Abstract
A biomimetic mineralization method was used in the facile and rapid preparation of nanoflowers for immobilizing alcohol dehydrogenase (ADH). The method mainly uses ADH as an organic component and zinc phosphate as an inorganic component to prepare flower-like ADH/Zn3(PO4)2 organic-inorganic hybrid nanoflowers (HNFs) with the high specific surface area through a self-assembly process. The synthesis conditions of the ADH HNFs were optimized and its morphology was characterized. Under the optimum enzymatic reaction conditions, the Michaelis-Menten constant (Km) of ADH HNFs (β-NAD+ as substrate) was measured to be 3.54 mM, and the half-maximal inhibitory concentration (IC50) of the positive control ranitidine (0.2–0.8 mM) was determined to be 0.49 mM. Subsequently, the inhibitory activity of natural medicine Penthorum chinense Pursh and nine small-molecule compounds on ADH was evaluated using ADH HNFs. The inhibition percentage of the aqueous extract of P. chinense is 57.9%. The vanillic acid, protocatechuic acid, gallic acid, and naringenin have obvious inhibitory effects on ADH, and their percentages of inhibition are 55.1%, 68.3%, 61.9%, and 75.5%, respectively. Moreover, molecular docking analysis was applied to explore the binding modes and sites of the four most active small-molecule compounds to ADH. The results of this study can broaden the application of immobilized enzymes through biomimetic mineralization, and provide a reference for the discovery of ADH inhibitors from natural products.
1. Introduction
Alcohol dehydrogenase (ADH), which is a key enzyme for the metabolism of short-chain alcohols in organisms, is abundant in human and animal liver, and plant and microbial cells [1,2,3,4]. As a substrate-specific zinc-containing metalloenzyme [5], each enzyme subunit in the ADH structure binds to two Zn2+ that maintain the structure and the catalytically active center of the enzyme. ADH plays an important role in many physiological processes of human body. ADH catalyzes the oxidation of primary and secondary alcohols to produce corresponding aldehydes and ketones [6], in which the cofactor nicotinamide adenine dinucleotide (NAD+) is reduced to NADH [7]. Studies have reported that in alcoholism or cirrhosis, ADH activity is significantly increased in serum [8,9]. The initial activity of ADH is positively correlated with liver damage-related indicators [10]. ADH inhibitors can reduce the production of aldehydes in alcohol metabolism by inhibiting the activity of ADH, thereby acting as the alcoholism antidote and hepatoprotector [11]. Currently, the mainly used antidotes in clinical practice are synthetic ADH inhibitors, such as 4-methylpyrazole, cimetidine, and formamides [7]. The 4-methylpyrazole can also prevent acetaminophen-induced acute kidney injury [12]. However, the 4-methylpyrazole is not indicated for patients with metabolic acidemia [13], and may cause several adverse reactions, including headache, nausea, and dizziness [14]. Several natural medicines possess specific anti-alcoholism and hepatoprotective effects, which have significant clinical curative efficacy with few side effects [15,16]. Thus, screening of ADH inhibitors from natural medicines becomes a new approach to developing alcoholism antidotes.
Penthorum chinense Pursh (P. chinense, Saxifragaceae) is a traditional Chinese medicine, which is named ‘GanHuanCao’ in Chinese [17]. It has the effects of clearing heat and detoxification, reducing icteric and dampness, activating blood circulation, and dissolving stasis and swelling, being used in the treatment of jaundice, edema, trauma, cholecystitis, and liver disease [18]. Furthermore, several previous studies reported that the aqueous extract of P. chinense can protect against both acute and chronic alcohol-induced liver injury [19], and displays antioxidative and free radical scavenging activities [20]. Therefore, the screening of ADH inhibitors from P. chinense is significant for exploring its pharmacological activity such as the protective effect on alcohol-induced liver injury.
Ge et al. [21] first reported a method for preparing protein-inorganic hybrid nanoflowers with certain morphology and structure using protein as an organic component and copper phosphate (Cu3(PO4)2) as an inorganic component. It was found that the laccase and carbonic anhydrase nanoflowers have higher catalytic activity than their corresponding free enzymes. Different from the traditional enzyme immobilization method, this biomimetic mineralization method can be realized by simply adding organic biological macromolecules to the inorganic metal ion solution, forming organic-inorganic hybrid nanoflowers. Therefore, the application of protein-inorganic hybrid nanoflowers for the immobilization of enzymes has received dominant attention owing to its simple and efficient synthesis method [22]. In recent years, organic-inorganic hybrid nanoflowers using lipase [23], horseradish peroxidase [24], glucose oxidase [25], α-amylase [26], urease [27], trypsin [28], chymotrypsin [29], papain [30], α-glucosidase [31], β-galactosidase [32], sucrose phosphorylase [33], and DNA [34] as organic components have been reported, and their catalytic activity and stability are higher than those of their corresponding free enzymes.
In this study, ADH-zinc phosphate hybrid nanoflowers were prepared through biomimetic mineralization and applied in the screening of ADH inhibitors from P. chinense (Figure 1). The ADH hybrid nanoflowers (HNFs) were prepared using ADH as an organic component and Zn3(PO4)2 as an inorganic component, in which the synthesis of nanoflower carriers and the fixation of ADH were completed simultaneously, greatly simplifying the immobilization process of enzymes. The enzyme immobilization conditions were optimized, and the enzymatic reaction conditions and the kinetic performance of immobilized ADH were studied. Subsequently, the ADH inhibitory activity of aqueous extract and nine small-molecule compounds from P. chinense were evaluated using the prepared ADH HNFs. Finally, molecular docking of ADH with small-molecule compounds of good inhibitory activity was performed to predict the binding site of ligands to the enzyme.
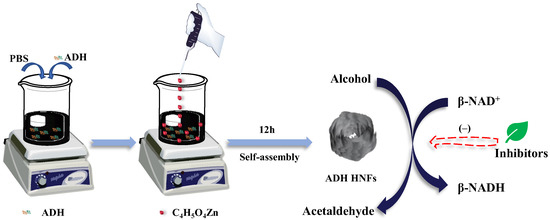
Figure 1.
Schematic illustration of the fabrication process of ADH HNFs and the principle of catalysis.
2. Results and Discussion
2.1. Monitoring the Enzyme Activity of ADH HNFs by UV Analysis
To investigate the feasibility of monitoring the enzyme activity by the design principle, the UV absorbances of five solutions at 240–400 nm were measured. As shown in Figure 2, both ethanol and HNFs + ethanol solutions (Figure 2A,C) have no UV absorption peaks at 240–400 nm. In the conversion of ethanol to acetaldehyde catalyzed by ADH, coenzyme I (β-NAD+) is involved to generate β-NADH. Both the β-NAD+ and β-NADH have absorption peaks at 260 nm (Figure 2B,D,E), but only the β-NADH shows a characteristic absorption peak at 340 nm (Figure 2E). Therefore, the enzymatic activity of ADH HNFs can be assessed by monitoring the absorbance of β-NADH at 340 nm.
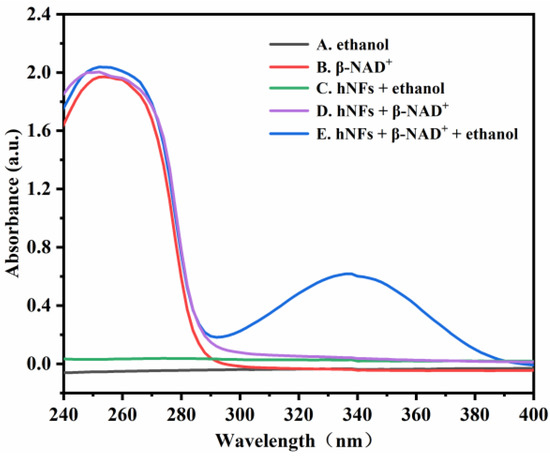
Figure 2.
UV absorption spectra of five different solutions. (A) ethanol; (B) β-NAD+; (C) HNFs + ethanol; (D) HNFs + β-NAD+; (E) HNFs + β-NAD+ + ethanol. Conditions: the volumes of ADH HNFs, tris-HCl (pH = 7.0), and ethanol are 25, 200, and 50 μL, respectively; β-NAD+ final concentration, 3 mM; enzymatic reaction at 35 °C for 2 min and centrifuged for 1.5 min, 60 μL of supernatant was taken for measurement.
2.2. Optimization of Preparation Conditions of ADH HNFs
For the growth of hybrid nanoflower, there are four main steps, including crystallization and coordination, in situ precipitation, self-assembly, and size growth (Figure 3) [35]. The primary crystals of zinc phosphate are first formed. The complexes of enzyme molecules with Zn2+ are formed predominantly through the coordination facility of amide groups in the ethanol dehydrogenase backbone and nitrogen atoms in amino acid residues. These complexes in turn provide nucleation sites for the primary crystals. Next, the growth of Zn3(PO4)2 crystals at the Zn2+ binding site on the surface of the complex allows the ethanol dehydrogenase molecule and the primary crystals to gradually grow into large agglomerates, resulting in the formation of individual nanosheets. Then, the self-assembly occurs between nanosheets to form a flower-like structure, and finally, anisotropic growth results in the complete formation of ADH HNFs. In this proposed growth process, alcohol dehydrogenase plays an important role in regulating the nucleation of Zn3(PO4)2 crystals to form the scaffold for the petals and serves as a ‘glue’ to bind the nanosheets together [21].
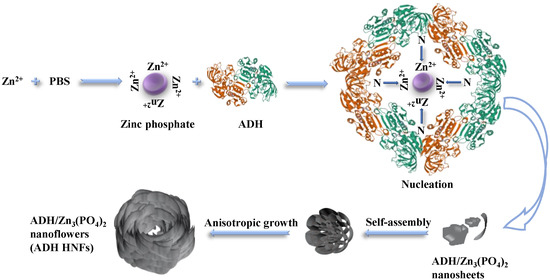
Figure 3.
Schematic diagram of the proposed mechanism for the formation of ADH HNFs through the biomineralization method.
To obtain ADH HNFs with suitable size and morphology, as well as good catalytic activity and storage stability, the preparation method, enzyme amount, and immobilization time were investigated. The magnetic stirring equipment used in this experiment cannot be temperature controlled and the room temperature was about 25 °C during the synthesis, the effect of different temperatures on the preparation of nanoflowers is not considered here. Furthermore, according to the previous reports [36,37,38], phosphate organic-inorganic hybrid nanoflowers are usually fully formed within 6 h, therefore, 6 h was used firstly as the time to immobilize ADH. The reproducibility of the immobilized enzyme can be used to appraise whether the immobilization method is stable and reliable. Three batches of ADH HNFs were prepared through different preparation methods (thermostatic oscillation for 6 h at 30 °C or magnetic stirring for 6 h at room temperature of 30 °C) and evaluated by measuring the absorbance of generated β-NADH at 340 nm. The relative standard deviation (RSD) of batch-to-batch (n = 3) for magnetic stirring and thermostatic oscillation is 4.1% and 9.4%, respectively, indicating that the ADH HNFs prepared by magnetic stirring have better reproducibility.
As shown in Figure 4A–I, the hybrid nanoflowers prepared through different methods have obviously different morphologies and sizes. Figure 4A–E shows that ADH HNFs prepared through the magnetic stirring method have a tight structure, approximating the structure of a ‘flower bud’. Figure 4F–I shows that ADH HNFs prepared through the constant temperature oscillation method have sparse petals and loose structures. The overall size is larger than that of ADH HNFs prepared through the magnetic stirring method. Based on these SEM images, the poor reproducibility of the ADH HNFs prepared by the thermostatic oscillation method may be due to the non-uniform flower structure of the nanoflowers prepared in different batches.
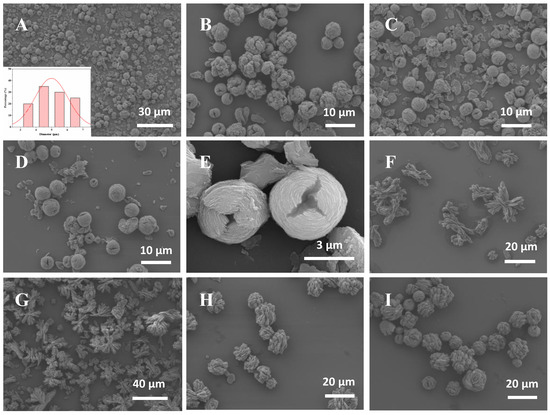
Figure 4.
The SEM images of ADH HNFs that were prepared through magnetic stirring for 6 h at room temperature (A–E) and thermostatically oscillation for 6 h at 30 °C (F–I). The enzyme amounts of (A,B,C,D,E,F,G,H,I) are 6, 4, 6, 8, 6, 2, 4, 6, and 8 mg, respectively. The insert in Figure (A) shows the particle size distribution of the nanoflower prepared with 6 mg of enzyme amount.
The SEM images of ADH HNFs synthesized through magnetic stirring under the optimum conditions (6 mg of enzyme amount and 12 h of immobilization time) are shown in Figure 4A,C,E. The nanoflowers are uniformly dispersed with similar particle sizes within the field of view, and no large number of agglomerates are found. Furthermore, it can be seen in Figure 4A that in addition to the structures bound as nanoflowers, there are also some incompletely assembled nanosheets, which confirms that the formation of nanoflowers is a self-assembly and gradual growth process. From the SEM image shown in Figure 4E and particle size distribution diagram (Figure 4A), the nanoflowers are spheres in shape with a diameter size of 4–6 µm, and the petals are arranged in a wrapped shape.
Then, the effect of enzyme amount on the catalytic activity of ADH HNFs prepared through magnetic stirring was investigated. The relative catalytic activity of ADH HNFs increased significantly in the range of 2–6 mg of the enzyme amount and decreased slightly to 8 mg (Figure 5A). In consideration of the cost-saving and good catalytic activity, the enzyme amount of 6 mg was chosen for the following studies. Furthermore, the relative catalytic activity of ADH HNFs is gradually decreased with the increase of immobilization time (Figure 5B), but the storage stability of the nanoflowers immobilized for 12 h is better than 6 h (Figure 5C). Therefore, 12 h was chosen as the optimum immobilization time. In addition, the relationship between immobilization time and relative enzyme activity confirms that it is reasonable to choose 6 h as the synthesis time when comparing different immobilization methods.
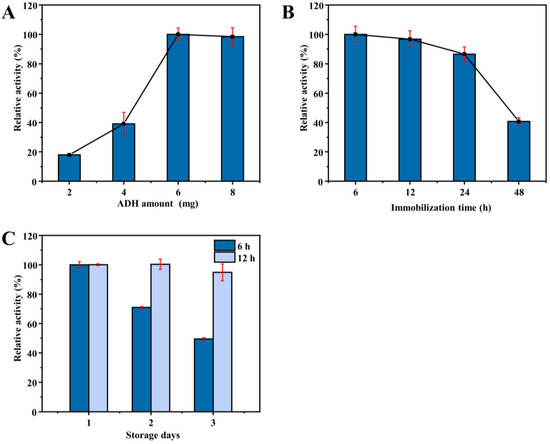
Figure 5.
Effect of enzyme amount (A) and immobilization time (B) on the catalytic activity of ADH HNFs prepared through magnetic stirring; effect of immobilization time (6 and 12 h) on the storage stability of ADH HNFs (C). Conditions: the volume of ADH HNFs, tris-HCl (pH 7.0), and ethanol are 15, 160, and 50 µL, respectively; β-NAD+ concentration, 3 mM; reaction temperature, 35 °C; reaction time, 2 min; centrifuged for 1.5 min, 60 μL of supernatant was taken for measurement at 340 nm.
2.3. Optimization of Enzymatic Reaction Conditions
To obtain the optimum reaction conditions for ADH HNFs, several experimental parameters were investigated, including reaction temperature (25, 35, and 45 °C), buffer pH (6.5, 7.0, and 7.5), and reaction time (2.0–10.0 min).
Results indicate that the relative catalytic activity of ADH HNFs reached its maximum at pH 7.0, while free ADH was leveling off at pH 7.5 (Figure 6A). At the same time, as the reaction temperature increased from 25 to 45 °C, the catalytic activity of both free ADH and ADH HNFs peaked at 35 °C (Figure 6B), which was chosen as the optimum temperature. Moreover, with the increase in reaction time (2, 4, 6, 8, 10, and 12 min), the catalytic activity of ADH HNFs is increased and reaches its maximum at 8 min, and free ADH reaches its maximum at 10 min (Figure 6C). Thus, 8 min and 10 min of reaction time for ADH HNFs and free ADH were used in the following experiments, respectively.
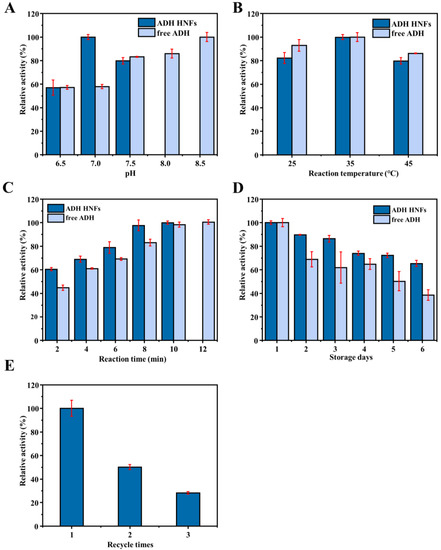
Figure 6.
Effect of buffer pH (A), reaction temperature (B), reaction time (C), and the storage stability (D) on the relative catalytic activity of ADH HNFs and free ADH, and reusability (E) of ADH HNFs. ADH HNFs conditions: the volume of ADH HNFs, tris-HCl, and ethanol are 15, 160, and 50 µL, respectively; β-NAD+ concentration, 3 mM; buffer pH, 7.0; reaction temperature, 35 °C; reaction time, 2 min for (A,B), 8 min for (D,E); centrifuged for 1.5 min, 60 μL of supernatant was taken for measurement at 340 nm. Free ADH conditions: the volume of tris-HCl and ethanol are 160 µL and 15 µL, respectively; free ADH concentration, 0.015 mg/mL for (A–C), 0.01 mg/mL for (D); β-NAD+ concentration, 3 mM; buffer pH, 7.5; reaction temperature, 35 °C; reaction time, 2 min for (A,B), 8 min for (D); 60 μL of supernatant was taken for measurement at 340 nm.
The storage stability and reusability of ADH HNFs were also evaluated and the results are shown in Figure 6D,E, respectively. Within six days of storage, the catalytic activity of the free enzyme decreased to 45.1%, but ADH HNFs remained at 65.3% (Figure 6D). Furthermore, as shown in Figure 6E, ADH HNFs can retain a certain level of catalytic activity after three times of usage. These results indicate that the immobilization of ADH through the formation of nanoflowers can improve the operational stability of the enzyme.
2.4. Kinetics Study of ADH HNFs
The steady-state kinetic study of ADH HNFs was carried out. Based on the varied β-NAD+ concentrations (0.38, 0.75, 1.51, 2.26, and 3.0 mM), the linear regression equations of the Lineweaver–Burk plot of ADH HNFs and free ADH are obtained through Equation (1) as y = 13.231x + 3.8107 (R2 = 0.993) and y = 6.729x + 1.2614 (R2 = 0.9981), respectively, where y and x are the reaction velocity and reciprocal of substrate (β-NAD+) concentration, respectively (Figure 7A). The Vmax of ADH HNFs and free ADH are 15.72 and 47.58 μM·min−1, and Km values are 3.47 and 5.33 mM, respectively, in which the Km value of ADH HNFs is comparable to that of free ADH. In addition, as can be seen in Table 1, the Km values obtained from different studies are different, which can be attributed to the source of ADH, the concentration of the substrate, and the conditions of the enzymatic reaction. Moreover, the Kcat value of free ADH (0.9366 min−1) was found to be a little higher than that of ADH HNFs (0.4546 min−1). Because Kcat demonstrates the maximum converted substrate molecule number by an enzyme per unit of time, a decrease in Kcat value reflects the reduced activity of the enzyme [39].
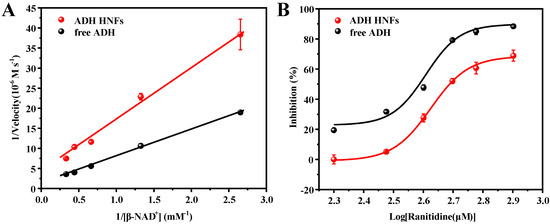
Figure 7.
Michaelis–Menten double reciprocal curve (A) and inhibition plot of ranitidine on ADH (B). ADH HNFs conditions: the volume of ADH HNFs, tris-HCl, and ethanol are 15, 160, and 50 µL, respectively; β-NAD+ concentration, 3 mM; buffer pH, 7.0; reaction temperature, 35 °C; reaction time, 8 min. β-NAD+ concentrations are from 0.375 to 3 mM for (A). Ranitidine concentrations are from 0.2 to 0.8 mM for (B); centrifuged for 1.5 min, 60 μL of supernatant was taken for measurement at 340 nm. Free ADH conditions: The volume of tris-HCl and ethanol are 160 and 50 µL, respectively; free ADH concentration, 0.015 mg/mL; buffer pH, 7.5; reaction temperature, 35 °C; reaction time, 10 min, 60 μL of supernatant was taken for measurement at 340 nm.

Table 1.
Comparison of present work with the previously reported methods for immobilization of ADH.
The change in the amounts of product produced with different ranitidine concentrations was measured and the inhibition rate at each concentration was calculated through Equation (2). Then, the inhibition plots of ranitidine against ADH HNFs were obtained through the dose–response nonlinear regression equation using Origin 2018 64 c software (Figure 7B). The IC50 value of positive control ranitidine against ADH HNFs and free ADH are calculated to be 0.49 and 0.39 mM according to the fitted equation.
2.5. Screening of Inhibitors and Molecular Docking
The inhibitory activity of the aqueous extract of P. chinense (8 mg/mL) and nine pure compounds (1.5 mM) on ADH HNFs were evaluated. Compared with the blank group without inhibitor, the inhibitory activity of each sample was calculated through Equation (2), and the results are summarized in Table 2. The inhibition percentage of the aqueous extract is 57.9%, indicating that P. chinense possesses a moderate inhibitory effect on ADH. The process of ADH alcohol metabolism is associated with increased oxidative stress, which may lead to liver inflammation and cellular damage [46]. The aqueous extract of P. chinense can inhibit the activity of ADH and reduce the production of aldehydes in alcohol metabolism, thereby playing the role of alcoholism antidote and hepatoprotector. Among the nine small-molecule compounds from P. chinense, vanillic acid, protocatechuic acid, gallic acid, and naringenin show obvious inhibitory effects on ADH with the % of inhibition of 55.1%, 68.3%, 61.9%, and 75.5%, respectively. Therefore, these active compounds may be responsible for the inhibitory activity of P. chinense on ADH, and the immobilized enzyme (ADH HNFs) has the potential application prospect in inhibitor screening.

Table 2.
The percentage of inhibition of nine compounds and P. chinense aqueous extracts on ADH (n = 3).
The interactions between ADH and the active compounds (potential inhibitors) were investigated through molecular docking, including ranitidine, vanillic acid, protocatechuic acid, naringenin, and gallic acid. Figure 8 and Figure 9 display the interaction diagrams of the best-docked conformations. Table 3 summarizes the amino acid residues, hydrogen bonds, and binding energy of the interactions between the small-molecule compounds and ADH. The molecular docking results show that ranitidine, vanillic acid, protocatechuic acid, naringenin, and gallic acid are all located in the active site cavity of ADH, and their binding energy to the docking region of the enzyme is below −5 Kcal/mol, indicating that these four small molecules may be potential ADH inhibitors [47]. Furthermore, hydrogen bonds that exist between vanillic acid, gallic acid, and PHE321, THR48; protocatechuic acid and VAL296, THR48; naringenin and HIS47, ALA298, LYS299; play important roles in the binding of ligands to ADH. Compared with the positive control ranitidine, vanillic acid, gallic acid, and protocatechuic acid have some similar binding amino acid residues. Particularly, the benzene rings of these potential inhibitors make close contacts with CYS46, CYS178, HIS67, ILE311, THR319, PHE93, PHE320, ZN380, and SER182, which are the residues lining the inner and middle part of the substrate-binding pocket together with the catalytic zinc and the zinc ligands [48]. However, naringenin exhibits a different binding site from the other three small-molecule compounds and mainly interacts with HIS47, ALA298, and LYS299, which can be used to explain its higher inhibitory activity than the other three ones. Therefore, these compounds may be effective ADH inhibitors, and the docking results correspond to that of inhibitor screening.
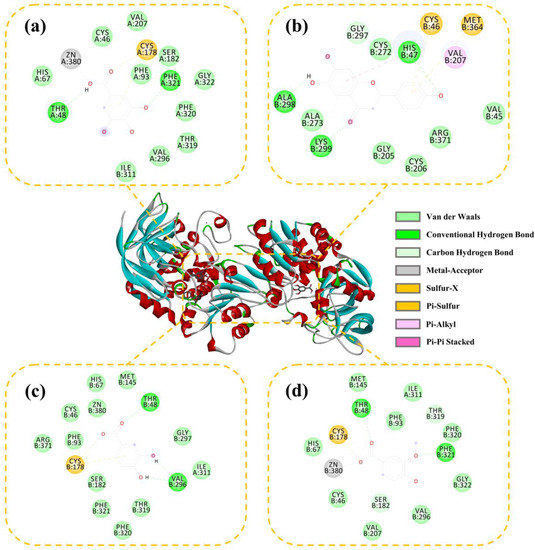
Figure 8.
Three-dimensional (3D) and 2D docking images of ADH with gallic acid (a), naringenin (b), protocatechuic acid (c), and vanillic acid (d).
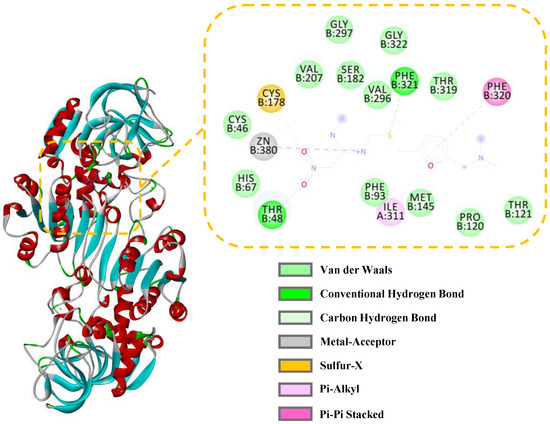
Figure 9.
Three-dimensional (3D) and 2D docking images of ADH with ranitidine.

Table 3.
Docking results of ranitidine and four small-molecule compounds with ADH.
3. Materials and Methods
3.1. Chemicals and Materials
Alcohol dehydrogenase (ADH, 300 U/mg, from yeast) was obtained from Shanghai Yingxin Laboratory Equipment Co., Ltd. (Shanghai, China). Zinc acetate dihydrate (C4H5O4Zn·2H2O) and sodium phosphate dibasic dodecahydrate (Na2HPO4·12H2O) were purchased from Shanghai Titan Scientific Co., Ltd. (Shanghai, China). Oxidative coenzyme I (β-NAD, ≥98%) and ranitidine hydrochloride (≥98%) were obtained from Shanghai Yuanye Biotechnology Co., Ltd. (Shanghai, China). Potassium dihydrogen phosphate (KH2PO4), potassium chloride (KCl), and sodium chloride (NaCl) were purchased from Chengdu Chron Chemicals Co., Ltd. (Chengdu, China). Hydrochloric acid (HCl) was purchased from Sichuan Xilong Science Co., Ltd. (Chengdu, China). Tris (hydroxymethyl) aminomethane (C4H11NO3) was obtained from Guangdong Guanghua Sci-Tech Co., Ltd. (Shantou, China). Vanillic acid (≥98%), (+)-catechin hydrate (>98%), protocatechuic acid (≥97%), ellagic acid (≥96%), and brilliant blue G were obtained from Shanghai Aladdin Biochemical Technology Co., Ltd. (Shanghai, China). Naringenin (>98%), gallic acid (≥98%), apigenin (>98%), and epicatechin (>98%) were purchased from Chengdu PureChem-Standard Co., Ltd. (Chengdu, China). Quercetin (≥98%) was obtained from Sinopharm Chemical Reagent Co., Ltd. (Shanghai, China). Penthorum chinense Pursh was obtained from Anguo Guangsheng Trading Co., Ltd. (Anguo, China).
3.2. Instruments
A DF-101S collector constant temperature magnetic stirrer (Zheng Great Wall Science Industry and Trade Co., Ltd., Zhengzhou, China) and a DHG-9146A electric heating constant temperature blast drying shaker (Shanghai Longyue Instrument Equipment Co., Ltd., Shanghai, China) were used for the synthesis of the materials. The SEM images for the characterization of material were obtained through a field-emission SEM (JSM-7600F, JEOL Ltd., Tokyo, Japan). The pH of solutions was measured through a FE 28 pH meter (Mettler-Toledo Instruments, Shanghai, China). A UV-5500 PC spectrophotometer (Shanghai Metash Instruments Co., Ltd., Shanghai, China) was used for the UV-Vis analysis. The tabletop low-speed centrifuge L420 and the ultrasonic cleaner used in this study were obtained from Hunan Xiang Yi Laboratory Instrument Development Co., Ltd. (Changsha, China) and Kunshan Jielimei Ultrasonic Instrument Co., Ltd. (Kunshan, China), respectively. The ultrapure water prepared through a water purification system (ATSelem 1820A, Antesheng Environmental Protection Equipment, Chongqing, China) was used for all experiments.
3.3. Synthesis of ADH HNFs
Phosphate (PBS, pH = 7.13) buffer for ADH HNFs was prepared by dissolving 0.56 g Na2HPO4·12H2O, 0.14 g KH2PO4, 0.098 g NaCl, and 0.017 g KCl in ultrapure water, respectively, and fixing the volume to 100 mL [49]. To prepare ADH/Zn3(PO4)2 hybrid nanoflowers, 6 mg of ADH was dissolved in 5 mL of PBS buffer. Afterward, 0.4 mL of zinc acetate solution was slowly added to the buffer and stirred magnetically for 12 h (Figure 1). The suspension was then centrifuged at 3000 rpm for 5 min and the supernatant was removed and the precipitate was washed twice with PBS buffer to remove the unfixed enzyme and the unreacted zinc acetate. Finally, the prepared ADH HNFs were suspended in 4 mL of PBS buffer and stored in the refrigerator at 4 °C.
3.4. Optimization of Preparation Conditions of ADH HNFs
Different enzyme amounts of 2, 4, 6, and 8 mg were introduced and reacted under constant temperature shaking (30 °C, 160 rpm) or magnetic stirring (room temperature of 30 °C) for 6 h, respectively. The unit of alcohol dehydrogenase activity is defined as the amount of 1 μmol NADH·min−1 consumed by alcohol dehydrogenase under specified conditions [50]. Subsequently, field emission scanning electron microscopy (SEM) was used to observe the morphology and size of the ADH HNFs synthesized by using different enzyme amounts and preparation methods. Finally, the enzyme concentration was determined using the Bradford method [51].
3.5. Determination of ADH HNFs Enzymatic Activity
The conversion of coenzyme I (β-NAD+) to β-NADH during the catalysis of ethanol to acetaldehyde by ADH displays a characteristic absorption peak at 340 nm. In a 0.5 mL centrifuge tube, 15 μL of ADH HNFs (dispersed in PBS, pH = 7.13), 160 μL of tris-HCl (prepared in 10 mM tris solution and the required pH was adjusted by 1 M HCl), 50 μL of anhydrous ethanol, and 25 μL of 3 mM of β-NAD+ (dispersed in tris-HCl, pH = 7.0) were mixed well, and the mixture was incubated at 35 °C for 8 min. After being centrifuged for 1.5 min by a portable mini centrifuge, the absorbance changes at 340 nm (ε = 6200 M−1 cm−1 [41]) of 60 μL of supernatant was measured. Each experiment was repeated three times.
3.6. Determination of Free ADH Enzymatic Activity
In a 0.5 mL centrifuge tube, 15 μL of free ADH (0.25 mM, dispersed in PBS, pH = 7.13), 160 μL of tris-HCl (prepared in 10 mM tris solution and the required pH was adjusted by 1 M HCl), 50 μL of anhydrous ethanol, and 25 μL of 3 mM of β-NAD+ (dispersed in tris-HCl, pH = 7.5) were mixed well, and the mixture was incubated at 35 °C for 10 min. The absorbance (340 nm) of 60 μL of supernatant was measured. Each experiment was repeated three times.
3.7. Enzyme Kinetics Assay
For an enzyme kinetic reaction, the Km (Michaelis–Menten constant) is an important parameter that can reflect the affinity between the substrate and enzyme, which is calculated through the Lineweaver–Burk Equation (1) [52].
where [S] is the concentration of the substrate (β-NAD+), Vmax and V are the maximum and reaction rate of the enzyme reaction, respectively. The reaction velocity of ADH HNFs was monitored through the absorbance (340 nm) of the product (β-NADH). Experiments were performed by altering the concentration of β-NAD+ under optimized conditions.
Ranitidine is a representative inhibitor of ADH [53], which was used as a model compound for evaluating the inhibition kinetics of ADH. IC50 was measured by varying the concentration of ranitidine (0.2–0.8 mM) with a fixed β-NAD+ concentration at 3 mM. The % of inhibition (I%) can be calculated through Equation (2) [52]:
where Ai and A0 are the absorbances of the reaction product with and without the inhibitor, respectively. I(%) is the inhibitory percentage. The dose–response nonlinear regression equation was used for the construction of an inhibition plot using Origin 2018.
3.8. Evaluation of ADH Inhibitory Activity of P. chinense and Its Small-Molecule Compounds
The preparation of an aqueous extract of P. chinense was referred to as the previously reported method [54]. In brief, 10.0 g of crushed P. chinense was soaked in 150 mL of ultrapure water in a 250 mL round-bottomed flask for 2 h, and condensed and refluxed at 100 °C for 1 h, then it was filtered. After that, 100 mL of ultrapure water was added to the residue, which was condensed and refluxed at 100 °C for 40 min, and it was filtered. After combining the two filtrates, a solution equivalent to 0.04 g/mL of the original drug was prepared.
Vanillic acid, (+)-catechin hydrate, gallic, epicatechin, and protocatechuic acid solutions were prepared by directly adding purified water to dissolve the corresponding substance. The ellagic acid, apigenin, and quercetin solutions were prepared by dissolving them in a small volume of lye (NaOH, 1 M) in ultrapure water. The naringenin solution was prepared in ethanol. The final concentrations of all solutions are 1.5 M.
The prepared ADH HNFs were used in the evaluation of the inhibitory activity of the aqueous extract and the monomers of P. chinense. In brief, 15 μL of ADH HNFs suspension was added to a 0.5 mL centrifuge tube, followed by 50 μL of the aqueous extract of P. chinense or 50 μL of the monomers, 110 μL of tris-HCl, and 50 μL of anhydrous ethanol, which were mixed with 25 μL of 3 mM of β-NAD+ and reacted at 35 °C for 8 min. Then, the reaction mixture was centrifuged for 1.5 min, and the absorbance of the product β-NADH at 340 nm was recorded in 60 μL of supernatant. The inhibition percentage of each sample was calculated according to Equation (2) based on the absorbance of enzymatic reaction product β-NADH with and without the addition of an inhibitor. At least three parallel experiments were carried out for each sample.
3.9. Molecular Docking
For the molecular docking analysis, the crystal structure of ADH (PDB ID: 1E3L) was obtained from the Protein Data Bank. Before molecular docking, the NAD ligand and H2O molecules in the ADH structure were removed in AutoDock 1.5.6, followed by the addition of polar hydrogen, calculation of point charge, and selection of atom type. Then, the processed ADH was output in PDBQT format. On the other hand, the molecular structures of the small-molecule compound were drawn in ChemDraw 20.0 and imported into Chem3D for energy minimization, and the results were saved in PDB format. Subsequently, the docking site and range of the enzyme were selected, and the processed ligands were docked with ADH using the Lamarckian genetic algorithm (LGA). Finally, the optimum conformations of ligands and ADH interactions were observed by Discovery Studio 2019.
4. Conclusions
In this study, ADH/Zn3(PO4)2 hybrid nanoflowers were successfully prepared through a simple biomimetic mineralization method. The SEM characterizations demonstrate the successful synthesis of ADH HNFs, and the kinetics of the enzymatic reaction of the nanoflowers shows that the immobilization method can effectively improve the affinity of the enzyme to the substrate. Furthermore, the catalytic activity of the immobilized enzyme shows better storage stability (6 days) and reusability (3 times) as compared to that of the free enzyme. In addition, the good inhibitory activity on ADH of nine small-molecule compounds from P. chinense has been discovered, and the molecular docking further confirmed the interaction between the enzyme and the four most active compounds, namely vanillic acid, protocatechuic acid, gallic acid, and naringenin. In short, the results of the present study provide references for the immobilization of enzymes through simple methods and the discovery of ADH inhibitors from natural products.
Author Contributions
M.-L.L.: conceptualization, methodology, investigation, writing—original draft. H.C.: investigation, funding acquisition. G.-Y.C.: conceptualization, methodology, investigation. S.W.: supervision, project administration. Y.W.: supervision. F.-Q.Y.: writing—review and editing, supervision, project administration, funding acquisition. All authors have read and agreed to the published version of the manuscript.
Funding
The research outputs of this Open Research Project Programme are funded by the Science and Technology Development Fund (SKL-QRCM(UM)-2020-2022) and the State Key Laboratory of Quality Research in Chinese Medicine, University of Macau (No. SKL-QRCM-OP21005).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are contained within the article.
Conflicts of Interest
All the authors declare that they have no conflict of interest.
Sample Availability
Samples of the compounds are available from the authors.
References
- Wolszczak-Biedrzycka, B.; Zasimowicz-Majewska, E.; Bieńkowska, A.; Biedrzycki, G.; Dorf, J.; Jelski, W. Activity of total alcohol dehydrogenase, alcohol dehydrogenase isoenzymes and aldehyde dehydrogenase in the serum of patients with alcoholic fatty liver disease. Medicina 2022, 58, 25. [Google Scholar] [CrossRef]
- Shah, A.M.; Mohamed, H.; Fazili, A.B.A.; Yang, W.; Song, Y. Investigating the effect of alcohol dehydrogenase gene knockout on lipid accumulation in mucor circinelloides WJ11. J. Fungi 2022, 8, 917. [Google Scholar] [CrossRef]
- Langley, C.; Tatsis, E.; Hong, B.; Nakamura, Y.; Paetz, C.; Stevenson, C.E.M.; Basquin, J.; Lawson, D.M.; Caputi, L.; O’Connor, S.E. Expansion of the catalytic repertoire of alcohol dehydrogenases in plant metabolism. Angew. Chem. Int. Ed. 2022, 61, e202210934. [Google Scholar] [CrossRef]
- Tran, S.; Nowicki, M.; Facciol, A.; Chatterjee, D.; Gerlai, R. Ethanol-induced ADH activity in zebrafish: Differential concentration-dependent effects on high-versus low-affinity ADH enzymes. Zebrafish Apr. 2016, 75–78. [Google Scholar] [CrossRef]
- Arigos, P.; Garavito, R.M.; Eventoff, W.; Rossmann, M.G.; Brändén, C.I. Similarities in active center geometries of zinc-containing enzymes, proteases, and dehydrogenases. J. Mol. Biol. 1978, 126, 141–158. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.G.; Yin, H.H.; Yu, D.F.; Chen, X.; Tang, X.L.; Zhang, X.J.; Xue, Y.P.; Wang, Y.J.; Liu, Z.Q. Recent advances in biotechnological applications of alcohol dehydrogenases. Appl. Microbiol. Biotechnol. 2017, 101, 987–1001. [Google Scholar] [CrossRef]
- Di, L.; Balesano, A.; Jordan, S.; Shi, S.M. The role of alcohol dehydrogenase in drug metabolism: Beyond ethanol oxidation. AAPS J. 2021, 23, 20. [Google Scholar] [CrossRef]
- Wolszczak-Biedrzycka, B.; Bieńkowska, A.; Zasimowicz, E.; Biedrzycki, G.; Dorf, J.; Jelski, W. An assessment of the serum activity of ADH and ALDH in patients with primary biliary cholangitis. Arch. Immunol. Ther. Exp. 2023, 71, 2. [Google Scholar] [CrossRef] [PubMed]
- Zhao, T.; Han, Z.; Zhang, J.; Ding, Y.; Chen, J.; Qiao, H.; Gao, N. Effect of ADHI on hepatic stellate cell activation and liver fibrosis in mice. Biochem. Biophys. Res. Commun. 2023, 651, 98–106. [Google Scholar] [CrossRef] [PubMed]
- Haseba, T.; Duester, G.; Shimizu, A.; Yamamoto, I.; Kameyama, K.; Ohno, Y. In vivo contribution of Class III alcohol dehydrogenase (ADH3) to alcohol metabolism through activation by cytoplasmic solution hydrophobicity. Biochim. Biophys. Acta 2006, 1762, 276–283. [Google Scholar] [CrossRef]
- Chen, M.; Liu, L.L.; Chen, X.Q. Rapid screening and analysis of alcohol dehydrogenase binders from Glycyrrhiza uralensis root extract using functionalized magnetic nanoparticles coupled with HPLC–MS/MS. Can. J. Chem. 2013, 91, 1147–1154. [Google Scholar] [CrossRef]
- Akakpo, J.Y.; Ramachandran, A.; Orhan, H.; Curry, S.C.; Rumack, B.H.; Jaeschke, H. 4-methylpyrazole protects against acetaminophen-induced acute kidney injury. Toxicol. Appl. Pharmacol. 2020, 409, 115317. [Google Scholar] [CrossRef]
- Beaulieu, J.; Roberts, D.M.; Gosselin, S.; Hoffman, R.S.; Lavergne, V.; Hovda, K.E.; Megarbane, B.; Lung, D.; Thanacoody, R.; Ghammoum, M. Treating ethylene glycol poisoning with alcohol dehydrogenase inhibition, but without extracorporeal treatments: A systematic review. Clin. Toxicol. 2022, 60, 784–797. [Google Scholar] [CrossRef]
- Mégarbane, B.; Borron, S.W.; Baud, F.J. Current recommendations for treatment of severe toxic alcohol poisonings. Intensive Care Med. 2005, 31, 189–195. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Wang, J.; Zhang, D.; Liu, J.; Wu, Q.; Chen, J.; Tan, P.; Xing, B.; Han, Y.; Zhang, P.; et al. Mechanism of drug-induced liver injury and hepatoprotective effects of natural drugs. Chin. Med. 2021, 16, 135. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Luo, P.; Zheng, L.; Chen, J.; Zhang, J.; Tang, H.; Liu, D.; He, X.; Shi, Q.; Gu, L.; et al. 18beta-glycyrrhetinic acid induces ROS-mediated apoptosis to ameliorate hepatic fibrosis by targeting PRDX1/2 in activated HSCs. J. Pharm. Anal. 2022, 12, 570–582. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.W.; Guo, W.W.; Guo, J.F.; Wang, X.; Chen, X.Q.; Wu, X. Three new flavonoids from Penthorum chinense Pursh and their docking studies. Nat. Prod. Res. 2021, 35, 49–56. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Zhang, S.; Luo, C.; Sun, Y.; Lu, Q.; Huang, L.; Chen, F.; Tang, L. Functional teas from the stems of Penthorum chinense Pursh.: Phenolic constituents, antioxidant and hepatoprotective activity. Plant Foods Hum. Nutr. 2019, 74, 83–90. [Google Scholar] [CrossRef]
- Wang, M.; Zhang, X.J.; Feng, R.; Jiang, Y.; Zhang, D.Y.; He, C.; Li, P.; Wan, J.B. Hepatoprotective properties of Penthorum chinense Pursh against carbon tetrachloride-induced acute liver injury in mice. Chin. Med. 2017, 12, 32. [Google Scholar] [CrossRef]
- Ding, Q.; Jin, Z.; Dong, J.; Wang, Z.; Jiang, K.; Ye, Y.; Dou, X.; Ding, B. Bioactivity evaluation of pinocembrin derivatives from Penthorum chinense Pursh stems. Nat. Prod. Commun. 2019, 14, 1934578X–1987589X. [Google Scholar] [CrossRef]
- Ge, J.; Lei, J.D.; Zare, R.N. Protein-inorganic hybrid nanoflowers. Nat. Nanotechnol. 2012, 7, 428–432. [Google Scholar] [CrossRef]
- Songa, E.A.; Okonkwo, J.O. Recent approaches to improving selectivity and sensitivity of enzyme-based biosensors for organophosphorus pesticides: A review. Talanta 2016, 155, 289–304. [Google Scholar] [CrossRef]
- Lee, H.R.; Chung, M.; Kim, M.I.; Ha, S.H. Preparation of glutaraldehyde-treated lipase-inorganic hybrid nanoflowers and their catalytic performance as immobilized enzymes. Enzym. Microb. Technol. 2017, 105, 24–29. [Google Scholar] [CrossRef]
- Somturk, B.; Hancer, M.; Ocsoy, I.; Özdemir, N. Synthesis of copper ion incorporated horseradish peroxidase-based hybrid nanoflowers for enhanced catalytic activity and stability. Dalton Trans. 2015, 44, 13845–13852. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Ge, J.; Liu, W.; Lan, M.; Zhang, H.; Wang, P.; Wang, Y.; Niu, Z. Multi-enzyme co-embedded organic-inorganic hybrid nanoflowers: Synthesis and application as a colorimetric sensor. Nanoscale 2014, 6, 255–262. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, Y.; He, R.; Zhuang, A.; Wang, X.; Zheng, J.; Hou, J.H. A new nanobiocatalytic system based on allosteric effect with dramatically enhanced enzymatic performance. J. Am. Chem. Soc. 2013, 135, 1272–1275. [Google Scholar] [CrossRef]
- Smoturk, B.; Yilmaz, I.; Altinkaynak, C.; Karatepe, A.; Özdemir, N.; Ocsoy, I. Synthesis of urease hybrid nanoflowers and their enhanced catalytic properties. Enzyme Microb. Technol. 2016, 86, 134–142. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Xiao, Y.; Wang, L.; Yin, Y.Q.; Zheng, J.N.; Yang, H.H.; Chen, G.N. Facile synthesis of enzyme-inorganic hybrid nanoflowers and their application as an immobilized trypsin reactor for highly efficient protein digestion. RSC Adv. 2014, 4, 13888–13891. [Google Scholar] [CrossRef]
- Yin, Y.; Xiao, Y.; Lin, G.; Xiao, Q.; Lin, Z.; Cai, Z. An enzyme-inorganic hybrid nanoflower based immobilized enzyme reactor with enhanced enzymatic activity. J. Mater. Chem. B 2015, 3, 2295–2300. [Google Scholar] [CrossRef]
- Liang, L.; Fei, X.; Li, Y.; Tian, J.; Xu, L.; Wang, X.; Wang, Y. Hierarchical assembly of enzyme-inorganic composite materials with extremely high enzyme activity. RSC Adv. 2015, 5, 96997–97002. [Google Scholar] [CrossRef]
- Lu, M.; Zhang, H.; Wang, X.; Jiang, H.; Hu, G.; Yang, F. Preparation of phytic acid modified α-glucosidase/Cu3(PO4)2·3H2O hybrid nanoflower and its application. Enzyme Microb. Technol. 2021, 146, 109776. [Google Scholar] [CrossRef]
- Li, S.; Zhang, S.; Tao, Y.; Chen, Y.; Yang, Y.; Liang, X.; Li, Q. Construction of β-galactosidase-inorganic hybrid nanoflowers through biomimetic mineralization for lactose degradation. Biochem. Eng. J. 2023, 197, 108980. [Google Scholar] [CrossRef]
- Xu, H.; Liang, H. Chitosan-regulated biomimetic hybrid nanoflower for efficiently immobilizing enzymes to enhance stability and by-product tolerance. Int. J. Biol. Macromol. 2022, 220, 124–134. [Google Scholar] [CrossRef]
- Hu, R.; Zhang, X.B.; Zhao, Z.L.; Zhu, G.Z.; Chen, T.; Fu, T.; Tan, W.H. DNA nanoflowers for multiplexed cellular imaging and traceable targeted drug delivery. Angew. Chem. Int. Ed. 2014, 126, 5931–5936. [Google Scholar] [CrossRef]
- Zhang, B.; Li, P.; Zhang, H.; Wang, H.; Li, X.; Tian, L.; Ali, N.; Ali, Z.; Zhang, Q. Preparation of lipase/Zn3(PO4)2 hybrid nanoflower and its catalytic performance as an immobilized enzyme. Chem. Eng. J. 2016, 291, 287–297. [Google Scholar] [CrossRef]
- Wang, Z.; Liu, P.; Fang, Z.; Jiang, H. Trypsin/Zn3(PO4)2 hybrid nanoflowers: Controlled synthesis and excellent performance as an immobilized enzyme. Int. J. Mol. Sci. 2022, 23, 11853. [Google Scholar] [CrossRef]
- Yang, H.; He, P.; Yin, Y.; Mao, Z.; Zhang, J.; Zhong, C.; Xie, T.; Wang, A. Succinic anhydride-based chemical modification making laccase@Cu3(PO4)2 hybrid nanoflowers robust in removing bisphenol a in wastewater. Bioproc. Biosyst. Eng. 2021, 44, 2061–2073. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Li, P.; Zhang, H.; Fan, L.; Wang, H.; Li, X.; Tian, L.; Ali, N.; Ali, Z.; Zhang, Q. Papain/Zn3(PO4)2 hybrid nanoflower: Preparation, characterization and its enhanced catalytic activity as an immobilized enzyme. RSC Adv. 2016, 6, 46702–46710. [Google Scholar] [CrossRef]
- Uygun, D.A.; Akduman, B.; Uygun, M.; Akgöl, S.; Denizli, A. Immobilization of alcohol dehydrogenase onto metal-chelated cryogels. J. Biomater. Sci. Polym. Ed. 2015, 26, 446–457. [Google Scholar] [CrossRef]
- Jiang, X.; Lu, T.; Liu, C.; Ling, X.; Zhuang, M.; Zhang, J.; Zhang, Y. Immobilization of dehydrogenase onto epoxy-functionalized nanoparticles for synthesis of (R)-mandelic acid. Int. J. Biol. Macromol. 2016, 88, 9–17. [Google Scholar] [CrossRef]
- Xia, H.; Li, Z.; Zhong, X.; Li, B.; Jiang, Y.; Jiang, Y. HKUST-1 catalyzed efficient in situ regeneration of NAD+ for dehydrogenase mediated oxidation. Chem. Eng. Sci. 2019, 203, 43–53. [Google Scholar] [CrossRef]
- Shakir, M.; Nasir, Z.; Khan, M.S.; Lutfullah; Alam, M.F.; Younus, H.; Al-Resayes, S.I. Study on immobilization of yeast alcohol dehydrogenase on nanocrystalline Ni-Co ferrites as magnetic support. Int. J. Biol. Macromol. 2015, 72, 1196–1204. [Google Scholar] [CrossRef]
- Alam, M.F.; Laskar, A.A.; Zubair, M.; Baig, U.; Younus, H. Immobilization of yeast alcohol dehydrogenase on polyaniline coated silver nanoparticles formed by green synthesis. J. Mol. Catal. B Enzym. 2015, 119, 78–84. [Google Scholar] [CrossRef]
- Baig, U.; Gondal, M.A.; Alam, M.F.; Laskar, A.A.; Alam, M.; Younus, H. Enzyme immobilization and molecular modeling studies on an organic-inorganic polypyrrole-titanium(iv) phosphate nanocomposite. New J. Chem. 2015, 39, 6976–6986. [Google Scholar] [CrossRef]
- Ghannadi, S.; Abdizadeh, H.; Miroliaei, M.; Saboury, A.A. Immobilization of alcohol dehydrogenase on titania nanoparticles to enhance enzyme stability and remove substrate inhibition in the reaction of formaldehyde to methanol. Ind. Eng. Chem. Res. 2019, 58, 9844–9854. [Google Scholar] [CrossRef]
- Wegner, S.A.; Pollard, K.A.; Kharazia, V.; Darevsky, D.; Perez, L.; Roychowdhury, S.; Xu, A.; Ron, D.; Nagy, L.E.; Hopf, F.W. Limited excessive voluntary alcohol drinking leads to liver dysfunction in mice. Alcohol. Clin. Exp. Res. 2017, 41, 345–358. [Google Scholar] [CrossRef]
- Wang, W.F.; Yang, J.L. Advances in screening enzyme inhibitors by capillary electrophoresis. Electrophoresis 2019, 40, 2075–2083. [Google Scholar] [CrossRef]
- Svensson, S.; Höög, J.; Schneider, G.; Sandalova, T. Crystal structures of mouse class II alcohol dehydrogenase reveal determinants of substrate specificity and catalytic efficiency. J. Mol. Biol. 2000, 302, 441–453. [Google Scholar] [CrossRef]
- Zheng, L.; Xie, X.N.; Wang, Z.; Zhang, Y.X.; Wang, L.; Cui, X.Y.; Huang, H.; Zhuang, H. Fabrication of a nano-biocatalyst for regioselective acylation of arbutin. Green Chem. Lett. Rev. 2018, 11, 55–61. [Google Scholar] [CrossRef]
- Kaya, N.; Aktaş Uygun, D.; Akgöl, S.; Denizli, A. Purification of alcohol dehydrogenase from saccharomyces cerevisiae using magnetic dye-ligand affinity nanostructures. Appl. Biochem. Biotechnol. 2013, 169, 2153–2164. [Google Scholar] [CrossRef]
- Yang, M.; Shi, D.; Wang, Y.; Ebadi, A.G.; Toughani, M. Study on interaction of coomassie brilliant blue g-250 with bovine serum albumin by multispectroscopic. Int. J. Pept. Res. Ther. 2021, 27, 421–431. [Google Scholar] [CrossRef]
- Chen, G.Y.; Qian, Z.M.; Yin, S.J.; Zhou, X.; Yang, F.Q. A Sensitive and selective colorimetric method based on the acetylcholinesterase-like activity of zeolitic imidazolate framework-8 and its applications. Molecules 2022, 27, 7491. [Google Scholar] [CrossRef] [PubMed]
- El-Bakary, A.A.; El-Dakrory, S.A.; Attalla, S.M.; Hasanein, N.A.; Malek, H.A. Ranitidine as an alcohol dehydrogenase inhibitor in acute methanol toxicity in rats. Hum. Exp. Toxicol. 2010, 29, 93–101. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Wu, Z.; Zhang, H.; Yin, S.; Xia, F.; Zhang, Q.; Wan, J.; Gao, J.; Yang, F. LC–MS-based multivariate statistical analysis for the screening of potential thrombin/factor Xa inhibitors from Radix Salvia Miltiorrhiza. Chin. Med. 2020, 15, 38. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).