Smartphone-Based Electrochemical Biosensor for On-Site Nutritional Quality Assessment of Coffee Blends
Abstract
1. Introduction
2. Results and Discussions
3. Materials and Method
3.1. Reagents and Samples
3.2. Electrochemical and Surface Characterization Apparatus
3.3. Real Sample Preparation and Treatment
3.4. Folin–Ciocâlteu Assay
3.5. Specificity Studies
3.6. TvLac-Based Biosensor Fabrication
3.7. Electrochemical Measurements
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Younis, M.R.; Wang, C.; Younis, M.A.; Xia, X. Smartphone-Based Biosensors. Nanobiosensors Des. Appl. 2020, 357–387. [Google Scholar] [CrossRef]
- Xu, K.; Chen, Q.; Zhao, Y.; Ge, C.; Lin, S.; Liao, J. Cost-effective, wireless, and portable smartphone-based electrochemical system for on-site monitoring and spatial mapping of the nitrite contamination in water. Sens. Actuators B Chem. 2020, 319, 128221. [Google Scholar] [CrossRef]
- Sun, A.C.; Hall, D.A. Point-of-care smartphone-based electrochemical biosensing. Electroanalysis 2019, 31, 2–16. [Google Scholar] [CrossRef]
- Lu, Y.; Shi, Z.; Liu, Q. Smartphone-based biosensors for portable food evaluation. Curr. Opin. Food Sci. 2019, 28, 74–81. [Google Scholar] [CrossRef]
- Yousefi, H.; Su, H.-M.; Imani, S.M.; Alkhaldi, K.; M. Filipe, C.D.; Didar, T.F. Intelligent food packaging: A review of smart sensing technologies for monitoring food quality. ACS Sens. 2019, 4, 808–821. [Google Scholar] [CrossRef]
- Wang, M.; Yang, Y.; Min, J.; Song, Y.; Tu, J.; Mukasa, D.; Ye, C.; Xu, C.; Heflin, N.; McCune, J.S. A wearable electrochemical biosensor for the monitoring of metabolites and nutrients. Nat. Biomed. Eng. 2022, 6, 1225–1235. [Google Scholar] [CrossRef]
- Irannejad, N.; Rezaei, B. Electrochemical sensors for food adulterants. In Biosensing and Micro-Nano Devices: Design Aspects and Implementation in Food Industries; Springer: Berlin/Heidelberg, Germany, 2022; pp. 69–90. [Google Scholar]
- Wang, K.; Lin, X.; Zhang, M.; Li, Y.; Luo, C.; Wu, J. Review of Electrochemical Biosensors for Food Safety Detection. Biosensors 2022, 12, 959. [Google Scholar] [CrossRef]
- Viswanathan, V.; Krishnan, D.; Kalra, S.; Chawla, R.; Tiwaskar, M.; Saboo, B.; Baruah, M.; Chowdhury, S.; Makkar, B.M.; Jaggi, S. Insights on medical nutrition therapy for type 2 diabetes mellitus: An Indian perspective. Adv. Ther. 2019, 36, 520–547. [Google Scholar] [CrossRef]
- Nestel, P.J.; Beilin, L.J.; Clifton, P.M.; Watts, G.F.; Mori, T.A. Practical guidance for food consumption to prevent cardiovascular disease. Hear. Lung Circ. 2021, 30, 163–179. [Google Scholar] [CrossRef]
- Valko, M.; Leibfritz, D.; Moncol, J.; Cronin, M.T.D.; Mazur, M.; Telser, J. Free radicals and antioxidants in normal physiological functions and human disease. Int. J. Biochem. Cell Biol. 2007, 39, 44–84. [Google Scholar] [CrossRef]
- Chiva-Blanch, G.; Visioli, F. Polyphenols and health: Moving beyond antioxidants. J. Berry Res. 2012, 2, 63–71. [Google Scholar] [CrossRef]
- Ding, S.; Jiang, H.; Fang, J. Review Article Regulation of Immune Function by Polyphenols. J. Immunol. Res. 2018, 2018, 1264074. [Google Scholar] [CrossRef] [PubMed]
- Scalbert, A.; Johnson, I.T.; Saltmarsh, M. Polyphenols: Antioxidants and beyond. Am. J. Clin. Nutr. 2005, 81, 215–217. [Google Scholar] [CrossRef] [PubMed]
- Okazaki, Y.; Han, Y.; Kayahara, M.; Watanabe, T.; Arishige, H.; Kato, N. Consumption of curcumin elevates fecal immunoglobulin A, an index of intestinal immune function, in rats fed a high-fat diet. J. Nutr. Sci. Vitaminol. 2010, 56, 68–71. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Berezo, T.; Franch, A.; Castellote, C.; Castell, M.; Pérez-Cano, F.J. Mechanisms involved in down-regulation of intestinal IgA in rats by high cocoa intake. J. Nutr. Biochem. 2012, 23, 838–844. [Google Scholar] [CrossRef]
- Di Meo, F.; Lemaur, V.; Cornil, J.; Lazzaroni, R.; Duroux, J.-L.; Olivier, Y.; Trouillas, P. Free radical scavenging by natural polyphenols: Atom versus electron transfer. J. Phys. Chem. A 2013, 117, 2082–2092. [Google Scholar] [CrossRef]
- Milanković, V.; Tasić, T.; Pejčić, M.; Pašti, I.; Lazarević-Pašti, T. Spent Coffee Grounds as an Adsorbent for Malathion and Chlorpyrifos-Kinetics, Thermodynamics, and Eco-Neurotoxicity. Foods 2023, 12, 2397. [Google Scholar] [CrossRef]
- Król, K.; Gantner, M.; Tatarak, A.; Hallmann, E. The content of polyphenols in coffee beans as roasting, origin and storage effect. Eur. Food Res. Technol. 2020, 246, 33–39. [Google Scholar] [CrossRef]
- Kolb, H.; Kempf, K.; Martin, S. Health effects of coffee: Mechanism unraveled? Nutrients 2020, 12, 1842. [Google Scholar] [CrossRef]
- Tarigan, E.B.; Wardiana, E.; Hilmi, Y.S.; Komarudin, N.A. The changes in chemical properties of coffee during roasting: A review. IOP Conf. Ser. Earth Environ. Sci. 2022, 974, 012115. [Google Scholar] [CrossRef]
- Farah, A.; Monteiro, M.C.; Calado, V.; Franca, A.S.; Trugo, L.C. Correlation between cup quality and chemical attributes of Brazilian coffee. Food Chem. 2006, 98, 373–380. [Google Scholar] [CrossRef]
- Perdani, C.G.; Pranowo, D. Total phenols content of green coffee (Coffea arabica and Coffea canephora) in East Java. In Proceedings of the IOP Conference Series: Earth and Environmental Science; IOP Publishing: Bristol, UK, 2019; Volume 230, p. 12093. [Google Scholar]
- Kwak, H.S.; Ji, S.; Jeong, Y. The effect of air flow in coffee roasting for antioxidant activity and total polyphenol content. Food Control 2017, 71, 210–216. [Google Scholar] [CrossRef]
- Ludwig, I.A.; Clifford, M.N.; Lean, M.E.J.; Ashihara, H.; Crozier, A. Coffee: Biochemistry and potential impact on health. Food Funct. 2014, 5, 1695–1717. [Google Scholar] [CrossRef]
- Bunney, J.; Williamson, S.; Atkin, D.; Jeanneret, M.; Cozzolino, D.; Chapman, J.; Power, A.; Chandra, S. The Use of Electrochemical Biosensors in Food Analysis. Curr. Res. Nutr. Food Sci. 2017, 5, 183–195. [Google Scholar] [CrossRef]
- Favero, G.; Fusco, G.; Mazzei, F.; Tasca, F.; Antiochia, R. Electrochemical characterization of graphene and MWCNT screen-printed electrodes modified with AuNPs for laccase biosensor development. Nanomaterials 2015, 5, 1995–2006. [Google Scholar] [CrossRef] [PubMed]
- Unal, Y.D.; Pazarlioglu, N.K. Production and Gelatin Entrapment of Laccase from Trametes versicolor and its Application to Quantitative Determination of Phenolic Contents of Commercial Fruit Juices. Food Biotechnol. 2011, 25, 351–368. [Google Scholar] [CrossRef]
- Castrovilli, M.C.; Bolognesi, P.; Chiarinelli, J.; Avaldi, L.; Calandra, P.; Antonacci, A.; Scognamiglio, V. The convergence of forefront technologies in the design of laccase-based biosensors—An update. TrAC Trends Anal. Chem. 2019, 119, 115615. [Google Scholar] [CrossRef]
- Aroca, R.F.; Ross, D.J.; Domingo, C. Surface-Enhanced Infrared Spectroscopy. Appl. Spectrosc. 2004, 58, 324A–338A. [Google Scholar] [CrossRef]
- Romero-Arcos, M.; Garnica-Romo, M.G.; Martínez-Flores, H.E. Electrochemical study and characterization of an amperometric biosensor based on the immobilization of laccase in a nanostructure of TiO2 synthesized by the sol-gel method. Materials 2016, 9, 543. [Google Scholar] [CrossRef]
- Tavares, A.P.M.; Silva, C.G.; Dražić, G.; Silva, A.M.T.; Loureiro, J.M.; Faria, J.L. Laccase immobilization over multi-walled carbon nanotubes: Kinetic, thermodynamic and stability studies. J. Colloid Interface Sci. 2015, 454, 52–60. [Google Scholar] [CrossRef]
- Latif, A.; Maqbool, A.; Sun, K.; Si, Y. Immobilization of Trametes Versicolor laccase on Cu-alginate beads for biocatalytic degradation of bisphenol A in water: Optimized immobilization, degradation and toxicity assessment. J. Environ. Chem. Eng. 2022, 10, 107089. [Google Scholar] [CrossRef]
- Patro, T.U.; Wagner, H.D. Influence of graphene oxide incorporation and chemical cross-linking on structure and mechanical properties of layer-by-layer assembled poly(Vinyl alcohol)-Laponite free-standing films. J. Polym. Sci. Part B Polym. Phys. 2016, 54, 2377–2387. [Google Scholar] [CrossRef]
- García-Miranda Ferrari, A.; Foster, C.W.; Kelly, P.J.; Brownson, D.A.C.; Banks, C.E. Determination of the electrochemical area of screen-printed electrochemical sensing platforms. Biosensors 2018, 8, 53. [Google Scholar] [CrossRef]
- Tortolini, C.; Di Fusco, M.; Frasconi, M.; Favero, G.; Mazzei, F. Laccase-polyazetidine prepolymer-MWCNT integrated system: Biochemical properties and application to analytical determinations in real samples. Microchem. J. 2010, 96, 301–307. [Google Scholar] [CrossRef]
- Chen, X.; Li, D.; Li, G.; Luo, L.; Ullah, N.; Wei, Q.; Huang, F. Facile fabrication of gold nanoparticle on zein ultrafine fibers and their application for catechol biosensor. Appl. Surf. Sci. 2015, 328, 444–452. [Google Scholar] [CrossRef]
- Kizling, M.; Dzwonek, M.; Wieckowska, A.; Bilewicz, R. Gold nanoparticles in bioelectrocatalysis—The role of nanoparticle size. Curr. Opin. Electrochem. 2018, 12, 113–120. [Google Scholar] [CrossRef]
- Kizling, M.; Dzwonek, M.; Więckowska, A.; Bilewicz, R. Size Does Matter—Mediation of Electron Transfer by Gold Clusters in Bioelectrocatalysis. ChemCatChem 2018, 10, 1988–1992. [Google Scholar] [CrossRef]
- Pankratov, D.; Sundberg, R.; Suyatin, D.B.; Sotres, J.; Barrantes, A.; Ruzgas, T.; Maximov, I.; Montelius, L.; Shleev, S. The influence of nanoparticles on enzymatic bioelectrocatalysis. RSC Adv. 2014, 4, 38164–38168. [Google Scholar] [CrossRef]
- Silveira, C.M.; Zumpano, R.; Moreira, M.; de Almeida, M.P.; Oliveira, M.J.; Bento, M.; Montez, C.; Paixão, I.; Franco, R.; Pereira, E.; et al. Star-Shaped Gold Nanoparticles as Friendly Interfaces for Protein Electrochemistry: The Case Study of Cytochrome c. ChemElectroChem 2019, 6, 4696–4703. [Google Scholar] [CrossRef]
- Mohamad, N.R.; Marzuki, N.H.C.; Buang, N.A.; Huyop, F.; Wahab, R.A. An overview of technologies for immobilization of enzymes and surface analysis techniques for immobilized enzymes. Biotechnol. Biotechnol. Equip. 2015, 29, 205–220. [Google Scholar] [CrossRef]
- Zhou, W.; Zhang, W.; Cai, Y. Laccase immobilization for water purification: A comprehensive review. Chem. Eng. J. 2021, 403, 126272. [Google Scholar] [CrossRef]
- Hanefeld, U.; Gardossi, L.; Magner, E. Understanding enzyme immobilisation. Chem. Soc. Rev. 2009, 38, 453–468. [Google Scholar] [CrossRef] [PubMed]
- Ashkan, Z.; Hemmati, R.; Homaei, A.; Dinari, A.; Jamlidoost, M.; Tashakor, A. Immobilization of enzymes on nanoinorganic support materials: An update. Int. J. Biol. Macromol. 2021, 168, 708–721. [Google Scholar] [CrossRef] [PubMed]
- Ciogli, L.; Zumpano, R.; Poloznikov, A.A.; Hushpulian, D.M.; Tishkov, V.I.; Andreu, R.; Gorton, L.; Mazzei, F.; Favero, G.; Bollella, P. Highly Sensitive Hydrogen Peroxide Biosensor Based on Tobacco Peroxidase Immobilized on p-Phenylenediamine Diazonium Cation Grafted Carbon Nanotubes: Preventing Fenton-like Inactivation at Negative Potential. ChemElectroChem 2021, 8, 2495–2504. [Google Scholar] [CrossRef]
- Lavín, Á.; de Vicente, J.; Holgado, M.; Laguna, M.F.; Casquel, R.; Santamaría, B.; Maigler, M.V.; Hernández, A.L.; Ramírez, Y. On the determination of uncertainty and limit of detection in label-free biosensors. Sensors 2018, 18, 2038. [Google Scholar] [CrossRef]
- Knopp, S.; Bytof, G.; Selmar, D. Influence of processing on the content of sugars in green Arabica coffee beans. Eur. Food Res. Technol. 2006, 223, 195–201. [Google Scholar] [CrossRef]
- Bhagat, A.R.; Delgado, A.M.; Issaoui, M.; Chammem, N.; Fiorino, M.; Pellerito, A.; Natalello, S. Review of the Role of Fluid Dairy in Delivery of Polyphenolic Compounds in the Diet: Chocolate Milk, Coffee Beverages, Matcha Green Tea, and Beyond. J. AOAC Int. 2019, 102, 1365–1372. [Google Scholar] [CrossRef]
- Thangaraj, R.; Manjula, N.; Kumar, A.S. Rapid simultaneous electrochemical sensing of tea polyphenols. Anal. Methods 2012, 4, 2922–2928. [Google Scholar] [CrossRef]
- Bounegru, A.V.; Apetrei, C. Simultaneous Determination of Caffeic Acid and Ferulic Acid Using a Carbon Nanofiber-Based Screen-Printed Sensor. Sensors 2022, 22, 4689. [Google Scholar] [CrossRef] [PubMed]
- Dybkowska, E.; Sadowska, A.; Rakowska, R.; Dębowska, M.; Świderski, F.; Świąder, K. Assessing polyphenols content and antioxidant activity in coffee beans according to origin and the degree of roasting. Rocz. Panstw. Zakl. Hig. 2017, 68, 347–353. [Google Scholar] [PubMed]
- Várady, M.; Hrušková, T.; Popelka, P. Effect of preparation method and roasting temperature on total polyphenol content in coffee beverages. Czech J. Food Sci. 2020, 38, 417–421. [Google Scholar] [CrossRef]
- Król, K.; Gantner, M.; Tatarak, A.; Hallmann, E. The effect of roasting, storage, origin on the bioactive compounds in organic and conventional coffee (Caffea arabica). Eur. Food Res. Technol. 2019, 246, 33–39. [Google Scholar] [CrossRef]
- Sunarharum, W.B.; Yuwono, S.S.; Aziza, O.F. Study on the effect of roasting temperature on antioxidant activity of early-roasted Java coffee powder (Arabica and Robusta). IOP Conf. Ser. Earth Environ. Sci. 2019, 230, 012045. [Google Scholar] [CrossRef]
- Martín, M.J.; Pablos, F.; González, A.G. Discrimination between arabica and robusta green coffee varieties according to their chemical composition. Talanta 1998, 46, 1259–1264. [Google Scholar] [CrossRef]
- Cho, A.R.; Park, K.W.; Kim, K.M.; Kim, S.Y.; Han, J. Influence of Roasting Conditions on the Antioxidant Characteristics of C olombian Coffee (C offea arabica L.) Beans. J. Food Biochem. 2014, 38, 271–280. [Google Scholar] [CrossRef]
- Somporn, C.; Kamtuo, A.; Theerakulpisut, P.; Siriamornpun, S. Effects of roasting degree on radical scavenging activity, phenolics and volatile compounds of Arabica coffee beans (Coffea arabica L. cv. Catimor). Int. J. Food Sci. Technol. 2011, 46, 2287–2296. [Google Scholar] [CrossRef]
- Farah, A.; Donangelo, C.M. Phenolic compounds in coffee. Brazilian J. Plant Physiol. 2006, 18, 23–36. [Google Scholar] [CrossRef]
- Bicho, N.C.; Lidon, F.C.; Ramalho, J.C.; Leitão, A.E. Quality assessment of Arabica and Robusta green and roasted coffees–A review. Emir. J. Food Agric. 2013, 25, 945–950. [Google Scholar] [CrossRef]
- Bobková, A.; Hudáček, M.; Jakabová, S.; Belej, Ľ.; Capcarová, M.; Čurlej, J.; Bobko, M.; Árvay, J.; Jakab, I.; Čapla, J.; et al. The effect of roasting on the total polyphenols and antioxidant activity of coffee. J. Environ. Sci. Health Part B Pestic. Food Contam. Agric. Wastes 2020, 55, 495–500. [Google Scholar] [CrossRef]
- Folin, O.; Denis, W. on Phosphotungstic-Phosphomolybdic Compounds As Color Reagents. J. Biol. Chem. 1912, 12, 239–243. [Google Scholar] [CrossRef]
- A Agbor, G.; Vinson, J.A.; Donnelly, P.E. Folin-Ciocalteau Reagent for Polyphenolic Assay. Int. J. Food Sci. Nutr. Diet. 2014, 3, 147–156. [Google Scholar] [CrossRef]
- Ovemair Barbosa; Claudia Ortiz; Angel Berenguer-Murcia; Rodrigo Torres; Rafael, C.; Rodrigues, R.F.-L. Glutaraldehyde in Bio-Catalysts Design: A useful crosslinker and a versatile tool in enzyme immobilization. RSC Adv. 2014, 4, 1583–1600. [Google Scholar] [CrossRef]
- Migneault, I.; Dartiguenave, C.; Bertrand, M.J.; Waldron, K.C. Glutaraldehyde: Behavior in aqueous solution, reaction with proteins, and application to enzyme crosslinking. BioTecniques 2004, 37, 790–802. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Qu, X.; Guo, H.; Chen, H.; Liu, B.; Dong, S. Facile preparation of amperometric laccase biosensor with multifunction based on the matrix of carbon nanotubes–chitosan composite. Biosens. Bioelectron. 2006, 21, 2195–2201. [Google Scholar] [CrossRef] [PubMed]

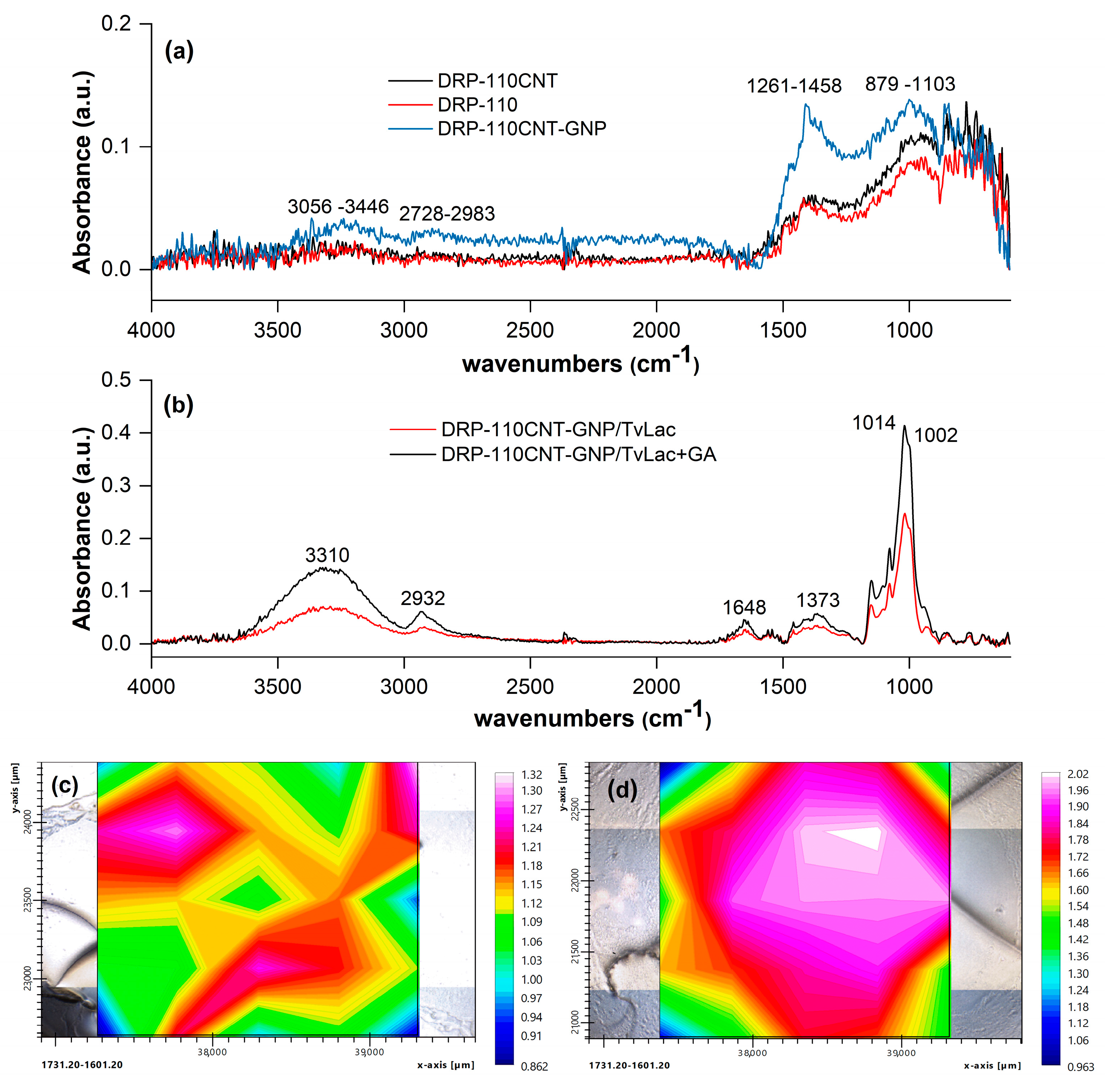

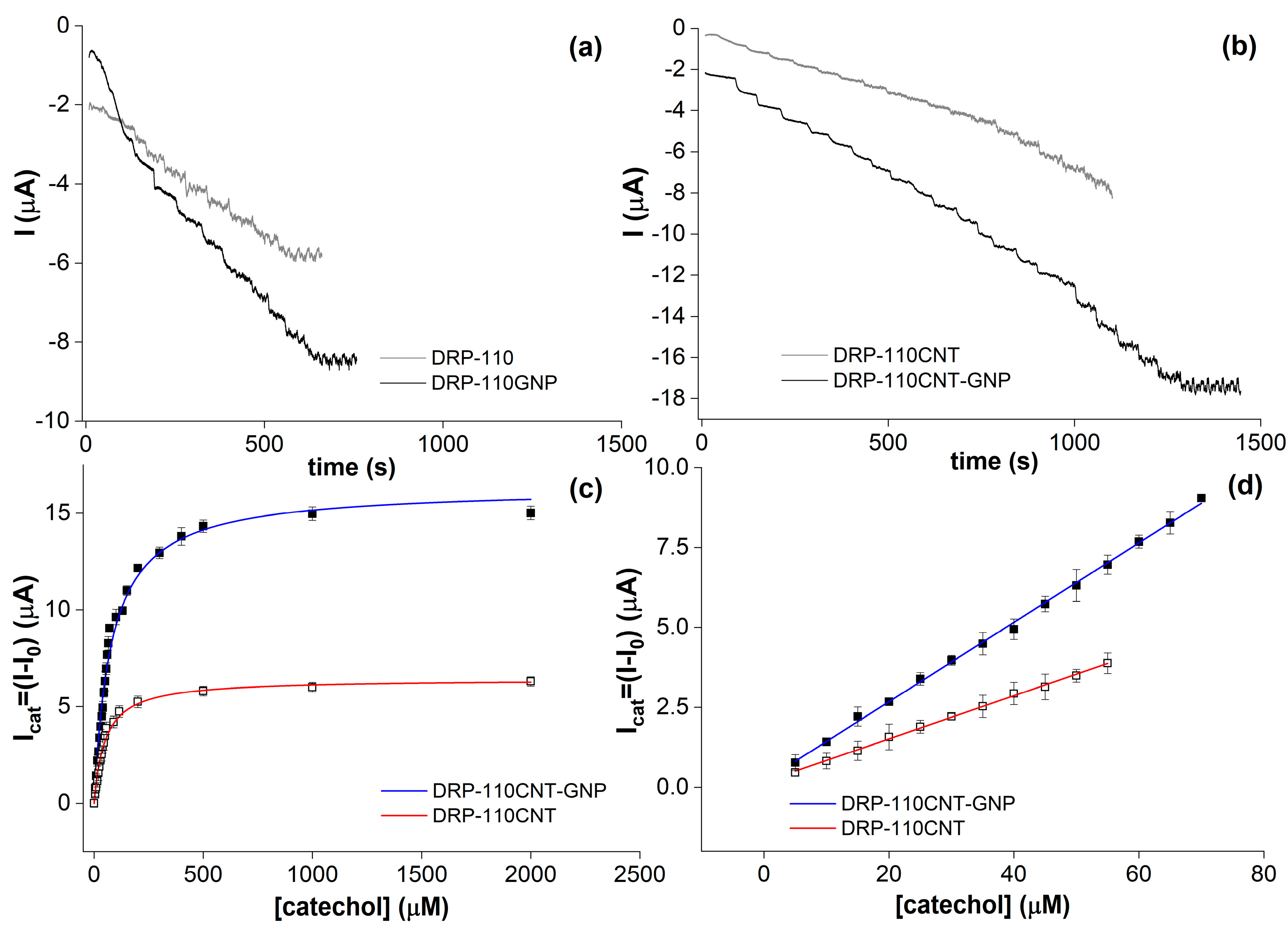

| Coffee Blend | Variety | Roasting Degree | [Phs] (mM) Folin–Cioc. | [Phs] (mM) Biosensor | [Phs] (mg/g) Biosensor | Recovery |
|---|---|---|---|---|---|---|
| I | 100% a. | Dark | 33.0 | 35.1 ± 0.3 | 170.0 ± 1.5 | 94% |
| II | 100% a. | Medium | 36.2 | 37.5 ± 0.2 | 180.2 ± 1.2 | 96% |
| III | 100% a. | Light | 41.1 | 44.8 ± 0.5 | 210.8 ± 2.3 | 91% |
| IV | 80% r. 20% a. | Medium | 49.7 | 52.0 ± 0.3 | 250.3 ± 1.3 | 95% |
| V | 60% r. 40% a. | Medium | 44.4 | 46.6 ± 0.3 | 227.7 ± 1.5 | 95% |
| VI | 100% a. | Medium | 34.3 | 36.1 ± 0.2 | 170.5 ± 1.0 | 95% |
| VII | 98% a. 2% r. | Medium | 40.8 | 42.1 ± 0.2 | 200.5 ± 0.8 | 97% |
| Roasting Degree | Oven t (°C) | Roasting Time (min) | [Phs] (mM) Folin–Cioc. | [Phs] (mM) Biosensor | [Phs] (mg/g) Biosensor | Recovery |
|---|---|---|---|---|---|---|
| unroasted | / | / | 22.7 | 22.0 ± 0.3 | 105.0 ± 0.3 | 97% |
| Light | 180 | 10 | 32.9 | 31.4 ± 0.1 | 152.6 ± 0.5 | 95% |
| Light | 180 | 15 | 16.2 | 17.0 ± 0.1 | 82.6 ± 0.6 | 95% |
| Light | 180 | 20 | 8.8 | 9.3 ± 0.2 | 45.4 ± 1.0 | 94% |
| Medium | 200 | 10 | 24.2 | 25.5 ± 0.1 | 124.0 ± 0.5 | 95% |
| Medium | 200 | 15 | 16.9 | 18.1 ± 0.2 | 80.8 ± 1.1 | 93% |
| Medium | 200 | 20 | 9.9 | 9.1 ± 0.1 | 44.4 ± 0.7 | 93% |
| Dark | 250 | 10 | 16.9 | 18.1 ± 0.2 | 80.8 ± 1.1 | 93% |
| Dark | 250 | 15 | 11.1 | 10.3 ± 0.2 | 50.0 ± 0.9 | 93% |
| Dark | 250 | 20 | 9.7 | 10.0 ± 0.2 | 40.9 ± 1.0 | 96% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
D’Agostino, C.; Chillocci, C.; Polli, F.; Surace, L.; Simonetti, F.; Agostini, M.; Brutti, S.; Mazzei, F.; Favero, G.; Zumpano, R. Smartphone-Based Electrochemical Biosensor for On-Site Nutritional Quality Assessment of Coffee Blends. Molecules 2023, 28, 5425. https://doi.org/10.3390/molecules28145425
D’Agostino C, Chillocci C, Polli F, Surace L, Simonetti F, Agostini M, Brutti S, Mazzei F, Favero G, Zumpano R. Smartphone-Based Electrochemical Biosensor for On-Site Nutritional Quality Assessment of Coffee Blends. Molecules. 2023; 28(14):5425. https://doi.org/10.3390/molecules28145425
Chicago/Turabian StyleD’Agostino, Cristine, Claudia Chillocci, Francesca Polli, Luca Surace, Federica Simonetti, Marco Agostini, Sergio Brutti, Franco Mazzei, Gabriele Favero, and Rosaceleste Zumpano. 2023. "Smartphone-Based Electrochemical Biosensor for On-Site Nutritional Quality Assessment of Coffee Blends" Molecules 28, no. 14: 5425. https://doi.org/10.3390/molecules28145425
APA StyleD’Agostino, C., Chillocci, C., Polli, F., Surace, L., Simonetti, F., Agostini, M., Brutti, S., Mazzei, F., Favero, G., & Zumpano, R. (2023). Smartphone-Based Electrochemical Biosensor for On-Site Nutritional Quality Assessment of Coffee Blends. Molecules, 28(14), 5425. https://doi.org/10.3390/molecules28145425











