The Expression of Antibacterial Peptide Turgencin A in Pichia pastoris and an Analysis of Its Antibacterial Activity
Abstract
1. Introduction
2. Results and Discussion
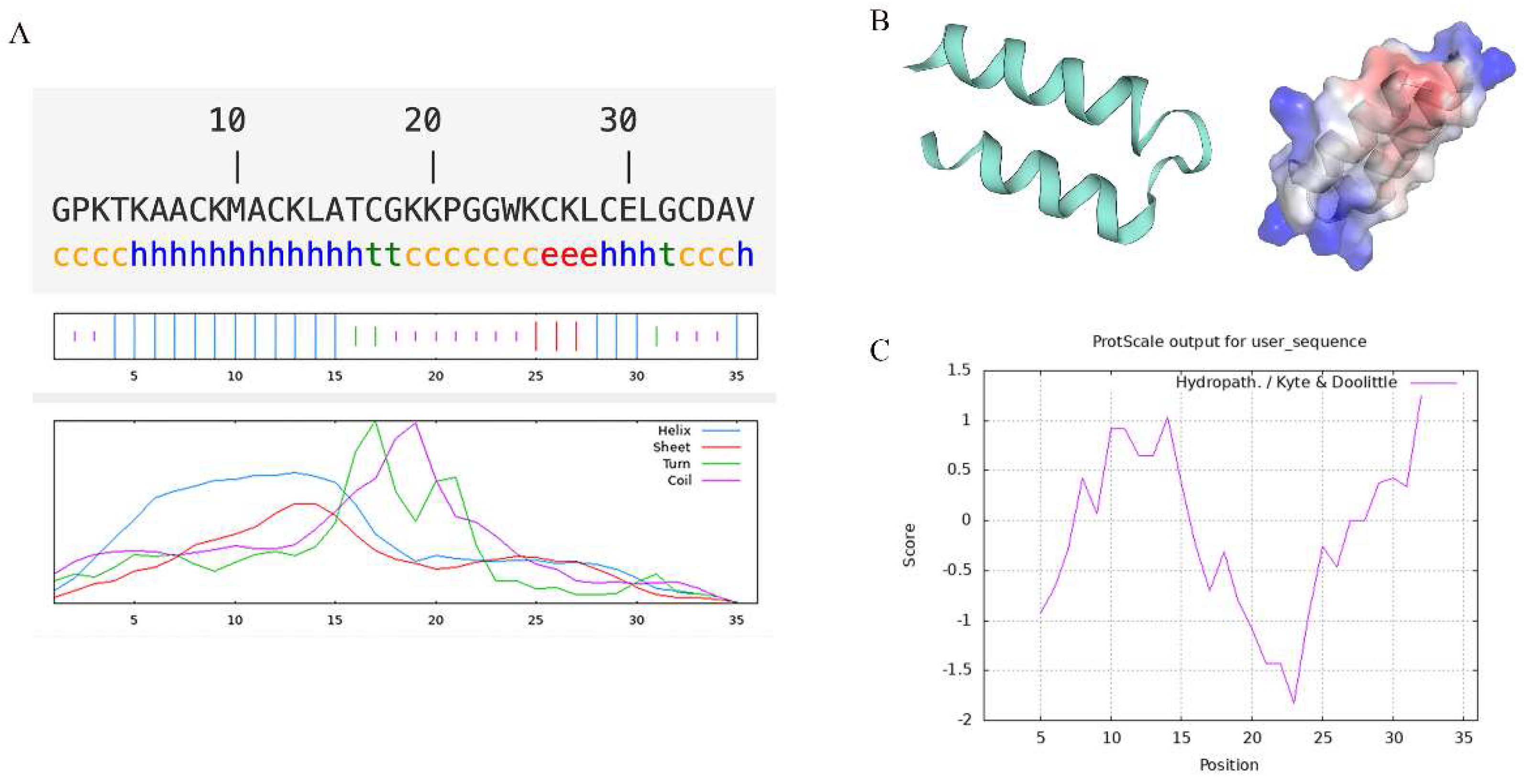
2.1. Structural Analysis of Recombinant Turgencin A Strains
2.2. Construction and Recombinant Expression of Positive Yeast Strains with High Expression of Recombinant Turgencin A
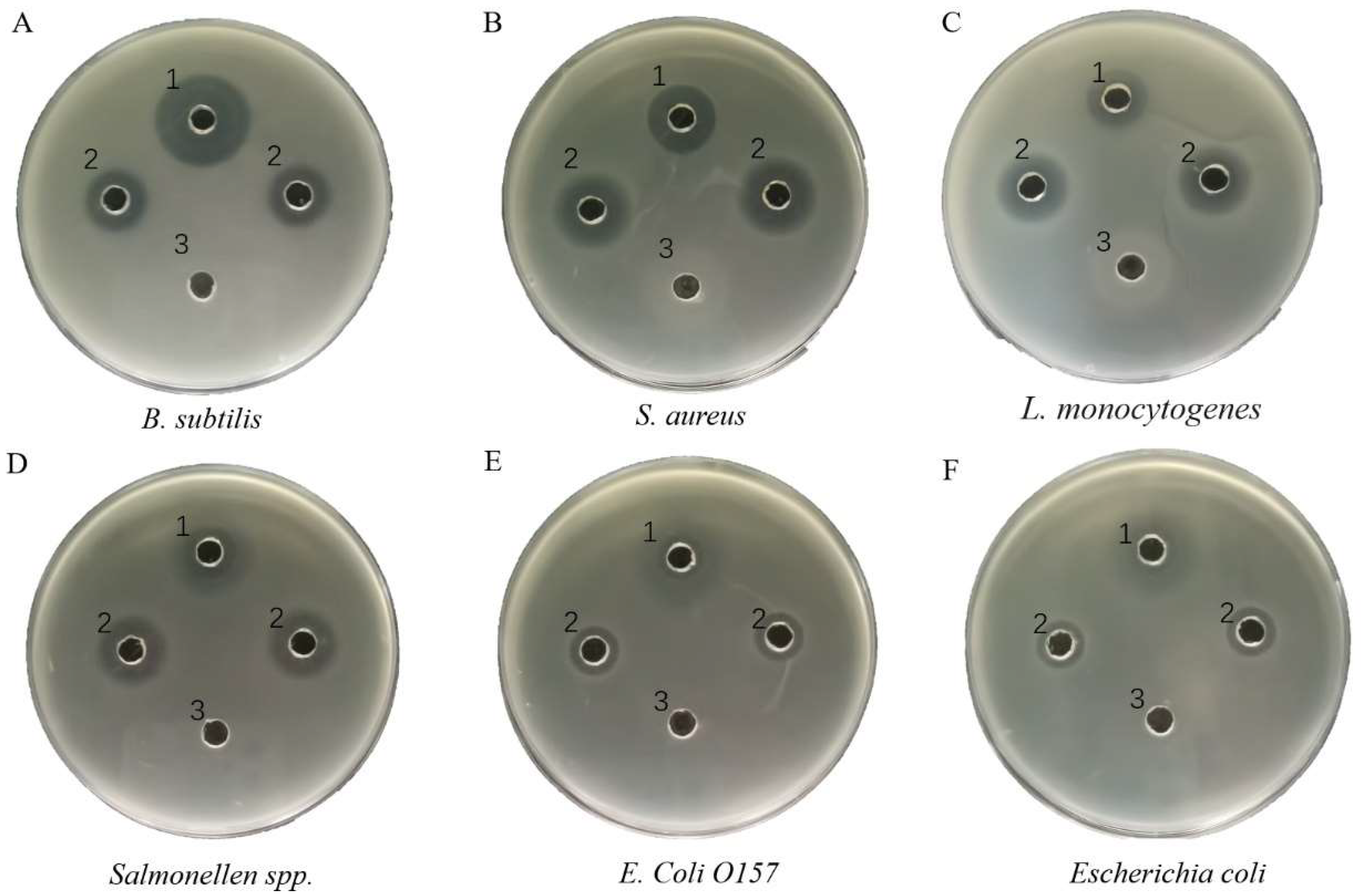
2.3. Determining Antibacterial Activity of Recombinant Turgencin A
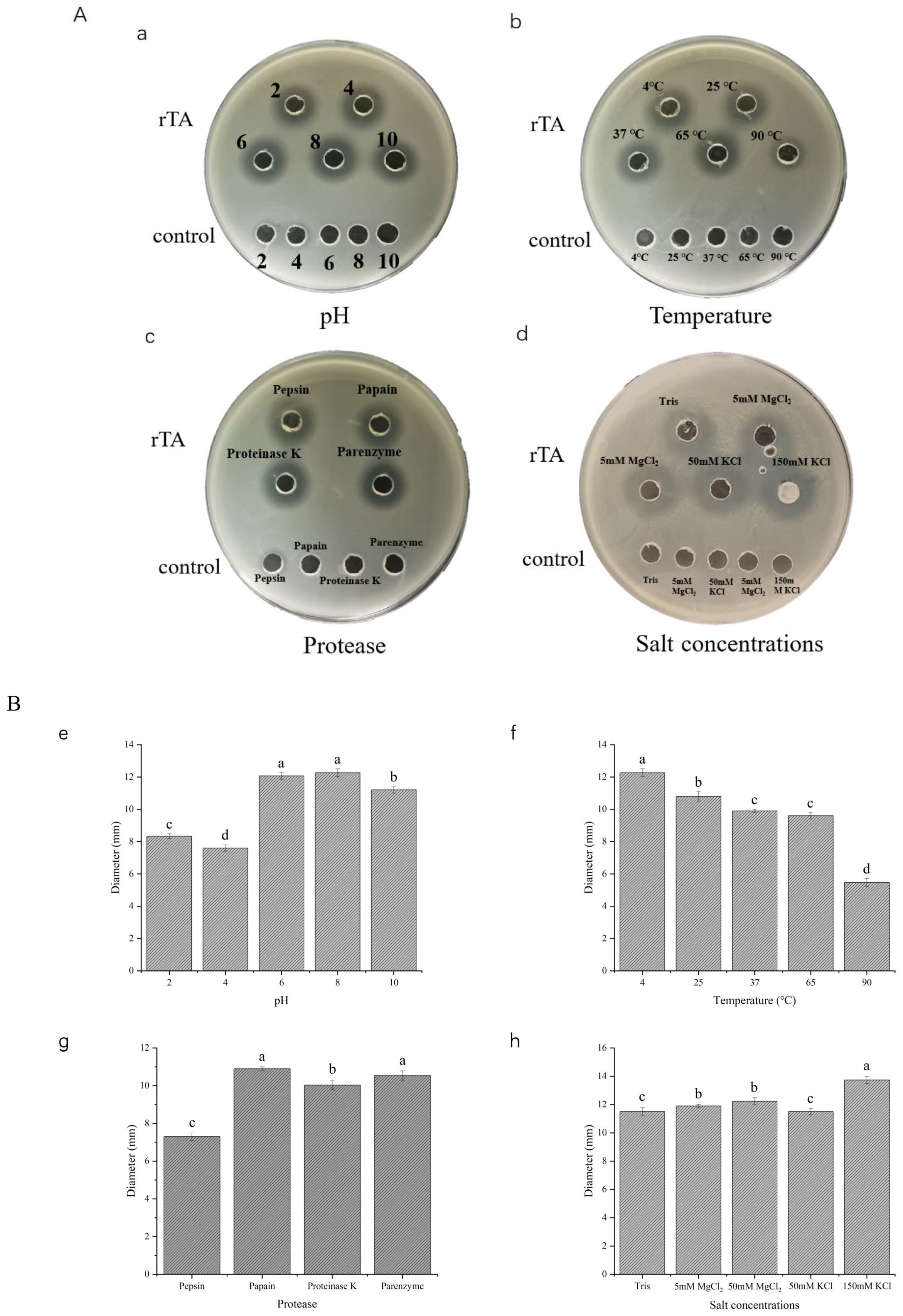
2.4. Stability of Recombinant Turgencin A
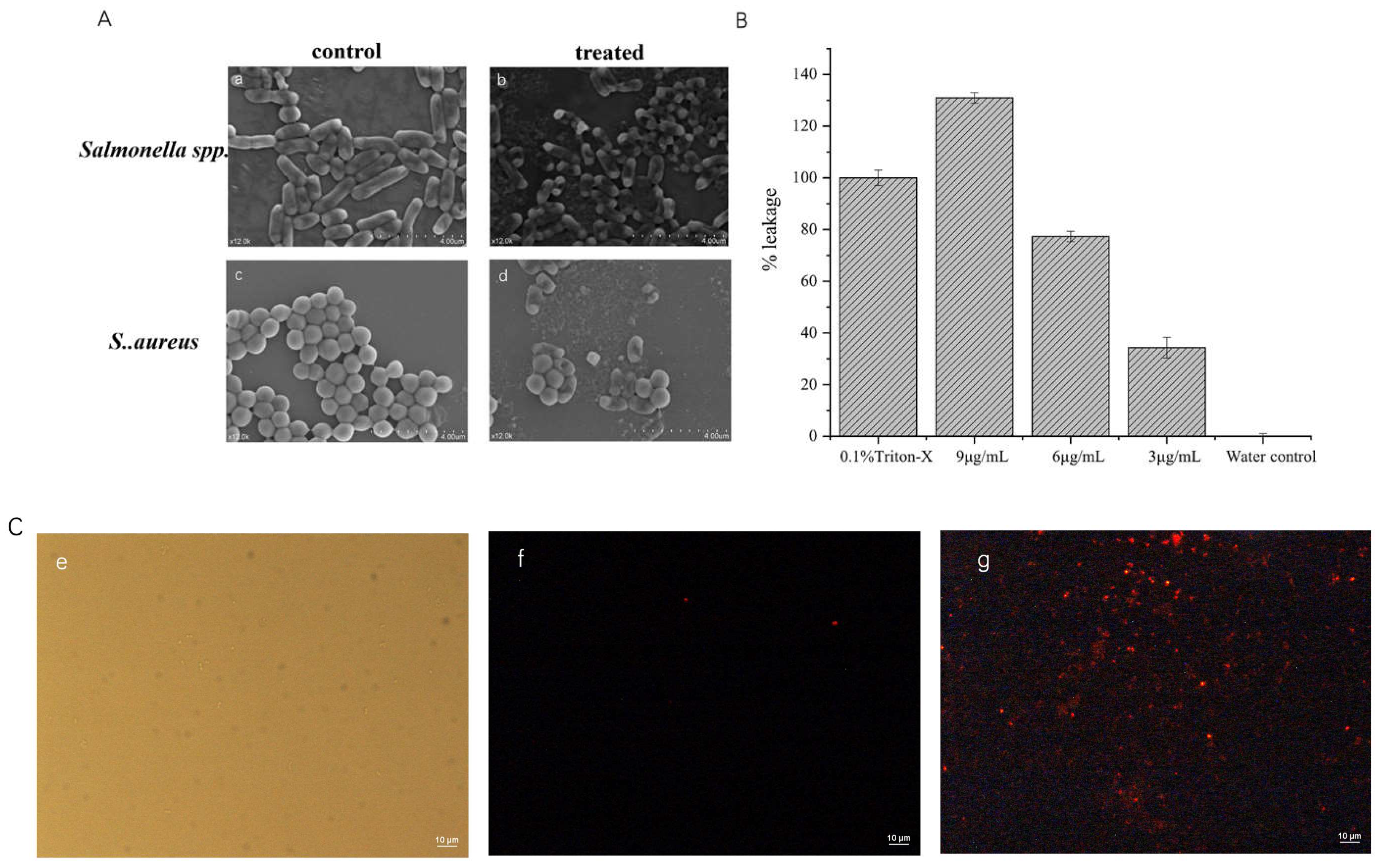
2.5. Mechanism Studies
2.5.1. Scanning Electron Microscope
2.5.2. Total Nucleotide Leakage
2.5.3. Cell Permeation Experiments
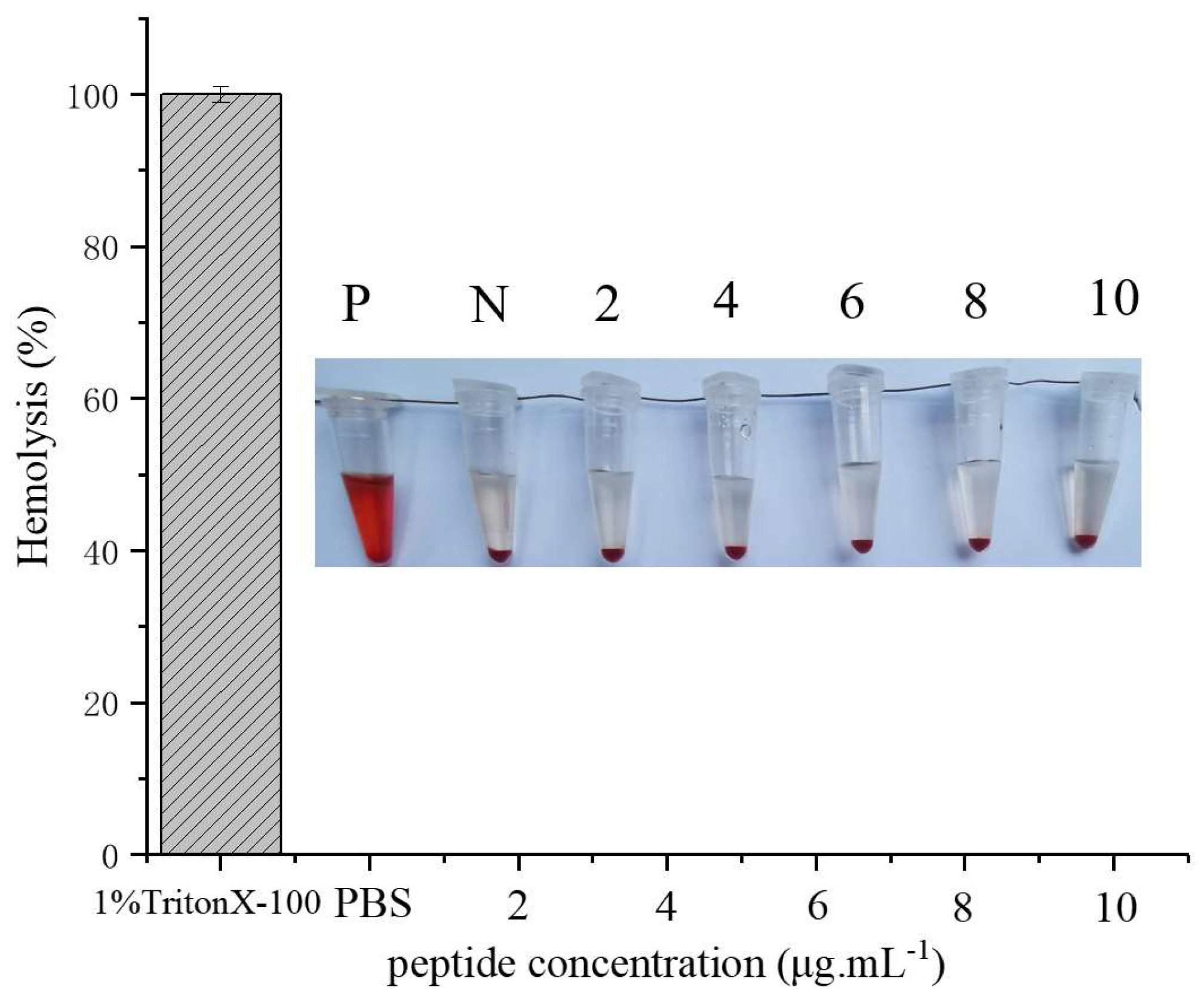
2.6. Hemolysis Assay
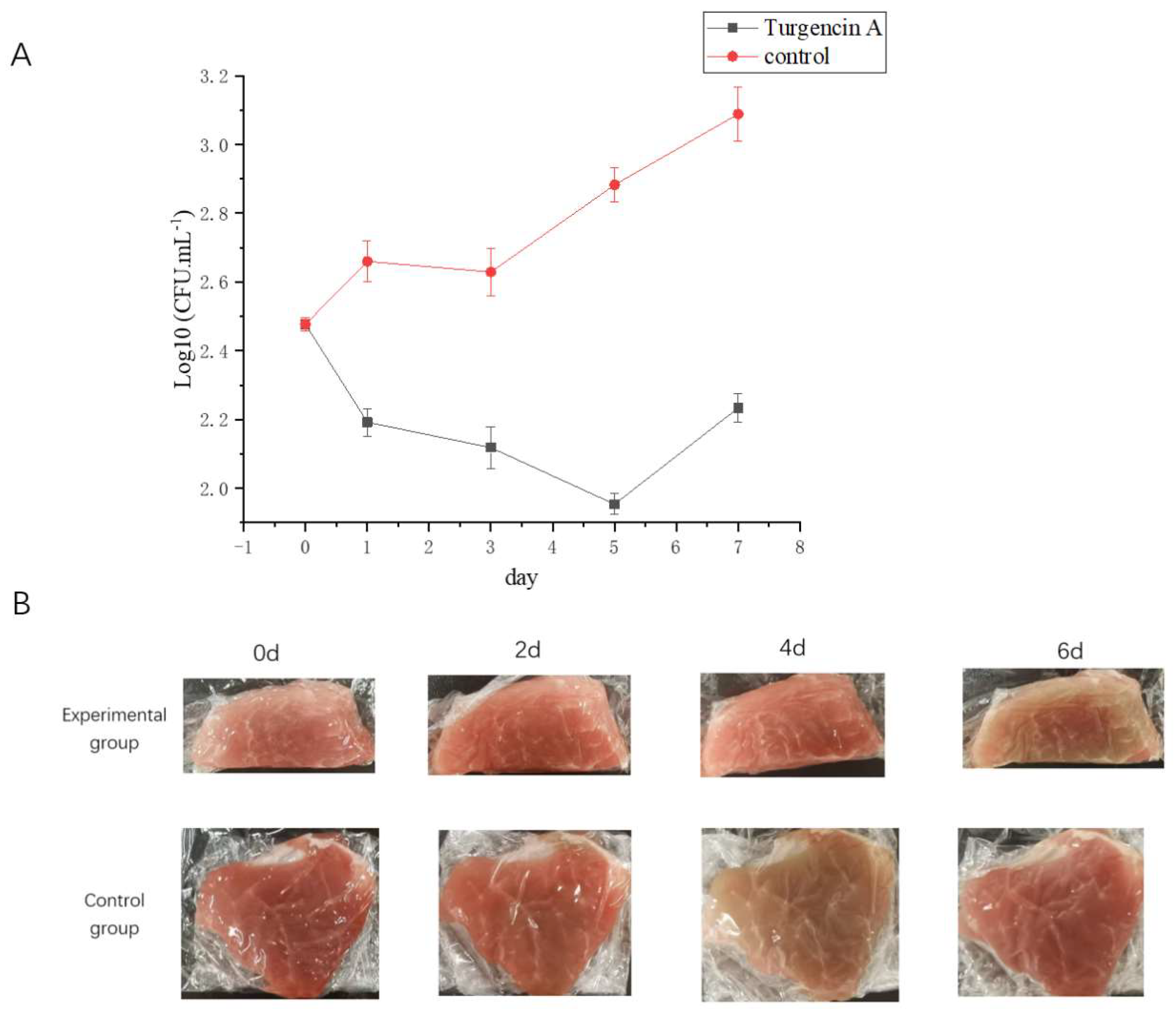
2.7. Application of Turgencin A as Preservative
2.7.1. Total Number of Bacteria
2.7.2. Flesh Color
3. Materials and Methods
3.1. Strains and Plasmids
3.2. Structural Prediction
3.3. Transformation of Pichia pastoris GS115
3.4. Induction of Expression and Detection of Peptides Obtained from Recombinant Pichia pastoris
3.5. Purification of Recombinant Antibacterial Peptides
3.6. Determination of Turgencin A Concentration: ELISA
3.7. Antibacterial Activity
3.7.1. Agar Well Diffusion Assay
3.7.2. Minimum Inhibitory Concentration (MIC) Determination
3.8. Stability Study of Recombinant Antimicrobial Peptides
3.8.1. Study of Temperature Stability
3.8.2. Study of pH Stability
3.8.3. Study of Protease Stability
3.8.4. Study on the Stability of Salt Concentration
3.9. Effect of Turgencin A on Cell Membrane
3.9.1. Total Nucleotide Leakage
3.9.2. Scanning Electron Microscopy Observations
3.9.3. Cell Permeability Assay
3.10. Hemolytic Analysis of Turgencin A
3.11. Pork Preservation Experiment
3.12. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Al-Tawfiq, J.A.; Momattin, H.; Al-Ali, A.Y.; Eljaaly, K.; Tirupathi, R.; Haradwala, M.B.; Areti, S.; Alhumaid, S.; Rabaan, A.A.; Al Mutair, A.; et al. Antibiotics in the pipeline: A literature review (2017–2020). Infection 2022, 50, 553–564. [Google Scholar] [CrossRef]
- Kumar, M.; Sarma, D.K.; Shubham, S.; Kumawat, M.; Verma, V.; Nina, P.B.; Devraj, J.P.; Kumar, S.; Singh, B.; Tiwari, R.R. Futuristic Non-antibiotic Therapies to Combat Antibiotic Resistance: A Review. Front. Microbiol. 2021, 12, 609459. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.F.; Battalapalli, D.; Draz, M.S.; Zhang, P.F.; Ruan, Z. The Application of Cell-Penetrating-Peptides in Antibacterial Agents. Curr. Med. Chem. 2021, 28, 5896–5925. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.Q.; Yu, X.; Yang, Q.L.; Zhao, Y.; Wu, W. Aptasensors for Staphylococcus aureus Risk Assessment in Food. Front. Microbiol. 2021, 12, 714265. [Google Scholar] [CrossRef] [PubMed]
- Thwala, T.; Madoroba, E.; Basson, A.; Butaye, P. Prevalence and Characteristics of Staphylococcus aureus Associated with Meat and Meat Products in African Countries: A Review. Antibiotics 2021, 10, 1108. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.T.; Chen, E.Z.; Yang, L.; Peng, C.; Wang, Q.; Xu, Z.B.; Chen, D.Q. Emerging resistance mechanisms for 4 types of common anti-MRSA antibiotics in Staphylococcus aureus: A comprehensive review. Microb. Pathog. 2021, 156, 104915. [Google Scholar] [CrossRef] [PubMed]
- Rima, M.; Rima, M.; Fajloun, Z.; Sabatier, J.M.; Bechinger, B.; Naas, T. Antimicrobial Peptides: A Potent Alternative to Antibiotics. Antibiotics 2021, 10, 1095. [Google Scholar] [CrossRef]
- Zhang, R.; Xu, L.; Dong, C. Antimicrobial peptides: An overview of their structure, function and mechanism of action. Protein Pept. Lett. 2022, 29, 641–650. [Google Scholar] [CrossRef]
- Browne, K.; Chakraborty, S.; Chen, R.X.; Willcox, M.; Black, D.S.; Walsh, W.R.; Kumar, N. A New Era of Antibiotics: The Clinical Potential of Antimicrobial Peptides. Int. J. Mol. Sci. 2020, 21, 7047. [Google Scholar] [CrossRef]
- Escobar-Salom, M.; Torrens, G.; Jordana-Lluch, E.; Oliver, A.; Juan, C. Mammals’ humoral immune proteins and peptides targeting the bacterial envelope: From natural protection to therapeutic applications against multidrug-resistant Gram-negatives. Biol. Rev. Camb. Philos. Soc. 2022, 97, 1005–1037. [Google Scholar] [CrossRef]
- Lin, D.N.; Sutherland, D.; Aninta, S.I.; Louie, N.; Nip, K.M.; Li, C.K.; Yanai, A.; Coombe, L.; Warren, R.L.; Helbing, C.C.; et al. Mining Amphibian and Insect Transcriptomes for Antimicrobial Peptide Sequences with rAMPage. Antibiotics 2022, 11, 952. [Google Scholar] [CrossRef]
- Saucedo-Vazquez, J.P.; Gushque, F.; Vispo, N.S.; Rodriguez, J.; Gudino-Gomezjurado, M.E.; Albericio, F.; Tellkamp, M.P.; Alexis, F. Marine Arthropods as a Source of Antimicrobial Peptides. Mar. Drugs 2022, 20, 501. [Google Scholar] [CrossRef] [PubMed]
- Azmiera, N.; Krasilnikova, A.; Sahudin, S.; Al-Talib, H.; Heo, C.C. Antimicrobial peptides isolated from insects and their potential applications. J. Asia-Pac. Entomol. 2022, 25, 101892. [Google Scholar] [CrossRef]
- Boparai, J.K.; Sharma, P.K. Mini Review on Antimicrobial Peptides, Sources, Mechanism and Recent Applications. Protein Pept. Lett. 2020, 27, 4–16. [Google Scholar] [CrossRef] [PubMed]
- Shwaiki, L.N.; Lynch, K.M.; Arendt, E.K. Future of antimicrobial peptides derived from plants in food application-A focus on synthetic peptides. Trends Food Sci. Technol. 2021, 112, 312–324. [Google Scholar] [CrossRef]
- Campagna, S.; Saint, N.; Molle, G.; Aumelas, A. Structure and mechanism of action of the antimicrobial peptide piscidin. Biochemistry 2007, 46, 1771–1778. [Google Scholar] [CrossRef]
- Li, X.; Zuo, S.Y.; Wang, B.; Zhang, K.Y.; Wang, Y. Antimicrobial Mechanisms and Clinical Application Prospects of Antimicrobial Peptides. Molecules 2022, 27, 2675. [Google Scholar] [CrossRef]
- Li, T.X.; Li, L.L.; Du, F.Y.; Sun, L.; Shi, J.C.; Long, M.; Chen, Z.L. Activity and Mechanism of Action of Antifungal Peptides from Microorganisms: A Review. Molecules 2021, 26, 3438. [Google Scholar] [CrossRef]
- Hou, X.Y.; Li, J.L.; Tang, H.Q.; Li, Q.Y.; Shen, G.H.; Li, S.S.; Chen, A.J.; Peng, Z.X.; Zhang, Y.; Li, C.W.; et al. Antibacterial Peptide NP-6 Affects Staphylococcus aureus by Multiple Modes of Action. Int. J. Mol. Sci. 2022, 23, 7812. [Google Scholar] [CrossRef]
- Wu, X.; Zhang, S.Q. Molecular mechanisms of antibacterial peptides against bacterium. Prog. Biochem. Biophys. 2005, 32, 1109–1113. [Google Scholar]
- Nicolas, P. Multifunctional host defense peptides: Intracellular-targeting antimicrobial peptides. FEBS J. 2009, 276, 6483–6496. [Google Scholar] [CrossRef]
- Malmsten, M. Antimicrobial peptides. Upsala J. Med. Sci. 2014, 119, 199–204. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Qu, L.K.; Lin, S.E.; Yang, Q.S.; Zhang, X.X.; Jin, L.B.; Dong, H.; Sun, D. Biological Functions and Applications of Antimicrobial Peptides. Curr. Protein Pept. Sc. 2022, 23, 226–247. [Google Scholar] [CrossRef]
- Raghavan, R.; Pannippara, M.A.; Kesav, S.; Mathew, A.; Bhat, S.G.; Rafeeq, C.M.; Elyas, K.K. Production Optimization and In Vitro Evaluation of Anti-proliferative, Anti-oxidant and Anti-inflammatory Potential of the Antibacterial Peptide MFAP9. Int. J. Pept. Res. Ther. 2022, 28, 139. [Google Scholar] [CrossRef]
- Rodriguez, A.A.; Otero-Gonzalez, A.; Ghattas, M.; Staendker, L. Discovery, Optimization, and Clinical Application of Natural Antimicrobial Peptides. Biomedicines 2021, 9, 1381. [Google Scholar] [CrossRef]
- Wu, R.; Patocka, J.; Nepovimova, E.; Oleksak, P.; Valis, M.; Wu, W.D.; Kuca, K. Marine Invertebrate Peptides: Antimicrobial Peptides. Front. Microbiol. 2021, 12, 785085. [Google Scholar] [CrossRef]
- Tong, J.R.; Zhang, Z.H.; Wu, Q.; Huang, Z.H.; Malakar, P.K.; Chen, L.B.; Liu, H.Q.; Pan, Y.J.; Zhao, Y. Antibacterial peptides from seafood: A promising weapon to combat bacterial hazards in food. Food Control. 2021, 125, 108004. [Google Scholar] [CrossRef]
- Hansen IK, Ø.; Isaksson, J.; Poth, A.G.; Hansen, K.Ø.; Andersen, A.J.C.; Richard, C.S.M.; Blencke, H.; Stensvåg, K.; Craik, D.J.; Haug, T. Isolation and Characterization of Antimicrobial Peptides with Unusual Disulfide Connectivity from the Colonial Ascidian Synoicum turgens. Mar. Drugs 2020, 18, 51. [Google Scholar] [CrossRef]
- Hansen, I.; Lovdahl, T.; Simonovic, D.; Hansen, K.O.; Andersen, A.; Devold, H.; Richard, C.; Andersen, J.H.; Strom, M.B.; Haug, T. Antimicrobial Activity of Small Synthetic Peptides Based on the Marine Peptide Turgencin A: Prediction of Antimicrobial Peptide Sequences in a Natural Peptide and Strategy for Optimization of Potency. Int. J. Mol. Sci. 2020, 21, 5460. [Google Scholar] [CrossRef]
- Arumugam, V.; Venkatesan, M.; Ramachandran, K.; Ramachandran, S.; Palanisamy, S.K.; Sundaresan, U. Purification, Characterization and Antibacterial Properties of Peptide from Marine Ascidian Didemnum sp. Int. J. Pept. Res. Ther. 2020, 26, 201–208. [Google Scholar] [CrossRef]
- Giuliani, A.; Rinaldi, A.C. Beyond natural antimicrobial peptides: Multimeric peptides and other peptidomimetic approaches. Cell. Mol. Life Sci. 2011, 68, 2255–2266. [Google Scholar] [CrossRef] [PubMed]
- Yaraksa, N.; Daduang, S. De novo Design and Synthesis of Potent Antimicrobial Peptide and Mode of Action. Chiang Mai J. Sci. 2021, 48, 444–459. [Google Scholar]
- Chen, Y.; Mant, C.T.; Farmer, S.W.; Hancock, R.E.; Vasil, M.L.; Hodges, R.S. Rational design of alpha-helical antimicrobial peptides with enhanced activities and specificity/therapeutic index. J. Biol. Chem. 2005, 280, 12316–12329. [Google Scholar] [CrossRef] [PubMed]
- Savini, F.; Loffredo, M.R.; Troiano, C.; Bobone, S.; Malanovic, N.; Eichmann, T.O.; Caprio, L.; Canale, V.C.; Park, Y.; Mangoni, M.L.; et al. Binding of an antimicrobial peptide to bacterial cells: Interaction with different species, strains and cellular components. Biochim. Biophys. Acta Biomembr. 2020, 1862, 183291. [Google Scholar] [CrossRef] [PubMed]
- Mandal, S.M.; Dey, S.; Mandal, M.; Sarkar, S.; Maria-Neto, S.; Franco, O.L. Identification and structural insights of three novel antimicrobial peptides isolated from green coconut water. Peptides 2009, 30, 633–637. [Google Scholar] [CrossRef]
- Lee, T.H.; Hall, K.N.; Aguilar, M.I. Antimicrobial Peptide Structure and Mechanism of Action: A Focus on the Role of Membrane Structure. Curr. Top. Med. Chem. 2016, 16, 25–39. [Google Scholar] [CrossRef]
- Gharsallaoui, A.; Oulahal, N.; Joly, C.; Degraeve, P. Nisin as a Food Preservative: Part 1: Physicochemical Properties, Antimicrobial Activity, and Main Uses. Crit. Rev. Food. Sci. Nutr. 2016, 56, 1262–1274. [Google Scholar] [CrossRef]
- Valizadeh, S.; Naseri, M.; Babaei, S.; Hosseini, S.M.H.; Imani, A. Development of bioactive composite films from chitosan and carboxymethyl cellulose using glutaraldehyde, cinnamon essential oil and oleic acid. Int. J. Biol. Macromol. 2019, 134, 604–612. [Google Scholar] [CrossRef]
- Karamucki, T.; Gardzielewska, J.; Rybarczyk, A.; Jakubowska, M.; Natalczyk-Szymkowska, W. Usefulness of Selected Methods of Colour Change Measurement for Pork Quality Assessment. Czech J. Food Sci. 2011, 29, 212–218. [Google Scholar] [CrossRef]
- Zamani, E.; Zargan, J.; Honari, H.; Hajizade, A.; Mohammadi, A.H.N.; Alikhani, H.K.; Heidari, A.; Pour, M.H. Immunological detection of AcAMP antimicrobial peptide secreted by Aspergillus clavatus. Iran. J. Microbiol. 2021, 13, 235–242. [Google Scholar] [CrossRef]
- Shwaiki, L.N.; Sahin, A.W.; Arendt, E.K. Study on the Inhibitory Activity of a Synthetic Defensin Derived from Barley Endosperm against Common Food Spoilage Yeast. Molecules 2021, 26, 165. [Google Scholar] [CrossRef] [PubMed]
- GB 4789.2-2022; National Food Safety Standard—Microbiological Examination Of Food: Aerobic Plate Count. Chinese Standard: Bejing, China, 2022.

| Peptide | Concentration (μg·mL−1) |
|---|---|
| Turgencin A | 11.23 |
| 200 mg·mL−1 TA fermentation supernatant freeze-dried | 13.71 |
| Bacteria Strain | MIC (μg·mL−1) |
|---|---|
| Gram−positive | |
| S. aureus (ATCC 25923) | 3 |
| B. subtilis (ATCC 6633) | 2 |
| L. monocytogenes (ATCC 21633) | 3 |
| Gram−negative | |
| Salmonella spp. (ATCC 10467) | 4 |
| E. coli O157 (ATCC 35150) | 5 |
| E. coli (ATCC 10305) | 5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dong, C.; Li, M.; Zhang, R.; Lu, W.; Xu, L.; Liu, J.; Chu, X. The Expression of Antibacterial Peptide Turgencin A in Pichia pastoris and an Analysis of Its Antibacterial Activity. Molecules 2023, 28, 5405. https://doi.org/10.3390/molecules28145405
Dong C, Li M, Zhang R, Lu W, Xu L, Liu J, Chu X. The Expression of Antibacterial Peptide Turgencin A in Pichia pastoris and an Analysis of Its Antibacterial Activity. Molecules. 2023; 28(14):5405. https://doi.org/10.3390/molecules28145405
Chicago/Turabian StyleDong, Chunming, Mengru Li, Rui Zhang, Weitao Lu, Lijun Xu, Jian Liu, and Xinlei Chu. 2023. "The Expression of Antibacterial Peptide Turgencin A in Pichia pastoris and an Analysis of Its Antibacterial Activity" Molecules 28, no. 14: 5405. https://doi.org/10.3390/molecules28145405
APA StyleDong, C., Li, M., Zhang, R., Lu, W., Xu, L., Liu, J., & Chu, X. (2023). The Expression of Antibacterial Peptide Turgencin A in Pichia pastoris and an Analysis of Its Antibacterial Activity. Molecules, 28(14), 5405. https://doi.org/10.3390/molecules28145405






