and BC1618 was evaluated in Huh-7 cells. The three compounds activated AMPK, determined by the increase in phosphorylated AMPK. However, of the three compounds, only 21 increased the protein abundance of GDF15 (Figure 1).
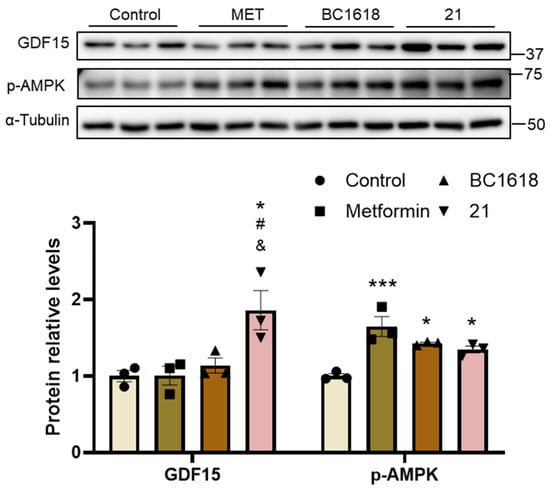
Figure 1.
Compound 21 increases GDF15 protein abundance. GDF15 and phosphorylated AMPK protein levels in Huh-7 human hepatic cells exposed to metformin (MET) (5 mM), BC1618 (10 µM), or 21 (10 µM) for 72 h. Data are presented as the mean ± SEM (n = 3). * p < 0.05, and *** p < 0.001 vs. control. # p < 0.05 vs. metformin-treated cells. & p < 0.05 vs. BC1618-treated cells. p-values determined by one-way ANOVA with Tukey’s post hoc test.
3. Discussion
There is conclusive evidence that GDF15 is an attractive target for the treatment of obesity and T2DM [1]. Indeed, recombinant GDF15 and its analogs reduce food intake and body weight and improve glucose intolerance in animal models of obesity [1,6,7]. In addition, metformin increases circulating GDF15 levels [8]. The metformin-mediated increase in GDF15 levels may mediate part of its effects on body weight and glucose metabolism [9,10,14]. Therefore, pharmacological modulation of endogenous GDF15 levels by small molecules offers promise for the treatment of obesity and T2DM. In this line, it has been reported that metformin increases GDF15 levels via AMPK [14], converting this kinase in a target to increase this cytokine. In this study, we synthesized new compounds based on the (1-dibenzylamino-3-propoxy)propan-2-ol group of the orphan ubiquitin E3 ligase subunit protein Fbxo48 inhibitor, BC1618 [15], to upregulate GDF15 levels. It is known that the potency of BC1618 activating AMPK exceeds that of metformin [15], but little was known about the capacity of this compound to increase GDF15.
Here, we show that BC1618 (10 µM) causes a strong increase in GDF15 mRNA levels in human Huh-7 cells compared with metformin (5 mM). In addition, we synthesized and fully characterized thirty-five new analogues of BC1618, and we evaluated their effects on GDF15 expression. All the new compounds increased GDF15 mRNA levels over the control, and a few of them were more potent than BC1618.
Compound 21, which features a bromine atom instead of the trifluoromethyl group of BC1618, caused a higher increase in GDF15 levels compared with BC1618 and was selected to evaluate its effects on GDF15 protein abundance compared with metformin and BC1618. The three compounds increased phosphorylated levels of AMPK. However, only 21 increased GDF15 protein levels. Although previous studies have reported that metformin administration increases serum GDF15 levels, the source tissue leading to the increase in this cytokine remains controversial [19]. Thus, two studies reported that metformin can induce GDF15 release from mouse primary hepatocytes [9,10], but one of these studies showed that metformin increased GDF15 levels in kidneys and intestines but not in the liver [9]. More recently, a new study demonstrated that acute metformin treatment elevated GDF15 levels in different tissues but not in the liver [11]. Therefore, it is likely that due to the slight increase in GDF15 expression caused by metformin, longer exposures are needed to detect an increase in GDF15 protein abundance in hepatic cells. However, BC1618 caused a higher increase in GDF15 mRNA levels compared to metformin, but this increase did not result in an elevation of the protein abundance of this cytokine. By contrast, 21 treatment increased GDF15 protein levels. These findings suggest that the substitution of the trifluoromethyl group by a bromine atom may contribute to increase GDF15 protein abundance. Although the reasons for this effect are unknown, several factors may contribute. For instance, a bigger atom such as bromine may affect the ability to interact with target proteins and receptors. In fact, a study investigating the effects of 3,3′-diindolylmethane and its synthetic halogenated derivatives found that a bromide derivative caused a higher activation of AMPK [20]. These findings suggest that the presence of bromide atoms in 21 could potentiate or sustain AMPK activation, leading to an increase in GDF15 protein levels. Although we did not observe a greater increase in phosphorylated AMPK following 21 treatment compared with BC1618, we only examined a single time point. Therefore, transient increases might have contributed to achieve a higher potency in AMPK activation. Further studies are needed to elucidate the mechanisms that contribute to increasing GDF15 protein levels.
Overall, the findings of this study show that there is no correlation between GDF15 mRNA and protein levels and that the presence of a bromine atom in the structure of molecules activating AMPK and increasing GDF15 expression allows to increase the protein abundance of this stress cytokine in hepatic cells. Moreover, we describe that new compound 21 has a unique capacity to increase GDF15 protein levels in human hepatic cells compared with metformin and BC1618.
4. Materials and Methods
4.1. Chemical Synthesis
4.1.1. General Methods
Reagents, solvents, and starting products were acquired from commercial sources. When indicated, the reaction products were purified by “flash” chromatography on silica gel (35–70 μm) with the indicated solvent system. The melting points were measured in a MFB 59510M Gallenkamp instruments. IR spectra were performed in a spectrophotometer Nicolet Avantar 320 FTR-IR or in a Spectrum Two FT-IR Spectrometer, and only noteworthy IR absorptions (cm−1) are listed. NMR spectra were recorded in CDCl3 at 400 MHz (1H) and 101 MHz (13C), and chemical shifts are reported in δ values downfield from TMS or relative to residual chloroform (7.26 ppm, 77.0 ppm) as an internal standard. Data are reported in the following manner: chemical shift, multiplicity, coupling constant (J) in hertz (Hz), and integrated intensity and assignment (when possible). Multiplicities are reported using the following abbreviations: s, singlet; d, doublet; dd, doublet of doublets; t, triplet; q, quadruplet; m, multiplet; br s, broad signal; app, apparent. Assignments and stereochemical determinations are given only when they are derived from definitive dimensional NMR experiments (g-HSQC). The accurate mass analyses were carried out using a LC/MSD-TOF spectrophotometer. HPLC-MS (Agilent 1260 Infinity II) analysis was conducted on a Poroshell 120 EC-C15 (4.6 mm × 50 mm, 2.7 μm) at 40 °C with mobile phase A (H2O + 0.05% formic acid) and B (ACN + 0.05% formic acid) using a gradient elution and flow rate 0.6 mL/min. The DAD detector was set at 254 nm, the injection volume was 5 μL, and the oven temperature was 40 °C.
General procedure for the preparation of final aryloxypropanolamines from aryloxyepoxydes and amines. The corresponding amines (1 equivalent) and the corresponding oxiranes (1 equivalent) were stirred under N2 atmosphere at 70 °C for 24 h. The reaction mixture was concentrated, and the resulting residue was purified by column chromatography to afford pure products.
4.1.2. 1-(Dibenzylamino)-3-[4-(trifluoromethyl)phenoxy]propan-2-ol (BC1618)
Following the general procedure, a mixture of dibenzylamine (CAS 103-49-1) (0.09 mL, 0.47 mmol) and 2-{[4-(trifluoromethyl)phenoxy]methyl}oxirane (100 mg, 0.46 mmol) afforded BC1618 (149 mg, 78%) as a white solid after column chromatography (hexane/DCM 9:1 to 8:2). IR (ATR) 3412, 3029, 2924, 1613, 1451, 1261, 1034, 840, 745, 698 cm−1. 1H NMR (500 MHz, CDCl3) δ 2.69 (d, J = 7.0 Hz, 2H, CH2N), 3.56 (d, J = 13.5 Hz, 2H, CH2Ph), 3.82 (d, J = 13.5 Hz, 2H, CH2Ph), 3.85–3.94 (m, 2H, CH2O), 4.06–4.14 (m, 1H, CH), 6.88 (d, J = 8.5 Hz, 2H, ArH), 7.24–7.30 (m, 2H, ArH), 7.31–7.35 (m, 8H, ArH), 7.51 (d, J = 8.5 Hz, 2H, ArH). 13C NMR (126 MHz, CDCl3) δ 56.0 (CH2N), 59.0 (2CH2Ph), 66.4 (CH), 70.6 (CH2O), 114.6 (2CHAr), 121.3 (CAr), 123.2 (q, J = 32.5 Hz, CF3), 125.6 (CAr), 127.0 (d, J = 4.0 Hz, 2CHAr), 127.6 (2CHAr), 128.7 (4CHAr), 129.1 (CHAr), 129.3 (2CHAr), 130.0 (CHAr), 138.4 (CAr), 161.2 (OCAr) [15]. Purity 96.90% (tR = 3.27 min).
4.1.3. 1-(Benzylamino)-3-[4-(trifluoromethyl)phenoxy]propan-2-ol (1)
Following the general procedure, benzylamine (CAS 100-46-9) (0.05 mL, 0.46 mmol) and 2-{[4-(trifluoromethyl)phenoxy]methyl}oxirane (100 mg, 0.46 mmol) afforded 1 (82 mg, 55%) as a white solid after column chromatography (hexane/EtOAc 9:1 to 8:2). Mp 102–105 °C (EtOAc). IR (ATR) 3266, 1614, 1520, 1335, 1256, 1153, 1109, 1035, 836, 698 cm−1. 1H NMR (400 MHz, CDCl3, HETCOR) δ 2.47 (s, 2H, OH, NH), 2.79 (dd, J = 12.0, 8.0 Hz, 1H, CH2N), 2.91 (dd, J = 12.0, 4.0, Hz, 1H, CH2N), 3.79–3.91 (m, 2H, CH2Ph), 4.02 (d, J = 5.5 Hz, 2H, CH2O), 4.05–4.13 (m, 1H, CH), 6.96 (d, J = 8.5 Hz, 2H, ArH), 7.25–7.38 (m, 5H, ArH), 7.54 (d, J = 8.5 Hz, 2H, ArH). 13C NMR (101 MHz, CDCl3) δ 51.1 (CH2N), 53.9 (CH2Ph), 68.3 (CH), 70.7 (CH2O), 114.7 (2CHAr), 121.4–125.4 (m, CF3), 127.0 (2CHAr), 127.4 (2CHAr), 128.3 (3CHAr), 128.7 (CAr), 139.8 (CAr), 161.2 (OCAr). HRMS C17H19F3NO2 [M + H]+ 326.1362; found 326.1366. Purity 95.23% (tR = 2.80 min).
4.1.4. 1-[Methyl(phenyl)amino]-3-[4-(trifluoromethyl)phenoxy]propan-2-ol (2)
Following the general procedure, N-benzylmethylamine (CAS 103-67-3) (0.06 mL, 0.46 mmol) and 2-{[4-(trifluoromethyl)phenoxy]methyl}oxirane (100 mg, 0.46 mmol) afforded 2 (73 mg, 47%) as a white solid after a reversed-phase column chromatography (H2O:ACN 95:5). Mp 53–56 °C (EtOAc). IR (ATR) 3432, 2843, 1615, 1519, 1327, 1258, 1153, 1107, 1068, 836, 699 cm−1. 1H NMR (400 MHz, CDCl3, HETCOR) δ 2.36 (s, 3H, CH3), 2.61 (dd, J = 12.5, 4.0, Hz, 1H, CH2), 2.71 (dd, J = 12.5, 10.0 Hz, 1H, CH2), 3.6 (br s, 1H, OH), 3.61 (d, J = 13.0 Hz, 1H, CH2Ph), 3.76 (d, J = 13.0 Hz, 1H, CH2Ph), 4.00 (dd, J = 5.0, 1.0 Hz, 2H, CH2), 4.12–4.21 (m, 1H, CH), 6.96 (d, J = 8.5 Hz, 2H, ArH), 7.34 (dd, J = 8.0, 2.0 Hz, 5H, ArH), 7.54 (d, J = 8.5 Hz, 2H, ArH). 13C NMR (101 MHz, CDCl3) δ 42.3 (CH3), 59.5 (CH2N), 62.6 (CH2Ph), 66.0 (CH), 70.5 (CH2O), 114.7 (2CHAr), 123.3 (q, J = 33.0 Hz, CF3), 127.0 (q, J = 4.0 Hz, 3CHAr), 127.8 (2CHAr), 128.7 (2CHAr), 129.4 (CAr), 137.3 (CAr), 161.2 (OCAr). HRMS C18H21F3NO2 [M + H]+ 340.1519; found 340.1523. Purity 100% (tR = 2.74 min).
4.1.5. 1-(Dimethylamino)-3-[4-(trifluoromethyl)phenoxy]propan-2-ol (3)
Following the general procedure, dimethylamine (CAS 124-40-3) (0.04 mL, 0.55 mmol) and 2-{[4-(trifluoromethyl)phenoxy]methyl}oxirane (120 mg, 0.55 mmol) afforded 3 (48 mg, 33%) as an oil after a reversed-phase column chromatography (H2O:ACN 95:5). IR (ATR) 3348, 2926, 1615, 1520, 1461, 1257, 1033, 835, 775, 691 cm−1. 1H NMR (400 MHz, CDCl3, HETCOR) δ 2.66 (s, 6H, 2CH3), 2.84 (m, 1H, CH2N), 2.98 (m, 1H, CH2N), 4.00 (dd, J = 9.5, 5.5 Hz, 1H, CH2O), 4.10 (dd, J = 9.5, 5.0 Hz, 1H, CH2O), 4.34 (m, 1H, CH), 6.97 (d, J = 8.5 Hz, 2H, ArH), 7.53 (d, J = 9.0 Hz, 2H, ArH). 13C NMR (101 MHz, CDCl3) δ 45.1 (2CH3), 62.0 (CH2N), 65.2 (CH), 70.1 (CH2O), 114.6 (2CHAr), 121.6–124.3 (m, CF3), 125.8 (CAr), 127.1 (d, J = 4.0 Hz, 2CAr), 160.9 (OCAr). HRMS C12H17F3NO2 [M + H]+ 264.1206; found 264.1210. Purity 98.36% (tR = 2.47 min).
4.1.6. 1-[Benzyl(phenyl)amino]-3-[4-(trifluoromethyl)phenoxy]propan-2-ol (4)
Following the general procedure, N-benzylaniline (CAS 103-32-2) (0.08 mL, 0.46 mmol) and 2-{[4-(trifluoromethyl)phenoxy]methyl}oxirane (100 mg, 0.46 mmol) afforded 4 (95 mg, 52%) as a white solid after column chromatography (hexane/EtOAc 9:1 to 8:2). Mp 82–86 °C (EtOAc). IR (ATR) 3445, 1616, 1521, 1336, 1259, 1153, 1108, 1025, 839, 692 cm−1. 1H NMR (400 MHz, CDCl3, HETCOR) δ 2.36 (br s, 1H, OH), 3.63 (dd, J = 12.0, 4.5 Hz, 2H, CH2N), 4.00 (dd, J = 12.0, 4.5 Hz, 2H, CH2Ph), 4.33 (br s, 1H, CH), 4.61 (d, J = 5.5 Hz, 2H, CH2O), 6.73 (t, J = 7.0 Hz, 1H, ArH), 6.81 (d, J = 8.0 Hz, 2H, ArH), 6.91 (d, J = 8.5 Hz, 2H, ArH), 7.17–7.24 (m, 5H, ArH), 7.26–7.28 (m, 2H, ArH), 7.52 (d, J = 9.0 Hz, 2H, ArH). 13C NMR (101 MHz, CDCl3) δ 54.2 (CH2N), 55.8 (CH2Ph), 68.2 (CH), 70.1 (CH2O), 113.4 (2CHAr), 114.7 (2CHAr), 117.8 (2CHAr), 122.0–125.2 (m, CF3), 126.9 (2CHAr), 127.2 (CAr), 128.8 (3CAr), 129.5 (3CAr), 138.4 (CAr), 148.8 (CAr), 161.0 (OCAr). HRMS C23H23F3NO2 [M + H]+ 402.1675; found 402.1673. Purity 98.66% (tR = 4.71 min).
4.1.7. 1-[Benzyl(pyridin-3-ylmethyl)amino]-3-[4-(trifluoromethyl)phenoxy]propan-2-ol (5)
Following the general procedure, N-benzyl-1-(pyridin-3-yl)methanamine (CAS 63361-56-8) (73 mg, 0.37 mmol) and 2-{[4-(trifluoromethyl)phenoxy]methyl}oxirane (80 mg, 0.37 mmol) afforded 5 (40 mg, 26%) as a white solid after column chromatography (hexane/EtOAc 9:1 to 8:2). Mp 95–97 °C (EtOAc). IR (ATR) 2920, 1453, 1325, 1257, 1158, 1109, 1068, 1028, 837, 701 cm−1. 1H NMR (400 MHz, CDCl3, HETCOR) δ 2.66–2.77 (m, 2H, CH2N), 3.62 (dd, J = 10.5, 13.5 Hz, 2H, CH2Ph, CH2Pyr), 3.82 (dd, J = 13.5, 5.0 Hz, 2H, CH2Ph, CH2Pyr), 3.91 (d, J = 5.0 Hz, 2H, CH2O), 4.08–4.16 (m, 1H, CH), 6.88 (d, J = 8.5 Hz, 2H, ArH), 7.26–7.37 (m, 6H, ArH), 7.52 (d, J = 8.5 Hz, 2H, ArH), 7.67 (d, J = 8.0 Hz, 1H, ArH), 8.53 (d J = 4.0 Hz, 1H, ArH), 8.57 (s, 1H, ArH). 13C NMR (101 MHz, CDCl3) δ 56.1 (CH2N), 56.3 (CH2Pyr), 59.0 (CH2Ph), 66.7 (CH), 70.4 (CH2O), 114.6 (2CHAr), 123.0–123.6 (m, CF3), 123.7 (CHAr), 125.6 (CAr) 127.0 (2CHAr), 127.9 (CHAr), 128.8 (2CHAr), 129.3 (2CHAr), 133.8 (CAr), 137.0 (CHAr), 137.7 (CAr), 149.0 (CHAr), 150.4 (CHAr), 161.1 (CAr). HRMS C23H24F3N2O2 [M + H]+ 417.1784; found, 417.1794. Purity 100% (tR = 2.43 min).
4.1.8. 1-(Dibenzylamino)-3-phenoxypropan-2-ol (6)
Following the general procedure, dibenzylamine (CAS 103-49-1) (0.57 mL, 2.96 mmol) and 2-(phenoxymethyl)oxirane (442 mg, 2.94 mmol) afforded 6 (698 mg, 68%) as an oil after column chromatography (hexane/DCM 9:1 to 8:2). IR (ATR) 3434, 3028, 1598, 1494, 1452, 1242, 1927, 814, 747, 691 cm−1. 1H NMR (400 MHz, CDCl3, HETCOR) δ 2.61–2.73 (m, 2H, CH2N), 3.54 (d, J = 13.5 Hz, 2H, CH2Ph), 3.80 (d, J = 13.5 Hz, 2H, CH2Ph), 3.87 (d, J = 5.5 Hz, 2H, CH2O), 4.05–4.16 (m, 1H, CH), 6.84 (d, J = 8.0 Hz, 2H, ArH), 6.94 (t, J = 7.5 Hz, 1H, ArH), 7.21–7.38 (m, 12H, ArH). 13C NMR (101 MHz, CDCl3) δ 56.3 (CH2N), 58.9 (2CH2Ph), 66.6 (CH), 70.4 (CH2O), 114.6 (2CHAr), 121.0 (CHAr), 127.5 (2CHAr), 128.6 (4CHAr), 129.2 (4CHAr), 129.5 (2CHAr), 138.6 (2CAr), 158.8 (OCAr). HRMS C23H26NO2 [M + H]+ 348.1958; found 348.1961. Purity 98.28% (tR = 3.16 min).
4.1.9. 1-(4-Chlorophenoxy)-3-(dibenzylamino)propan-2-ol (7)
Following the general procedure, dibenzylamine (CAS 103-49-1) (0.33 mL, 1.71 mmol) and 2-[(4-chlorophenoxy)methyl]oxirane (316 mg, 1.71 mmol) afforded 7 (624 mg, 96%) as an oil after column chromatography (hexane/DCM 9:1 to 8:2). IR (ATR) 3426, 3027, 1589, 1485, 1446, 1248, 1027, 827, 743, 697 cm−1. 1H NMR (400 MHz, CDCl3, HETCOR) δ 2.75 (d, J = 6.5 Hz, 2H, CH2N), 3.57 (d, J = 13.5 Hz, 2H, CH2), 3.81 (d, J = 13.5 Hz, 2H, CH2Ph), 3.89–3.99 (m, 2H, CH2O), 4.13 (m, 1H, CH), 6.81–6.93 (m, 2H, ArH), 7.18 (m, 1H, ArH), 7.22–7.30 (m, 2H, ArH), 7.30–7.36 (m, 9H, ArH). 13C NMR (101 MHz, CDCl3) δ 56.1 (CH2N), 59.0 (2CH2Ph), 66.6 (CH), 71.3 (CH2O), 113.7 (2CHAr), 121.7 (2CHAr), 123.2 (CAr), 127.4 (2CHAr), 127.8 (2CHAr), 128.6 (2CHAr), 129.2 (2CHAr), 130.3 (2CHAr), 138.6 (2CAr), 154.3 (OCAr). HRMS C23H25ClNO2 [M + H]+ 382.1568; found 382.1570. Purity 96.90% (tR = 3.27 min).
4.1.10. 1-(Dibenzylamino)-3-(p-tolyloxy)propan-2-ol (8)
Following the general procedure, dibenzylamine (CAS 103-49-1) (0.20 mL, 1.04 mmol) and 2-[(p-tolyloxy)methyl]oxirane (170 mg, 1.04 mmol) afforded 8 (300 mg, 80%) as a white solid after column chromatography (hexane/DCM 9:1 to 8:2). Mp 55–57 °C (EtOAc). IR (ATR) 3419, 3028, 2938, 2805, 1615, 1511, 1238, 1044, 803, 744, 697 cm−1. 1H NMR (400 MHz, CDCl3, HETCOR) δ 2.27 (s, 3H, CH3), 2.66 (d, J = 7.5 Hz, 2H, CH2N), 3.18 (s, 1H, OH), 3.53 (d, J = 13.5 Hz, 2H, CH2Ph), 3.79 (d, J = 13.5 Hz, 2H, CH2Ph), 3.84 (d, J = 5.5 Hz, 2H, CH2O), 4.04–4.14 (m, 1H, CH), 6.74 (d, J = 8.5 Hz, 2H, ArH), 7.04 (d, 2H, ArH), 7.32 (m, J = 3.0 Hz, 10H, ArH). 13C NMR (101 MHz, CDCl3) δ 20.6 (CH3), 56.3 (CH2N), 58.8 (2CH2Ph), 66.7 (CH), 70.6 (CH2O), 114.5 (2CHAr), 127.4 (2CHAr), 128.6 (4CHAr), 129.2 (4CHAr), 130.0 (CAr), 130.2 (2CHAr), 138.7 (2CAr), 156.7 (OCAr). HRMS C24H28NO2 [M + H]+ 362.2115; found 362.2115. Purity 97.23% (tR = 3.35 min).
4.1.11. 1-(Dibenzylamino)-3-(4-isopropylphenoxy)propan-2-ol (9)
Following the general procedure, dibenzylamine (CAS 103-49-1) (0.25 mL, 1.30 mmol) and 2-[(4-isopropylphenoxy)methyl]oxirane (253 mg, 1.32 mmol) afforded 9 (320 mg, 62%) as a white solid after column chromatography (hexane/DCM 9:1 to 8:2). Mp 70–72 °C (EtOAc). IR (ATR) 3449, 3027, 1609, 1513, 1455, 1245, 1039, 838, 745, 696 cm−1. 1H NMR (400 MHz, CDCl3, HETCOR) δ 1.21 (d, J = 7.0 Hz, 6H, CH3), 2.63–2.69 (m, 2H, CH2N), 2.85 (m, J = 7.0 Hz, 1H, CH), 3.17 (s, 1H, OH), 3.53 (d, J = 13.5 Hz, 2H, CH2Ph), 3.79 (d, J = 13.5 Hz, 2H, CH2Ph), 3.85 (d, J = 5.5 Hz, 2H, CH2O), 4.04–4.14 (m, 1H, CH), 6.77 (d, J = 9.0 Hz, 2H, ArH), 7.11 (d, J = 8.5 Hz, 2H, ArH), 7.25–7.36 (m, 10H, ArH). 13C NMR (101 MHz, CDCl3) δ 24.3 (2CH3), 33.4 (CH), 56.4 (CH2N), 58.9 (2CH2Ph), 66.7 (CH), 70.5 (CH2O), 114.4 (2CHAr), 127.3 (2CHAr), 127.4 (2CHAr), 128.6 (4CHAr), 129.2 (4CHAr), 138.7 (2CAr), 141.5 (CAr), 156.9 (OCAr). HRMS C26H32NO2 [M + H]+ 390.2428; found 390.2427. Purity 98.32% (tR = 3.63 min).
4.1.12. 1-[4-(Tert-butyl)phenoxy]-3-(dibenzylamino)propan-2-ol (10)
Following the general procedure, dibenzylamine (CAS 103-49-1) (0.24 mL, 1.25 mmol) and 2-{[4-(tert-butyl)phenoxy]methyl}oxirane (261 mg, 1.27 mmol) afforded 10 (375 mg, 74%) as a white solid after column chromatography (hexane/DCM 9:1 to 8:2). Mp 80–83 °C (EtOAc). IR (ATR) 3432, 3028, 2963, 1605, 1513, 1247, 1042, 828, 746, 698 cm−1. 1H NMR (400 MHz, CDCl3, HETCOR) δ 1.29 (s, 9H, 3CH3), 2.61–2.70 (m, 2H, CH2N), 3.16 (s, 1H, OH), 3.53 (d, J = 13.5 Hz, 2H, CH2Ph), 3.79 (d, J = 13.5 Hz, 2H, CH2Ph), 3.85 (d, J = 5.5 Hz, 2H, CH2O), 4.05–4.12 (m, 1H, CH), 6.78 (d, J = 9.0 Hz, 2H, ArH), 7.24 (m, 2H, ArH), 7.26–7.35 (m, 10H, ArH). 13C NMR (101 MHz, CDCl3) δ 31.7 (3CH3), 34.2 (C), 56.4 (CH2N), 58.9 (2CH2Ph), 66.7 (CH), 70.5 (CH2O), 114.1 (2CHAr), 126.3 (2CHAr), 127.4 (2CHAr), 128.6 (4CHAr), 129.2 (4CHAr), 138.7 (2CAr), 143.7 (CAr), 156.5 (OCAr). HRMS C27H34NO2 [M + H]+ 404.2584; found 404.2590. Purity 100% (tR = 3.62 min).
4.1.13. 1-(4-Cyclohexylphenoxy)-3-(dibenzylamino)propan-2-ol (11)
Following the general procedure, dibenzylamine (CAS 103-49-1) (0.21 mL, 1.09 mmol) and 2-[(4-cyclohexylphenoxy)methyl]oxirane (253 mg, 1.09 mmol) afforded 11 (340 mg, 73%) as a white solid after column chromatography (hexane/DCM 9:1 to 8:2). Mp 113–115 °C (EtOAc). IR (ATR) 3427, 3028, 2928, 1512, 1515, 1248, 1047, 809, 744, 696 cm−1. 1H NMR (400 MHz, CDCl3, HETCOR) δ 1.18–1.31 (m, 1H, CH2cycl), 1.31–1.46 (m, 4H, 2CH2cycl), 1.75 (m, 1H, CH2cycl), 1.78–1.91 (m, 4H, 2CH2cycl), 2.39–2.51 (m, 1H, CHcycl), 2.63–2.75 (m, 2H, CH2N), 3.18 (s, 1H, OH), 3.55 (d, J = 13.5 Hz, 2H, CH2Ph), 3.80 (d, J = 13.5 Hz, 2H, CH2 Ph), 3.86 (d, J = 5.5 Hz, 2H, CH2O), 4.05–4.15 (m, 1H, CH), 6.78 (d, J = 9.8 Hz, 2H, ArH), 7.11 (d, J = 6.5 Hz, 2H, ArH), 7.24–7.31 (m, 2H, ArH), 7.30–7.39 (m, 8H, ArH). 13C NMR (101 MHz, CDCl3) 26.3 (CH2cycl), 27.1 (2CH2cycl), 34.8 (2CH2cycl), 43.8 (CHcycl), 56.4 (CH2N), 58.9 (2CH2Ph), 66.7 (CH), 70.5 (CH2O), 114.4 (2CHAr), 127.4 (2CHAr), 127.7 (2CHAr), 128.6 (4CHAr), 129.2 (4CHAr), 138.7 (2CAr), 140.8 (CAr), 156.9 (OCAr). HRMS C29H36NO2 [M + H]+ 430.2741; found 430.2736. Purity 99.24% (tR = 3.88 min).
4.1.14. 1-(4-Cyclopentylphenoxy)-3-(dibenzylamino)propan-2-ol (12)
Following the general procedure, dibenzylamine (CAS 103-49-1) (0.23 mL, 1.19 mmol) and 2-[(4-cyclopentylphenoxy)methyl]oxirane (257 mg, 1.18 mmol) afforded 12 (295 mg, 60%) as a white solid after column chromatography (hexane/DCM 9:1 to 8:2). Mp 88–90 °C (EtOAc). IR (ATR) 3413, 2938, 1610, 1511, 1451, 1242, 1044, 824, 747, 698 cm−1. 1H NMR (400 MHz, CDCl3, HETCOR) δ1.48–1.60 (m, 2H, CH2cycl), 1.62–1.71 (m, 2H, CH2cycl), 1.73–1.83 (m, 2H, CH2cycl), 1.96–2.09 (m, 2H, CH2cycl), 2.63–2.69 (m, 2H, CH2N), 2.86–2.99 (m, 1H, CHcycl), 3.53 (d, J = 13.5 Hz, 2H, CH2Ph), 3.79 (d, J = 13.5 Hz, 2H, CH2Ph), 3.84 (d, J = 5.5 Hz, 2H, CH2O), 4.05–4.11 (m, 1H, CH), 6.73–6.80 (d, 2H, ArH), 7.09–7.16 (d, 2H, ArH), 7.22–7.37 (m, 10H, ArH). 13C NMR (101 MHz, CDCl3) δ 25.5 (2CH2cycl), 34.8 (2CH2cycl), 45.3 (CHcycl), 56.3 (CH2), 58.9 (2CH2Ph), 66.7 (CH), 70.6 (CH2), 114.4 (2CHAr), 127.4 (2CHAr), 128.0 (2CHAr), 128.6 (4CHAr), 129.2 (4CHAr), 138.7 (CAr), 139.0 (2CAr), 156.9 (OCAr). HRMS C28H34NO2 [M + H]+ 416.2584; found 416.2591. Purity 100% (tR = 3.66 min).
4.1.15. 1-(Dibenzylamino)-3-(3,4-dimethylphenoxy)propan-2-ol (13)
Following the general procedure, dibenzylamine (CAS 103-49-1) (0.37 mL, 1.92 mmol) and 2-[(3,4-dimethylphenoxy)methyl]oxirane (347 mg, 1.95 mmol) afforded 13 (555 mg, 76%) as an oil after column chromatography (hexane/DCM 9:1 to 8:2). IR (ATR) 3435, 3026, 2921, 1607, 1501, 1452, 1252, 1046, 735, 697 cm−1. 1H NMR (400 MHz, CDCl3, HETCOR) δ 2.18 (s, 3H, CH3), 2.21 (s, 3H, CH3), 2.62–2.73 (m, 2H, CH2N), 3.54 (d, J = 13.5 Hz, 2H, CH2Ph), 3.79 (d, J = 13.5 Hz, 2H, CH2Ph), 3.83 (d, J = 5.0 Hz, 2H, CH2O), 4.02–4.14 (m, 1H, CH), 6.58 (dd, J = 8.5, 3.0 Hz, 1H, ArH), 6.65 (d, J = 3.0 Hz, 1H, ArH), 7.00 (d, J = 8.3 Hz, 1H, ArH), 7.21–7.42 (m, 10H, ArH). 13C NMR (101 MHz, CDCl3) δ 18.9 (CH3), 20.1 (CH3), 56.3 (CH2N), 58.8 (2CH2Ph), 66.7 (CH), 70.6 (CH2O), 111.6 (CHAr), 116.2 (CHAr), 127.4 (2CHAr), 128.6 (4CHAr), 129.0 (CHAr), 129.2 (4CHAr), 130.4 (CAr), 137.8 (CAr), 138.7 (2CAr), 157.0 (OCAr). HRMS C25H30NO2 [M + H]+ 376.2271; found 376.2270. Purity 97.06% (tR = 3.35 min).
4.1.16. 1-(Dibenzylamino)-3-(4-methoxyphenoxy)propan-2-ol (14)
Following the general procedure, dibenzylamine (CAS 103-49-1) (0.13 mL, 0.70 mmol) and 2-[(4-methoxyphenoxy)methyl]oxirane (126 mg, 0.70 mmol) afforded 14 (170 mg, 64%) as a white solid after column chromatography (hexane/DCM 9:1 to 8:2). Mp 61–63 °C (EtOAc). IR (ATR) 3417, 2925, 1505, 1451, 1231, 1044, 1028, 822, 744, 697 cm−1. 1H NMR (400 MHz, CDCl3, HETCOR) δ 2.67 (d, J = 7.5 Hz, 2H, CH2N), 3.54 (d, J = 13.5 Hz, 2H, CH2Ph), 3.76 (s, 3H, CH3), 3.77 (d, J = 13.5 Hz, 2H, CH2Ph), 3.78–3.85 (m, 2H, CH2O), 4.08 (m, 1H, CH), 6.75–6.83 (m, 4H, ArH), 7.23–7.37 (m, 10H, ArH). 13C NMR (101 MHz, CDCl3) δ 55.9 (CH3), 56.3 (CH2N), 58.9 (2CH2Ph), 66.7 (CH), 71.2 (CH2O), 114.7 (2CHAr), 115.6 (2CHAr), 127.4 (2CHAr), 128.6 (4CHAr), 129.2 (4CHAr), 138.7 (2CAr), 153.0 (CAr), 154.1 (OCAr). HRMS C24H28NO3 [M + H]+ 378.2064; found 378.2062. Purity 98.43% (tR = 3.02 min).
4.1.17. 1-[4-(Benzyloxy)phenoxy]-3-(dibenzylamino)propan-2-ol (15)
Following the general procedure, dibenzylamine (CAS 103-49-1) (0.19 mL, 0.99 mmol) and 2-{[4-(benzyloxy)phenoxy]methyl}oxirane (247 mg, 0.96 mmol) afforded 15 (325 mg, 74%) as a white solid after column chromatography (hexane/DCM 9:1 to 8:2). Mp 87–88 °C (EtOAc). IR (ATR) 3454, 3027, 2795, 1602, 1509, 1235, 1041, 825, 731, 696 cm−1. 1H NMR (400 MHz, CDCl3, HETCOR) δ 2.63–2.68 (m, 2H, CH2N), 3.17 (s, 1H, OH), 3.53 (d, J = 13.5 Hz, 2H, CH2Ph), 3.75–3.84 (m, 4H, 2CH2Ph), 4.07 (m, J = 5.8 Hz, 1H, CH), 5.00 (s, 2H, PhCH2O), 6.73–6.80 (m, 2H, ArH), 6.83–6.90 (m, 2H, ArH), 7.22–7.44 (m, 15H, ArH). 13C NMR (101 MHz, CDCl3) δ 56.3 (CH2N), 58.9 (2CH2Ph), 66.7 (CH), 70.8 (PhCH2O), 71.1 (CH2O), 115.5 (2CHAr), 115.9 (2CHAr), 127.4 (2CHAr), 127.6 (2CHAr), 128.0 (CHAr), 128.6 (4CHAr), 128.7 (2CHAr), 129.2 (4CHAr), 137.4 (2CAr), 138.7 (CAr), 153.1 (CAr), 153.2 (OCAr). HRMS C30H32NO3 [M + H]+ 454.2377; found 454.2385. Purity 96.19% (tR = 3.43 min).
4.1.18. 1-(Dibenzylamino)-3-(4-propoxyphenoxy)propan-2-ol (16)
Following the general procedure, dibenzylamine (CAS 103-49-1) (0.46 mL, 2.38 mmol) and 2-[(4-propoxyphenoxy)methyl]oxirane (490 mg, 2.35 mmol) afforded 16 (620 mg, 65%) as a white solid after column chromatography (hexane/DCM 9:1 to 8:2). Mp 51–53 °C (EtOAc). IR (ATR) 3380, 3027, 2921, 1589, 1508, 1228, 1048, 827, 747, 697 cm−1. 1H NMR (400 MHz, CDCl3, HETCOR) δ 1.02 (t, J = 7.5 Hz, 3H, CH3), 1.71–1.84 (m, 2H, OCH2CH2), 2.63–2.69 (m, 2H, CH2N), 3.54 (d, J = 13.5 Hz, 2H, CH2Ph), 3.80 (d, J = 13.5 Hz, 2H, CH2Ph), 3.86 (t, J = 6.4 Hz, 2H, OCH2), 4.08 (m, 1H, CH), 6.73–6.84 (m, 4H, ArH), 7.26–7.37 (m, 10H, ArH).13C NMR (101 MHz, CDCl3) δ 10.7 (CH3), 22.8 (OCH2CH2), 56.3 (CH2N), 58.9 (2CH2Ph), 66.7 (CH), 70.3 (CH2O), 71.2 (OCH2), 115.5 (2CHAr), 115.5 (2CHAr), 127.4 (2CHAr), 128.6 (4CHAr), 129.2 (4CHAr), 138.7 (2CAr), 152.9 (CAr), 153.6 (OCAr). HRMS C26H32NO3 [M + H]+ 406.2377; found 406.2379. Purity 98.33% (tR = 3.33 min).
4.1.19. 1-(Dibenzylamino)-3-(3,4-dichlorophenoxy)propan-2-ol (17)
Following the general procedure, dibenzylamine (CAS 103-49-1) (0.45 mL, 2.35 mmol) and 2-[(3,4-dichlorophenoxy)methyl]oxirane (510 mg, 2.33 mmol) afforded 17 (882 mg, 91%) as an oil after column chromatography (hexane/DCM 9:1 to 8:2). IR (ATR) 3425, 3027, 2931, 1592, 1451, 1230, 1026, 839, 735, 698 cm−1. 1H NMR (400 MHz, CDCl3, HETCOR) δ 2.61–2.67 (m, 2H, CH2N), 3.17 (s, 1H, OH), 3.53 (d, J = 13.0 Hz, 2H, CH2Ph), 3.73–3.78 (m, 4H, CH2Ph, CH2O), 4.03 (m, 1H, CH), 6.67 (dd, J = 9.0, 3.0 Hz, 1H, ArH), 6.89 (d, J = 3.0 Hz, 1H, ArH), 7.22–7.28 (m, 1H, ArH), 7.28–7.36 (m, 10H, ArH). 13C NMR (101 MHz, CDCl3) δ 55.9 (CH2N), 59.0 (2CH2Ph), 66.4 (CH), 71.0 (CH2O), 114.7 (CHAr), 116.5 (CHAr), 124.3 (CAr), 127.6 (2CHAr), 128.6 (4CHAr), 129.2 (4CHAr), 130.7 (CHAr), 132.9 (CAr), 138.5 (2CAr), 157.9 (OCAr). HRMS C23H24Cl2NO2 [M + H]+ 416.1179; found 416.1176. Purity 96.24% (tR = 3.59 min).
4.1.20. 1-[4-Chloro-3-(trifluoromethyl)phenoxy]-3-(dibenzylamino)propan-2-ol (18)
Following the general procedure, dibenzylamine (CAS 103-49-1) (0.40 mL, 2.10 mmol) and 2-{[4-chloro-3-(trifluoromethyl)phenoxy]methyl}oxirane (529 mg, 2.10 mmol) afforded 18 (878 mg, 93%) as an oil after column chromatography (hexane/DCM 9:1 to 8:2). IR (ATR) 3433, 3028, 1606, 1482, 1423, 1239, 1027, 816, 747, 698 cm−1. 1H NMR (400 MHz, CDCl3, HETCOR) δ 2.63–2.69 (m, 2H, CH2N), 3.20 (s, 1H, OH), 3.54 (d, J = 13.0 Hz, 2H, CH2Ph), 3.85 (m, 2H, CH2O), 4.04 (m, 1H, CH), 6.90 (dd, J = 9.0, 3.0 Hz, 1H, ArH), 7.11 (d, J = 3.0 Hz, 1H, ArH), 7.22–7.38 (m, 11H, ArH). 13C NMR (101 MHz, CDCl3) δ 55.8 (CH2N), 59.0 (2CH2Ph), 66.4 (CH), 70.9 (CH2O), 114.2 (q, J = 5.5 Hz, CHAr), 118.7 (CHAr), 121.4 (CAr), 123.5 (CF3), 124.1 (CAr), 127.6 (2CHAr), 128.6 (4CHAr), 129.2 (4CHAr), 132.4 (CHAr), 138.5 (2CAr), 157.2 (OCAr). HRMS C24H24ClF3NO2 [M + H]+ 450.1442; found 450.1448. Purity 99.79% (tR = 3.72 min).
4.1.21. 1-(Dibenzylamino)-3-[4-nitro-3-(trifluoromethyl)phenoxy]propan-2-ol (19)
Following the general procedure, dibenzylamine (CAS 103-49-1) (0.45 mL, 2.34 mmol) and 2-{[4-nitro-3-(trifluoromethyl)phenoxy]methyl}oxirane (609 mg, 2.31 mmol) afforded 19 (619 mg, 58%) as an oil after column chromatography (hexane/DCM 9:1 to 8:2). IR (ATR) 3424, 2931, 1453, 1310, 1242, 1038, 1027, 834, 748, 698 cm−1. 1H NMR (400 MHz, CDCl3, HETCOR) δ 1H NMR (400 MHz, CDCl3) δ 2.56–2.75 (m, 2H, CH2N), 3.55 (d, J = 13.0 Hz, 2H, CH2Ph), 3.80 (d, J = 13.0 Hz, 2H, CH2Ph), 3.88–4.01 (m, 2H, CH2O), 4.01–4.10 (m, 1H, CH), 7.01 (dd, J = 9.0, 3.0 Hz, 1H, ArH), 7.18 (d, J = 2.5 Hz, 1H, ArH), 7.23–7.43 (m, 10H, ArH), 7.95 (d, J = 9.5 Hz, 1H, ArH). 13C NMR (101 MHz, CDCl3) δ 55.5 (CH2N), 59.1 (2CH2Ph), 66.3 (CH), 71.3 (CH2O), 115.0 (q, J = 6.0 Hz, CF3), 116.8 (CHAr), 120.6 (CHAr), 123.3 (CAr), 126.2 (q, J = 34.0 Hz, CF3), 127.6 (2CAr), 128.1 (CAr), 128.7 (4CAr), 129.2 (4CAr), 138.4 (2CAr), 141.1 (CAr), 161.9 (OCAr). HRMS C24H24F3N2O4 [M + H]+ 461.1683; found 461.1681. Purity 98.68% (tR = 3.50 min).
4.1.22. 1-(Dibenzylamino)-3-(2,4-dichlorophenoxy)propan-2-ol (20)
Following the general procedure, dibenzylamine (CAS 103-49-1) (0.48 mL, 2.49 mmol) and 2-[(2,4-dichlorophenoxy)methyl]oxirane (551 mg, 2.52 mmol) afforded 20 (642 mg, 61%) as an oil after column chromatography (hexane/DCM 9:1 to 8:2). IR (ATR) 3426, 3027, 2803, 1585, 1482, 1290, 1062, 802, 744, 697 cm−1. 1H NMR (400 MHz, CDCl3, HETCOR) δ 2.64–2.70 (m, 2H, CH2N), 3.49 (d, J = 13.5 Hz, 2H, CH2Ph), 3.74 (d, J = 13.5 Hz, 2H, CH2Ph), 3.79–3.90 (m, 2H, CH2O), 3.99–4.08 (m, 1H, CH), 6.69 (d, J = 9.0 Hz, 1H, ArH), 7.08 (dd, J = 9.0, 2.5 Hz, 1H, ArH), 7.16–7.31 (m, 11H, ArH). 13C NMR (101 MHz, CDCl3) δ 55.9 (CH2N), 59.0 (2CH2Ph), 66.5 (CH), 71.6 (CH2O), 114.4 (CHAr), 124.0 (CHAr), 126.1 (2CHAr), 127.5 (4CHAr), 127.6 (CAr), 128.6 (4CHAr), 129.2 (2CHAr), 130.0 (CAr), 138.6 (2CAr), 153.2 (OCAr). HRMS C23H24Cl2NO2 [M + H]+ 416.1179; found 416.1179. Purity 95.03% (tR = 3.58 min).
4.1.23. 1-(4-Bromophenoxy)-3-(dibenzylamino)propan-2-ol (21)
Following the general procedure, dibenzylamine (CAS 103-49-1) (0.42 mL, 2.21 mmol) and 2-[(4-bromophenoxy)methyl]oxirane (500 mg, 2.18 mmol) afforded 21 (800 mg, 86%) as a white solid after column chromatography (hexane/EtOAc 9:1 to 8:2). Mp 63–65 °C (EtOAc). IR (ATR) 3568, 3413, 3031, 2936, 1587, 1488, 1243, 1036, 830, 804, 744, 733, 698 cm−1. 1H NMR (500 MHz, CDCl3, HETCOR) δ 2.66 (d, J = 7.0 Hz, 2H, CH2N), 3.19 (s, 1H, OH), 3.54 (d, J = 13.5 Hz, 2H, CH2Ph), 3.80 (d, J = 13.5 Hz, 2H, CH2Ph), 3.83 (m, 2H, CH2O), 4.03–4.14 (m, 1H, CH), 6.68–6.74 (m, 2H, ArH), 7.23–7.30 (m, 2H, ArH), 7.30–7.42 (m, 10H, ArH). 13C NMR (101 MHz, CDCl3) δ 56.1 (CH2N), 58.9 (2CH2Ph), 66.5 (CH), 70.7 (CH2O), 113.2 (CAr), 116.5 (2CHAr), 127.5 (2CHAr), 128.6 (4CHAr), 129.2 (4CHAr), 132.3 (2CHAr), 138.6 (2CAr), 157.9 (OCAr). HRMS C23H25BrNO2 [M + H]+ 426.1063; found 426.1064. Purity 97.85% (tR = 4.17 min).
4.1.24. 1-(Dibenzylamino)-3-(4-nitrophenoxy)propan-2-ol (22)
Following the general procedure, dibenzylamine (CAS 103-49-1) (0.45 mL, 2.33 mmol) and 2-[(4-nitrophenoxy)methyl]oxirane (450 mg, 2.31 mmol) afforded 22 (610 mg, 67.4%) as an oil after column chromatography (hexane/EtOAc 9:1 to 8:2). IR (ATR) 3427, 3028, 2931, 1592, 1509, 1496, 1331, 1259, 1109, 1026, 842, 750, 697 cm−1. 1H NMR (500 MHz, CDCl3, HETCOR) δ 2.60–2.74 (m, 2H, CH2N), 3.25 (s, 1H, OH), 3.55 (d, J = 13.5 Hz, 2H, CH2Ph), 3.82 (d, J = 13.5 Hz, 2H, CH2Ph), 3.91 (dd, J = 10, 4.5 Hz, 1H, CH2O), 3.95 (dd, J = 10, 4.5 Hz, 1H, CH2O), 4.08(m, 1H, CH), 6.87 (m, 2H, ArH), 7.23 -7.41 (m, 10H, ArH), 8.11–8.22 (m, 2H, ArH). 13C NMR (126 MHz, CDCl3) δ 55.8 (CH2N), 59.0 (2CH2Ph), 66.3 (CH), 71.1 (CH2O), 114.7 (CHAr), 126.0 (2CHAr), 127.6 (2CHAr), 128.7 (4CHAr), 129.2 (4CHAr), 138.5 (2CAr), 141.8 (CAr), 163.8 (OCAr). HRMS C23H24N2O4 [M + H]+ 393.1809; found 393.1814. Purity 96.42% (tR = 4 min).
4.1.25. 1-(Dibenzylamino)-3-(4-iodophenoxy)propan-2-ol (23)
Following the general procedure, dibenzylamine (CAS 103-49-1) (0.33 mL, 1.7 mmol) and 2-[(4-iodophenoxy)methyl]oxirane (465 mg, 1.68 mmol) afforded 23 (525 mg, 65.8%) as a white solid after column chromatography (hexane/EtOAc 9:1 to 8:2). Mp 103–105 °C (EtOAc). IR (ATR) 3406, 3030, 2936, 1580, 1484, 1282, 1246, 1035, 828, 803, 744, 697 cm−1. 1H NMR (500 MHz, CDCl3, HETCOR) δ 2.66 (d, J = 7.0 Hz, 2H, CH2N), 3.19 (s, 1H, OH), 3.54 (d, J = 13.5 Hz, 2H, CH2Ph), 3.79 (d, J = 13.5 Hz, 2H, CH2Ph), 3.74–3.87 (m, 2H, CH2O), 4.07 (m, 1H, CH), 6.58–6.64 (m, 2H, ArH), 7.19–7.38 (m, 10H, ArH), 7.49–7.56 (m, 2H, ArH). 13C NMR (126 MHz, CDCl3) δ 56.1 (CH2N), 58.9 (2CH2Ph), 66.5 (CH), 70.5 (CH2O), 83.1 (CAr), 117.1 (2CHAr), 127.5 (2CHAr), 128.6 (4CHAr), 129.2 (4CHAr), 138.3 (2CAr), 138.6 (2CHAr), 158.7 (CAr). HRMS C23H24INO2 [M + H]+ 474.0924; found 474.0927. Purity 96.40% (tR = 4.26 min).
4.1.26. 1-(Dibenzylamino)-3-[4-(pentafluoro-λ6-sulfaneyl)phenoxy]propan-2-ol (24)
Following the general procedure, dibenzylamine (CAS 103-49-1) (0.45 mL, 2.33 mmol) and 2-[(4-(pentafluoro-λ6-sulfaneyl)phenoxy)methyl]oxirane (643 mg, 2.33 mmol) afforded 24 (657 mg, 59.6%) as a white solid after column chromatography (hexane/EtOAc 9:1 to 8:2). Mp 67–69⁰C (EtOAc). IR (ATR) 3414, 3028, 2938, 1597, 1504, 1451, 1309, 1264, 1101, 1037, 844, 826, 806, 745, 698, 595, 579 cm−1. 1H NMR (500 MHz, CDCl3, HETCOR) δ 2.67 (d, J = 6.0 Hz, 2H, CH2N), 3.21 (s, 1H, OH), 3.54 (d, J = 13.5 Hz, 2H, CH2Ph), 3.81 (d, J = 13.5 Hz, 2H, CH2Ph), 3.85–3.94 (m, 2H, CH2O), 4.03–4.10 (m, 1H, CH), 6.83 (d, J = 9.5 Hz, 2ArH), 7.19–7.38 (m, 10H, ArH), 7.61–7.68 (m, 2H, ArH).13C NMR (126 MHz, CDCl3) δ 56.0 (CH2N), 59.0 (2CH2Ph), 66.4 (CH), 70.8 (CH2O), 114.2 (2CHAr), 127.6 (4CHAr), 127.7 (m, C-SF5), 128.7 (4CHAr), 129.2 (4CHAr), 138.5 (2CAr), 160.6 (OCAr). HRMS C23H24F5NO2S [M + H]+ 474.1521; found 474.1524. Purity 95.25% (tR = 4.36 min).
4.1.27. 1-[Benzyl(4-chlorobenzyl)amino]-3-(4-chlorophenoxy)propan-2-ol (25)
Following the general procedure, N-benzyl-1-(4-chlorophenyl)methanamine (CAS 13541-00-9) (0.14 mL, 0.70 mmol) and 2-[(4-chlorophenoxy)methyl]oxirane (130 mg, 0.70 mmol) afforded 25 (182.5 mg, 62.3%) as an oil after column chromatography (hexane/EtOAc 9:1 to 8:2). IR (ATR) 3426, 3026, 2932, 1590, 1485, 1445, 1276, 1248, 1087, 1063, 1015, 801, 743, 697 cm−1. 1H NMR (400 MHz, CDCl3, HETCOR) δ 2.72 (dd, J = 6.5, 2.0 Hz, 2H, CH2N), 3.54 (dd, J = 13.5, 11 Hz, 2H, CH2Ph), 3.74 (t, J = 13.5 Hz, 2H, CH2Ph), 3.90–3.95 (m, 2H, CH2O), 4.06–4.13 (m, 1H, CH), 6.83 (dd, J = 8.5, 1.5 Hz, 1H, ArH), 6.88 (td, J = 7.5, 1.5 Hz, 1H, ArH), 7.18 (ddd, J = 8.5, 7.5, 1.5 Hz, 1H, ArH), 7.22–7.36 (m, 10H, ArH). 13C NMR (101 MHz, CDCl3) δ 56.1 (CH2N), 58.3 (CH2Ph), 59.0 (CH2Ph), 66.8 (CH), 71.2 (CH2O), 113.6 (CHAr), 121.8 (CHAr), 123.2 (CAr), 127.5 (CHAr), 127.8 (CHAr), 128.6 (2CHAr), 128.7 (2CHAr), 129.2 (2CHAr), 130.4 (CHAr), 130.5 (2CHAr), 133.2 (CAr), 137.3 (CAr), 138.5 (CAr), 154.2 (CAr). HRMS C23H23Cl2NO2 [M + H]+ 416.1179; found 4161191. Purity 95.59% (tR = 4.45 min).
4.1.28. 1-[Bis(4-chlorobenzyl)amino]-3-(4-chlorophenoxy)propan-2-ol (26)
Following the general procedure, bis(4-chlorobenzyl)amine (CAS 21913-13-3)(150 mg, 0.56 mmol) and 2-[(4-chlorophenoxy)methyl]oxirane (104 mg, 0.56 mmol) afforded 26 (194.2 mg, 76.4%) as an oil after column chromatography (hexane/EtOAc 9:1 to 8:2). IR (ATR) 3438, 3058, 2921, 1740, 1589, 1486, 1447, 1248, 1087, 1062, 1014, 807, 746 cm−1. 1H NMR (400 MHz, CDCl3) δ 2.63–2.77 (m, 2H, CH2N), 2.96 (s, 1H, OH), 3.52 (d, J = 13.5 Hz, 2H, CH2Ph), 3.69 (d, J = 13.5 Hz, 2H, CH2Ph), 3.90–3.94 (m, 2H, CH2O), 4.06–4.14 (m, 1H, CH), 6.82 (dd, J = 8.5, 1.5 Hz, 1H, ArH), 6.89 (td, J = 7.5, 1.5 Hz, 1H, ArH), 7.15–7.29 (m, 9H, ArH), 7.33 (dd, J = 8.0, 1.5 Hz, 1H, ArH). 13C NMR (101 MHz, CDCl3) δ 56.0 (CH2N), 58.3 (2CH2Ph), 66.9 (CH), 71.0 (CH2O), 113.6 (CHAr), 121.9 (CHAr), 123.1 (CAr), 127.9 (CHAr), 128.8 (4CHAr), 130.4 (CHAr), 130.4 (4CHAr), 133.3 (2CAr), 137.1 (2CAr), 154.1 (CAr). HRMS C23H22Cl3NO2 [M + H]+ 450.0789; found 450.9787. Purity 99.45% (tR = 5.24 min).
4.1.29. 1-[Benzyl(4-methylbenzyl)amino]-3-(4-chlorophenoxy)propan-2-ol (27)
Following the general procedure, N-benzyl-1-(p-tolyl)methanamine (CAS 55096-86-1) (148.8 mg, 0.70 mmol) and 2-[(4-chlorophenoxy)methyl]oxirane (130 mg, 0.70 mmol) afforded 27 (119,5 mg, 42.9%) as an oil after column chromatography (hexane/EtOAc 9:1 to 8:2). IR (ATR) 3436, 3028, 2924, 1589, 1485, 1446, 1277, 1249, 1061, 1028, 801, 744, 697 cm−1. 1H NMR (400 MHz, CDCl3, HETCOR) δ 2.33 (s, 3H, CH3), 2.73 (d, J = 6.5 Hz, 2H, CH2N), 3.53 (t, J = 13.0 Hz, 2H, CH2Ph), 3.80 (t, J = 14.0 Hz, 2H, CH2Ph), 3.91–4.00 (m, 2H, CH2O), 4.11 (m, 1H, CH), 6.84 (dd, J = 8.0, 1.5 Hz, 1H, ArH), 6.89 (td, J = 7.5, 1.5 Hz, 1H, ArH), 7.11–7.38 (m, 11H, ArH). 13C NMR (101 MHz, CDCl3) δ 21.3 (CH3), 56.0 (CH2N), 58.6 (CH2Ph), 58.9 (CH2Ph), 66.6 (CH), 71.3 (CH2O), 113.6 (CHAr), 121.7 (CHAr), 123.2 (CAr), 127.4 (CHAr), 127.8 (CHAr), 128.6 (4CHAr), 129.1–129.3 (4CHAr), 130.3 (CHAr), 135.5 (CAr), 137.0 (CAr), 138.8 (CAr), 154.3 (CAr). HRMS C24H26ClNO2 [M + H]+ 396.1729; found 396.1725. Purity 95.30% (tR = 4.12 min).
4.1.30. 1-[Bis(4-methylbenzyl)amino]-3-(4-chlorophenoxy)propan-2-ol (28)
Following the general procedure, bis(4-methylbenzyl)amine (CAS 98180-43-9) (100.0 mg, 0.43 mmol) and 2-[(4-chlorophenoxy)methyl]oxirane (81.9 mg, 0.48 mmol) afforded 28 (219.8 mg, 87.3%) as an oil after column chromatography (hexane/EtOAc 9:1 to 8:2). IR (ATR) 2920, 2843, 1590, 1510, 1485, 1445, 1278, 1253, 1061, 1036, 807, 741 cm−1. 1H NMR (400 MHz, CDCl3) δ 2.33 (s, 6H, 2CH3), 2.72 (d, J = 6.5 Hz, 2H, CH2N), 3.51 (d, J = 13.0 Hz, 2H, CH2Ph), 3.76 (d, J = 13.0 Hz, 2H, CH2Ph), 3.93 (d, J = 5.0 Hz, 2H, CH2O), 4.10 (m, 1H, CH), 6.84 (dd, J = 8.0, 1.5 Hz, 1H, ArH), 6.88 (td, J = 7.5, 1.5 Hz, 1H, ArH), 7.10–7.15 (m, 4H, ArH), 7.16–7.18 (m, 1H, ArH), 7.18–7.23 (m, 4H, ArH), 7.33 (dd, J = 8.0, 1.5 Hz, 1H, ArH). 13C NMR (101 MHz, CDCl3) δ 21.3 (2CH3), 55.9 (CH2N), 58.57 (2CH2Ph), 66.5 (CH), 71.25 (CH2O), 113.61 (2CHAr), 121.67 (2CHAr), 123.17 (CAr), 127.7 (2CHAr), 129.2 (2CHAr), 130.3 (4CHAr), 135.6 (2CAr), 137.0 (2CAr), 154.3 (CAr). HRMS C25H29ClNO2 [M + H]+ 464.0945; found 464.0947.
4.1.31. 1-[(4-Chlorobenzyl)(4-methylbenzyl)amino]-3-(4-chlorophenoxy)propan-2-ol (29)
Following the general procedure, N-(4-chlorobenzyl)-1-(p-tolyl)methanamine (150 mg, 0.61 mmol) and 2-[(4-chlorophenoxy)methyl]oxirane (112.7 mg, 0.61 mmol) afforded 29 (240 mg, 91.4%) as an oil after column chromatography (hexane/EtOAc 9:1 to 8:2). IR (ATR) 3445, 3024, 2924, 1736, 1588, 1485, 1446, 1277, 1247, 1062, 1015, 805, 746 cm−1. 1H NMR (400 MHz, CDCl3) δ 2.32 (s, 3H, CH3), 2.69–2.73 (m, 2H, CH2N), 3.42–3.55 (dd, J = 13.5, 2 Hz, 2H, CH2Ph), 3.72 (d, J = 13.5 Hz, 2H, CH2Ph), 3.92 (d, J = 5.0 Hz, 2H, CH2O), 4.06–4.13 (m, 1H, CH), 6.83 (dd, J = 8.5 Hz, 1.5 Hz, 1H, ArH), 6.88 (td, J = 7.5 Hz, 1.5 Hz, 1H, ArH), 7.10–7.29 (m, 10H, ArH), 7.33 (dd, J = 8.0, 1.5 Hz, 1H, ArH). 13C NMR (101 MHz, CDCl3) δ 21.3 (CH3), 56.0 (CH2N), 58.2 (CH2Ph), 58.6 (CH2Ph), 66.7 (CH), 71.1 (CH2O), 113.6 (CHAr), 121.8 (CHAr), 123.2 (CAr), 127.8 (CHAr), 128.7 (2CHAr), 129.2 (2CHAr), 129.3 (2CHAr), 130.4 (CHAr), 130.5 (2CHAr), 133.1 (CAr), 135.3 (CAr), 137.2 (CAr), 137.4 (CAr), 154.2 (CAr). HRMS C24H25Cl2NO2 [M + H]+ 430.1335; found 430.1341. Purity 94.16% (tR = 4.51 min).
4.1.32. 1-[Benzyl(3,4-dichlorobenzyl)amino]-3-(4-chlorophenoxy)propan-2-ol (30)
Following the general procedure, N-benzyl-1-(3,4-dichlorophenyl)methanamine (CAS 14502-37-5) (271.6 mg, 1.02 mmol) and 2-[(4-chlorophenoxy)methyl]oxirane (188 mg, 1.02 mmol) afforded 30 (402.5 mg, 87.5%) as an oil after column chromatography (hexane/EtOAc 9:1 to 8:2). IR (ATR) 3432, 3025, 2932, 1588, 1485, 1445, 1278, 1247, 1062, 1028, 812, 743, 698 cm−1.1H NMR (400 MHz, CDCl3) δ 2.68–2.79 (m, 2H, CH2N), 2.96 (s, 1H, OH), 3.51 (d, J = 13.5 Hz, 1H, CH2Ph), 3.57 (d, J = 13.5 Hz, 1H, CH2Ph), 3.70 (d, J = 13.5 Hz, 1H, CH2Ph), 3.74 (d, J = 13.5 Hz, 1H, CH2Ph), 3.98 (m, 2H, CH2O), 4.15 (m, 1H, CH), 6.82–6.95 (m, 2H, ArH), 7.13–7.23 (m, 2H, ArH), 7.25–7.43 (m, 8H, ArH).13C NMR (101 MHz, CDCl3) δ 56.1 (CH2N), 58.0 (CH2Ph), 59.1 (CH2Ph), 67.0 (CH), 71.1 (CH2O), 113.6 (CHAr), 121.9 (CHAr), 123.2 (CAr), 127.6 (CHAr), 127.9 (CHAr), 128.4 (CHAr), 128.7 (2CHAr), 129.2 (2CHAr), 130.4 (CHAr), 130.5 (CHAr), 131.0 (CHAr), 131.3 (CHAr), 132.6 (CAr), 138.2 (CAr), 139.3 (CAr), 154.2 (CAr). HRMS C23H22Cl3NO2 [M + H]+ 450.0789; found 450.0789. Purity 93.97% (tR = 5.36 min).
4.1.33. 1-[(4-Chlorobenzyl)(3,4-dichlorobenzyl)amino]-3-(4-chlorophenoxy)propan-2-ol (31)
Following the general procedure, N-(4-chlorobenzyl)-1-(3,4-dichlorophenyl)methan amine (150 mg, 0.5 mmol) and 2-[(4-chlorophenoxy)methyl]oxirane (92.1 mg, 0.5 mmol) afforded 31 (192.7 mg, 79.6%) as an oil after column chromatography (hexane/EtOAc 9:1 to 8:2). IR (ATR) 3438, 3058, 2934, 1740, 1589, 1485, 1277, 1248, 1062, 1029, 815, 746 cm−1. 1H NMR (400 MHz, CDCl3) δ 2.60–2.80 (m, 2H, CH2N), 2.88 (s, 1H, OH), 3.54 (dd, J = 13.5, 11.5 Hz, 2H, CH2), 3.69 (dd, J = 13.5, 11.5 Hz, 2H, CH2), 3.89–3.99 (m, 2H, CH2O), 4.12 (m, 1H, CH), 6.84 (dd, J = 8.0, 1.5 Hz, 1H, ArH), 6.91 (td, J = 7.5, 1.5 Hz, 1H, ArH), 7.12–7.25 (m, 4H, ArH), 7.27–7.40 (m, 5H, ArH). 13C NMR (101 MHz, CDCl3) δ 56.0 (CH2N), 58.0 (CH2Ph), 58.4 (CH2Ph), 67.2 (CH), 71.0 (CH2O), 113.6 (CHAr), 122.0 (CHAr), 123.1 (CAr), 127.9 (CHAr), 128.3 (CHAr), 128.9 (2CHAr), 130.4 (3CHAr), 130.6 (CHAr), 130.9 (CHAr), 131.5 (CAr), 132.7 (CAr), 133.4 (CAr), 136.8 (CAr), 139.1 (CAr), 154.1 (OCAr). HRMS C23H21Cl4NO2 [M + H]+ 484.0399; found 484.04. Purity 92.14% (tR = 5.93 min).
4.1.34. 1-(4-Chlorophenoxy)-3-[(3,4-dichlorobenzyl)(4-methylbenzyl)amino]propan-2-ol (32)
Following the general procedure, N-(3,4-dichlorobenzyl)-1-(p-tolyl)methanamine (151.8 mg, 0.54 mmol) and 2-[(4-chlorophenoxy)methyl]oxirane (100 mg, 0.54 mmol) afforded 32 (219.8 mg, 87.3%) as an oil after column chromatography (hexane/EtOAc 9:1 to 8:2). IR (ATR) 3414, 3019, 2928, 1737, 1588, 1485, 1445, 1247, 1061, 1029, 814, 802, 746 cm−1. 1H NMR (400 MHz, CDCl3) δ 2.34 (s, 3H, CH3), 2.67–2.78 (m, 2H, CH2N), 3.50 (d, J = 13.5 Hz, 1H, CH2Ph), 3.53 (d, J = 13.5 Hz, 1H, CH2Ph), 3.68 (d, J = 13.5 Hz, 1H, CH2Ph), 3.71 (d, J = 13.5 Hz, 1H, CH2Ph), 3.90–3.98 (m, 2H, CH2O), 4.11 (m, 1H, CH), 6.85 (dd, J = 8.5, 1.5 Hz, 1H, ArH), 6.90 (td, J = 7.5, 1.5 Hz, 1H, ArH), 7.10–7.22 (m, 6H, HAr), 7.31–7.41 (m, 3H, 3HAr). 13C NMR (101 MHz, CDCl3) δ 21.3 (CH3), 56.0 (CH2N), 57.9 (CH2Ph), 58.8 (CH2Ph), 66.9 (CH), 71.1 (CH2O), 113.6 (CHAr), 121.9 (CHAr), 123.1 (CAr), 127.8 (CHAr), 128.4 (CHAr), 129.1 (2CHAr), 129.4 (2CHAr), 130.4 (CHAr), 130.5 (CHAr), 130.9 (CHAr), 131.3 (CAr), 132.6 (CAr), 135.1 (CAr), 137.3 (CAr), 139.4 (CAr), 154.2 (OCAr). HRMS C24H24Cl3NO2 [M + H]+ 464.0945; found 464.0947. Purity 90.51% (tR = 5.34 min).
4.1.35. 1-[Benzyl(4-methoxybenzyl)amino]-3-(4-chlorophenoxy)propan-2-ol (33)
Following the general procedure, N-benzyl-1-(4-methoxyphenyl)methanamine (CAS 14429-02-8) (184.6 mg, 0.45 mmol) and 2-[(4-chlorophenoxy)methyl]oxirane (104 mg, 0.81 mmol) afforded 33 (283.1 mg, 84.6%) as an oil after column chromatography (hexane/EtOAc 9:1 to 8:2). IR (ATR) 2932, 2828, 1612, 1587, 1509, 1485, 1447, 1244, 1063, 1029, 810, 742, 697 cm−1. 1H NMR (400 MHz, CDCl3) δ 2.72 (d, J = 6.5 Hz, 2H, CH2N), 3.50 (d, J = 13.5 Hz, 1H, CH2Ph), 3.51 (d, J = 13.5 Hz, 1H, CH2Ph), 3.53 (d, J = 13.5 Hz, 1H, CH2Ph), 3.71 (d, J = 13.5 Hz, 1H, CH2Ph), 3.79 (d, J = 13.5 Hz, 1H, CH2Ph), 3.79 (s, 3H, CH3), 3.92 (dd, J = 5.0, 1.5 Hz, 2H, CH2), 4.10 (m, 1H, CH), 6.81–6.86 (m, 2H, ArH), 6.88 (dd, J = 7.5, 1.5 Hz, 2H, ArH), 7.17 (ddd, J = 8.0, 7.5, 1.5 Hz, 1H, ArH), 7.20–7.26 (m, 3H), 7.29–7.32 (m, 3H, ArH), 7.33 (d, J = 1.5 Hz, 2H, ArH). 13C NMR (101 MHz, CDCl3) δ 55.2 (CH3), 55.8 (CH2N), 58.1 (CH2Ph), 58.7 (CH2Ph), 66.4 (CH), 71.15 (CH2O), 113.5 (CHAr), 113.8 (CHAr), 121.6 (2CHAr), 123.0 (2CHAr), 127.3 (2CHAr), 127.6 (CHAr), 128.4 (CAr), 129.1 (CAr), 130.2 (CAr), 130.3 (CAr), 130.5 (2CAr), 138.6 (CAr), 154.2 (CAr), 158.8 (OCAr). HRMS C24H27ClNO3 [M + H]+ 484.0399; found 484.04. Purity 92.14% (tR = 5.93 min).
4.1.36. 1-[Bis(4-methoxybenzyl)amino]-3-(4-chlorophenoxy)propan-2-ol (34)
Following the general procedure, bis(4-methoxybenzyl)amine (CAS 17061-62-0) (278.8 mg, 1.08 mmol) and 2-[(4-chlorophenoxy)methyl]oxirane (200 mg, 1.08 mmol) afforded 34 (405.6 mg, 84.7%) as a white solid after column chromatography (hexane/EtOAc 9:1 to 8:2). Mp 112–114 °C (EtOAc). IR (ATR) 3409, 3005, 2932, 2834, 1586, 1510, 1486, 1446, 1249, 1232, 1060, 1026, 808, 746, 696 cm−1. 1H NMR (400 MHz, CDCl3) δ 2.71 (d, J = 6.5 Hz, 2H, CH2N), 3.48 (d, J = 13.5 Hz, 2H, CH2Ph), 3.72 (d, J = 13.5 Hz, 2H, CH2Ph), 3.80 (s, 4H, 2CH3), 3.92 (d, J = 5.0 Hz, 2H, CH2O), 4.05–4.12 (m, 1H, CH), 6.82–6.91 (m, 6H, ArH), 7.14–7.24 (m, 5H, ArH), 7.33 (dd, J = 8.0, 1.5 Hz, 1H, ArH). 13C NMR (101 MHz, CDCl3) δ 55.4 (2CH3), 55.7 (CH2N), 58.1 (2CH2Ph), 66.5 (CH), 71.3 (CH2O), 113.6 (C-ipso), 113.9 (4CHAr), 121.7 (CHAr), 123.2 (CAr), 127.8 (CHAr), 130.3 (CHAr), 130.4 (5CHAr), 130.7 (CAr), 154.3 (CAr), 158.9 (2CAr). HRMS C25H28ClNO4 [M + H]+ 442.178; found 442.1777. Purity 95.65% (tR = 4.02 min).
4.1.37. 1-(4-Chlorophenoxy)-3-[(3,4-dichlorobenzyl)(4-methoxybenzyl)amino]propan-2-ol (35)
Following the general procedure, N-(3,4-dichlorobenzyl)-1-(4-methoxyphenyl)methanamine (160.4 mg, 0.54 mmol) and 2-[(4-chlorophenoxy)methyl]oxirane (100 mg, 0.54 mmol) afforded 35 (251 mg, 96.4%) as an oil after column chromatography (hexane/EtOAc 9:1 to 8:2). IR (ATR) 3451, 3006, 2934, 1588, 1511, 1486, 1245, 1061, 1028, 819, 746 cm−1. 1H NMR (400 MHz, CDCl3) δ 2.65–2.79 (m, 2H, CH2N), 2.99 (s, 1H, OH), 3.51 (dd, J = 13.5, 3.0 Hz, 2H, CH2), 3.69 (d, J = 13.5 Hz, 2H, CH2), 3.80 (s, 3H, CH3), 3.87–4.03 (m, 2H, CH2O), 4.06–4.15 (m, 1H, CH), 6.83–6.93 (m, 4H, ArH), 7.12–7.23 (m, 4H, ArH), 7.31–7.40 (m, 3H, ArH). 13C NMR (101 MHz, CDCl3) δ 55.4 (CH3), 55.9 (CH2N), 57.8 (CH2Ph), 58.4 (CH2Ph), 66.9 (CH), 71.1 (CH2O), 113.6 (CHAr), 114.1 (2CHAr), 121.9 (CHAr), 123.1 (CAr), 127.8 (CHAr), 128.4 (CHAr), 130.1 (CAr), 130.4 (3CHAr), 130.5 (CHAr), 130.9 (CHAr), 131.3 (CAr), 132.6 (CAr), 139.4 (CAr), 154.2 (CAr), 159.1 (OCAr). HRMS C24H24Cl3NO3 [M + H]+ 480.0895; found 480.0894. Purity 96.56% (tR = 4.71 min).
4.2. Cell Culture
Human Huh-7 hepatoma cells (kindly donated by Dr. Mayka Sanchez from the Josep Carreras Leukemia Research Institute, Barcelona) were cultured in DMEM supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin, at 37 °C under 5% CO2.
4.3. Reverse Transcription-Polymerase Chain Reaction and Quantitative Polymerase Chain Reaction
Isolated RNA was reverse transcribed to obtain 1 μg of complementary DNA (cDNA) using Random Hexamers (Thermo Fisher Scientific, Waltham, MA, USA), 10 mM deoxynucleotide (dNTP) mix, and the reverse transcriptase enzyme derived from the Moloney murine leukemia virus (MMLV, Thermo Fisher Scientific). The experiment was run in a thermocycler (BioRad, Hercules, CA, USA) and consisted of a program with different steps and temperatures: 65 °C for 5 min, 4 °C for 5 min, 37 °C for 2 min, 25 °C for 10 min, 37 °C for 50 min, and 70 °C for 15 min. The relative levels of specific mRNAs were assessed by real-time RT-PCR in a Mini 48-Well T100™ thermal cycler (Bio-Rad), using the SYBR Green Master Mix (Applied Biosystems, Waltham, MA, USA), as previously described [13]. Briefly, samples had a final volume of 20 μL, with 20 ng of total cDNA, 0.9 μM of the primer mix, and 10 μL of 2× SYBR Green Master Mix. The thermal cycler protocol for real-time PCR included a first step of denaturation at 95 °C for 10 min, followed by 40 repeated cycles of 95 °C for 15 s, 60 °C for 30 s, and 72 °C for 30 s for denaturation, primer annealing, and amplification, respectively. Primer sequences were designed using the Primer-BLAST tool (NCBI), based on the full mRNA sequences to find the optimal primers for amplification, and evaluated with the Oligo-Analyzer Tool (Integrated DNA Technologies) to ensure an optimal melting temperature (Tm) and avoid the formation of homo/heterodimers or non-specific structures that can interfere with the interpretation of the results. The primer sequences were designed specifically to span the junction between the exons. Values were normalized to the glyceraldehyde 3-phosphate dehydrogenase (Gapdh) expression levels, and measurements were performed in triplicate. All changes in expression were normalized to the untreated control.
4.4. Immunoblotting
The isolation of total protein extracts was performed as described elsewhere [13]. Immunoblotting was performed with antibodies against α-tubulin (T6074 Sigma-Aldrich, St. Louis, MO, USA), phosphorylated AMPKT172 (2531, Cell Signaling Technology), and GDF15 (sc-515675, Santa Cruz Biotechnology Inc, Dallas, TX, USA). Signal acquisition was conducted using the Bio-Rad ChemiDoc apparatus, and quantification of the immunoblot signal was performed with the Bio-Rad Image Lab software. The results for protein quantification were normalized to the levels of a control protein (α-tubulin) to avoid unwanted sources of variation.
4.5. Statistical analysis
Results are expressed as the mean ± SEM. Significant differences were assessed by two-way ANOVA using the GraphPad Prism program (version 9.0.2) (GraphPad Software Inc., San Diego, CA, USA). When significant variations were found by ANOVA, Tukey’s post hoc test for multiple comparisons was performed only if F achieved a p-value < 0.05. Differences were considered significant at p < 0.05.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/molecules28145468/s1, General procedure for the preparation of epoxyde intermediates; 1H-NMR and 13C-NMR spectra; HPLC/MS analysis, Smiles, HETCOR, HMRS.
Author Contributions
Conceptualization, M.V.-C., C.E. and S.V.; methodology, M.V.-C., C.E., S.V., A.B. and M.Z.; investigation, A.B., D.M., E.B. and M.Z.; data curation, A.B. and M.Z.; writing—original draft preparation, M.V.-C. and C.E.; writing—review and editing, M.V.-C., C.E., E.B., X.P. and S.V.; supervision, M.V.-C. and C.E.; funding acquisition, M.V.-C. and S.V. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by grants PID2020-118127RB-I00 (to S.V.), PID2019-107991RB-100 (to C.E.) and PID2021-122116OB-I00 (to M.V.-C.) from MCIN/AEI/10.13039/501100011033 and “ERDF, A Way of Making Europe”. CIBER de Diabetes y Enfermedades Metabólicas Asociadas (CIBERDEM) is a Carlos III Health Institute project. Support was also received from the CERCA Programme/Generalitat de Catalunya. We thank the Generalitat de Catalunya (Grup de recerca 2021 SGR 00357). Zhang was supported by a grant from the China Scholarship Council (CSC) (202007565030).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The study data are available upon request.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Aguilar-Recarte, D.; Barroso, E.; Palomer, X.; Wahli, W.; Vázquez-Carrera, M. Knocking on GDF15′s door for the treatment of type 2 diabetes mellitus. Trends Endocrinol. Metab. 2022, 33, 741–754. [Google Scholar] [PubMed]
- Hsu, J.-Y.; Crawley, S.; Chen, M.; Ayupova, D.A.; Lindhout, D.A.; Higbee, J.; Kutach, A.; Joo, W.; Gao, Z.; Fu, D.; et al. Non-homeostatic body weight regulation through a brainstem-restricted receptor for GDF15. Nature 2017, 550, 255–259. [Google Scholar] [PubMed]
- Tran, T.; Yang, J.; Gardner, J.; Xiong, Y. GDF15 deficiency promotes high fat diet-induced obesity in mice. PLoS ONE 2018, 13, e0201584. [Google Scholar] [CrossRef] [PubMed]
- Emmerson, P.J.; Wang, F.; Du, Y.; Liu, Q.; Pickard, R.T.; Gonciarz, M.D.; Coskun, T.; Hamang, M.J.; Sindelar, D.K.; Ballman, K.K.; et al. The metabolic effects of GDF15 are mediated by the orphan receptor GFRAL. Nat. Med. 2017, 23, 1215–1219. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Chang, C.-C.; Sun, Z.; Madsen, D.; Zhu, H.; Padkjær, S.B.; Wu, X.; Huang, T.; Hultman, K.; Paulsen, S.J.; et al. GFRAL is the receptor for GDF15 and is required for the anti-obesity effects of the ligand. Nat. Med. 2017, 23, 1158–1166. [Google Scholar] [CrossRef] [PubMed]
- Benichou, O.; Coskun, T.; Gonciarz, M.D.; Garhyan, P.; Adams, A.C.; Du, Y.; Dunbar, J.D.; Martin, J.A.; Mather, K.J.; Pickard, R.T.; et al. Discovery, development, and clinical proof of mechanism of LY3463251, a long-acting GDF15 receptor agonist. Cell Metab. 2023, 35, 274–286.e10. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhao, X.; Dong, X.; Zhang, Y.; Zou, H.; Jin, Y.; Guo, W.; Zhai, P.; Chen, X.; Kharitonenkov, A. Activity-balanced GLP-1/GDF15 dual agonist reduces body weight and metabolic disorder in mice and non-human primates. Cell Metab. 2023, 35, 287–298.e4. [Google Scholar] [CrossRef] [PubMed]
- Gerstein, H.C.; Pare, G.; Hess, S.; Ford, R.J.; Sjaarda, J.; Raman, K.; McQueen, M.; Lee, S.; Haenel, H.; Steinberg, G.R. Growth Differentiation Factor 15 as a Novel Biomarker for Metformin. Diabetes Care 2017, 40, 280–283. [Google Scholar] [CrossRef] [PubMed]
- Coll, A.P.; Chen, M.; Taskar, P.; Rimmington, D.; Patel, S.; Tadross, J.A.; Cimino, I.; Yang, M.; Welsh, P.; Virtue, S.; et al. GDF15 mediates the effects of metformin on body weight and energy balance. Nature 2020, 578, 444–448. [Google Scholar] [PubMed]
- EDay, A.; Ford, R.J.; Smith, B.K.; Mohammadi-Shemirani, P.; Morrow, M.R.; Gutgesell, R.M.; Lu, R.; Raphenya, A.R.; Kabiri, M.; McArthur, A.G.; et al. Metformin-induced increases in GDF15 are important for suppressing appetite and promoting weight loss. Nat. Metab. 2019, 1, 1202–1208. [Google Scholar]
- Klein, A.B.; Nicolaisen, T.S.; Johann, K.; Fritzen, A.M.; Mathiesen, C.V.; Gil, C.; Pilmark, N.S.; Karstoft, K.; Blond, M.B.; Quist, J.S.; et al. The GDF15-GFRAL pathway is dispensable for the effects of metformin on energy balance. Cell Rep. 2022, 40, 111258. [Google Scholar] [CrossRef]
- Zhang, S.-Y.; Bruce, K.; Danaei, Z.; Li, R.J.; Barros, D.R.; Kuah, R.; Lim, Y.-M.; Mariani, L.H.; Cherney, D.Z.; Chiu, J.F.; et al. Metformin triggers a kidney GDF15-dependent area postrema axis to regulate food intake and body weight. Cell Metab. 2023, 35, 875–886. [Google Scholar] [CrossRef] [PubMed]
- He, L. Metformin and Systemic Metabolism. Trends Pharmacol. Sci. 2020, 41, 868–881. [Google Scholar] [PubMed]
- Aguilar-Recarte, D.; Barroso, E.; Zhang, M.; Rada, P.; Pizarro-Delgado, J.; Peña, L.; Palomer, X.; Valverde, M.; Wahli, W.; Vázquez-Carrera, M. A positive feedback loop between AMPK and GDF15 promotes metformin antidiabetic effects. Pharmacol. Res. 2022, 187, 106578. [Google Scholar] [PubMed]
- Liu, Y.; Jurczak, M.J.; Lear, T.B.; Lin, B.; Larsen, M.B.; Kennerdell, J.R.; Chen, Y.; Huckestein, B.R.; Nguyen, M.K.; Tuncer, F.; et al. A Fbxo48 inhibitor prevents pAMPKalpha degradation and ameliorates insulin resistance. Nat. Chem. Biol. 2021, 17, 298–306. [Google Scholar] [PubMed]
- Chen, B.; Mallampalli, R.K.; Liu, Y. WO2018067685A1; University of Pittsbugh, Department of Veterans Affairs: Pittsburgh, PA, USA, 2018. [Google Scholar]
- Xie, B.; Murali, A.; Vandevender, A.M.; Chen, J.; Silva, A.G.; Bello, F.M.; Chuan, B.; Bahudhanapati, H.; Sipula, I.; Dedousis, N.; et al. Hepatocyte-derived GDF15 suppresses feeding and improves insulin sensitivity in obese mice. iScience 2022, 25, 105569. [Google Scholar] [PubMed]
- Zarei, M.; Pujol, E.; Quesada-López, T.; Villarroya, F.; Barroso, E.; Vázquez, S.; Pizarro-Delgado, J.; Palomer, X.; Vázquez-Carrera, M. Oral administration of a new HRI activator as a new strategy to improve high-fat-diet-induced flucose intolerance, hepatic steatosis, and hypertriglyceridaemia through FGF21. Br. J. Pharmacol. 2019, 176, 2292–2305. [Google Scholar] [PubMed]
- Kincaid, J.W.R.; Coll, A.P. Metformin and GDF15: Where are we now? Nat. Rev. Endocrinol. 2023, 19, 6–7. [Google Scholar] [PubMed]
- Draz, H.; Goldberg, A.A.; Titorenko, V.I.; Guns, E.S.T.; Safe, S.H.; Sanderson, J.T. Diindolylmethane and its halogenated derivatives induce protective autophagy in human prostate cancer cells via induction of the oncogenic protein AEG-1 and activation of AMP-activated protein kinase (AMPK). Cell. Signal. 2017, 40, 172–182. [Google Scholar] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).