In Silico Screening and Identification of Antidiabetic Inhibitors Sourced from Phytochemicals of Philippine Plants against Four Protein Targets of Diabetes (PTP1B, DPP-4, SGLT-2, and FBPase)
Abstract
1. Introduction
2. Results
2.1. ADMET Profiling
2.2. DFT Optimization of Ligands
2.3. Molecular Docking
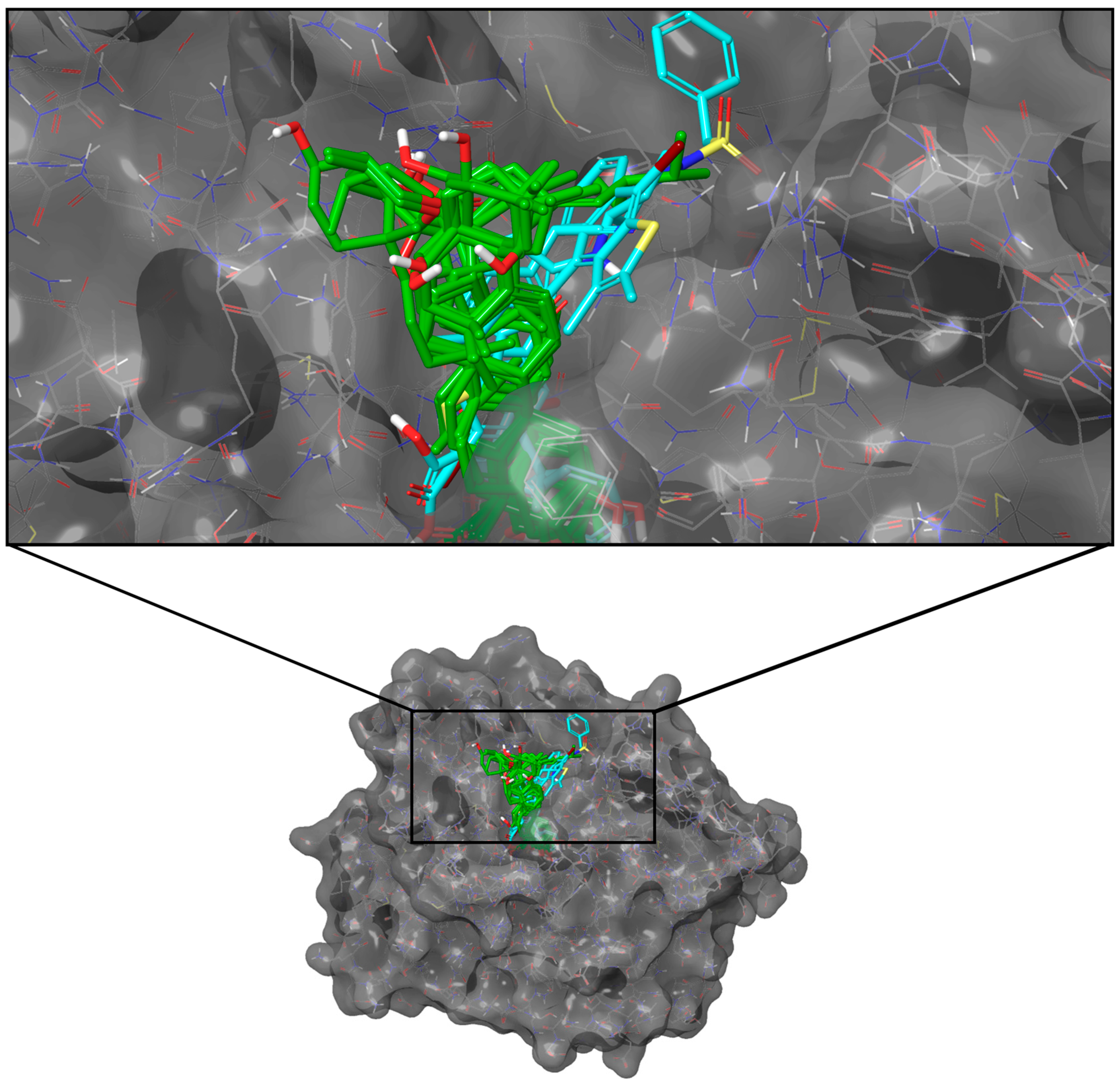
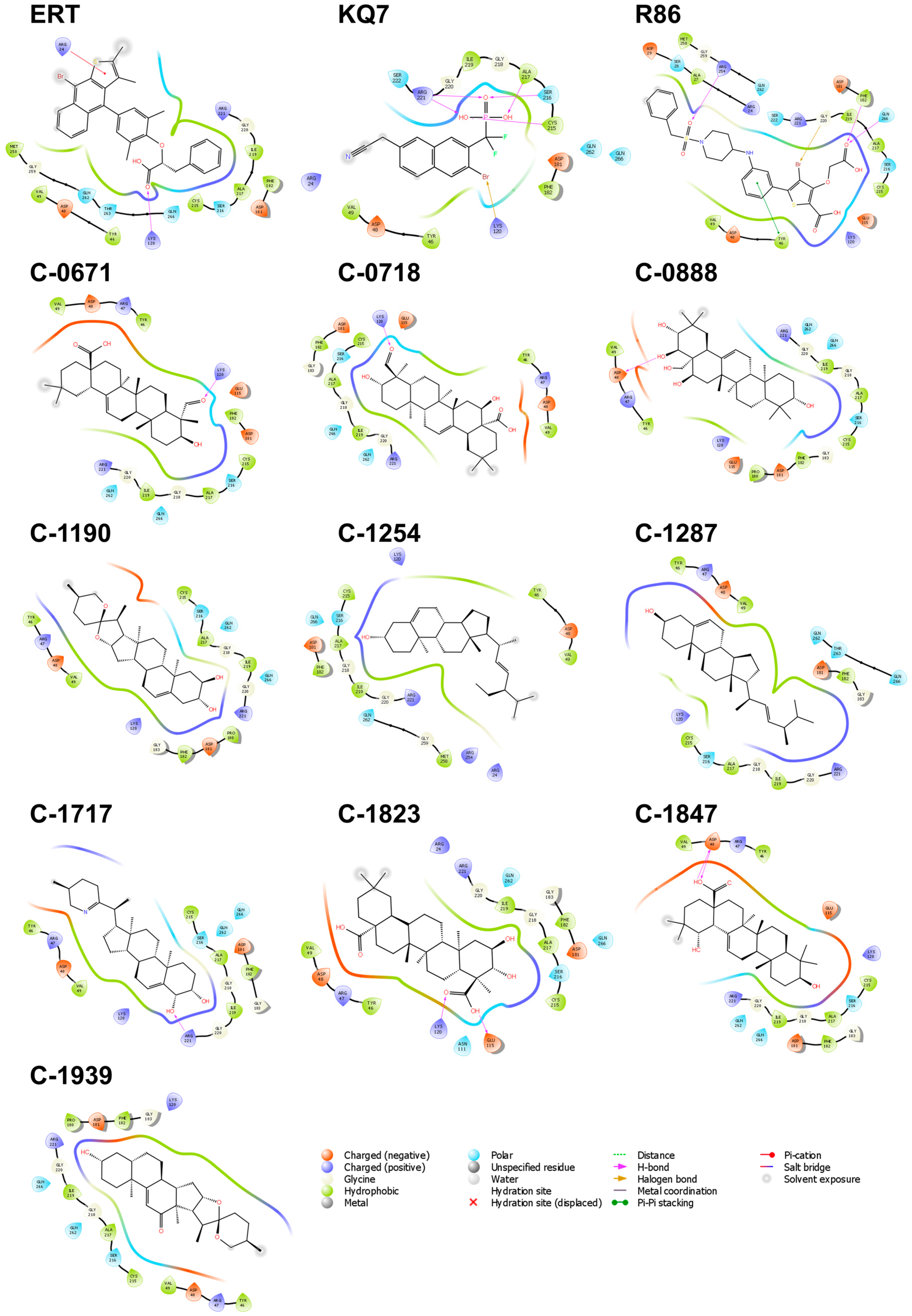
2.3.1. PTP1B
2.3.2. DPP-4
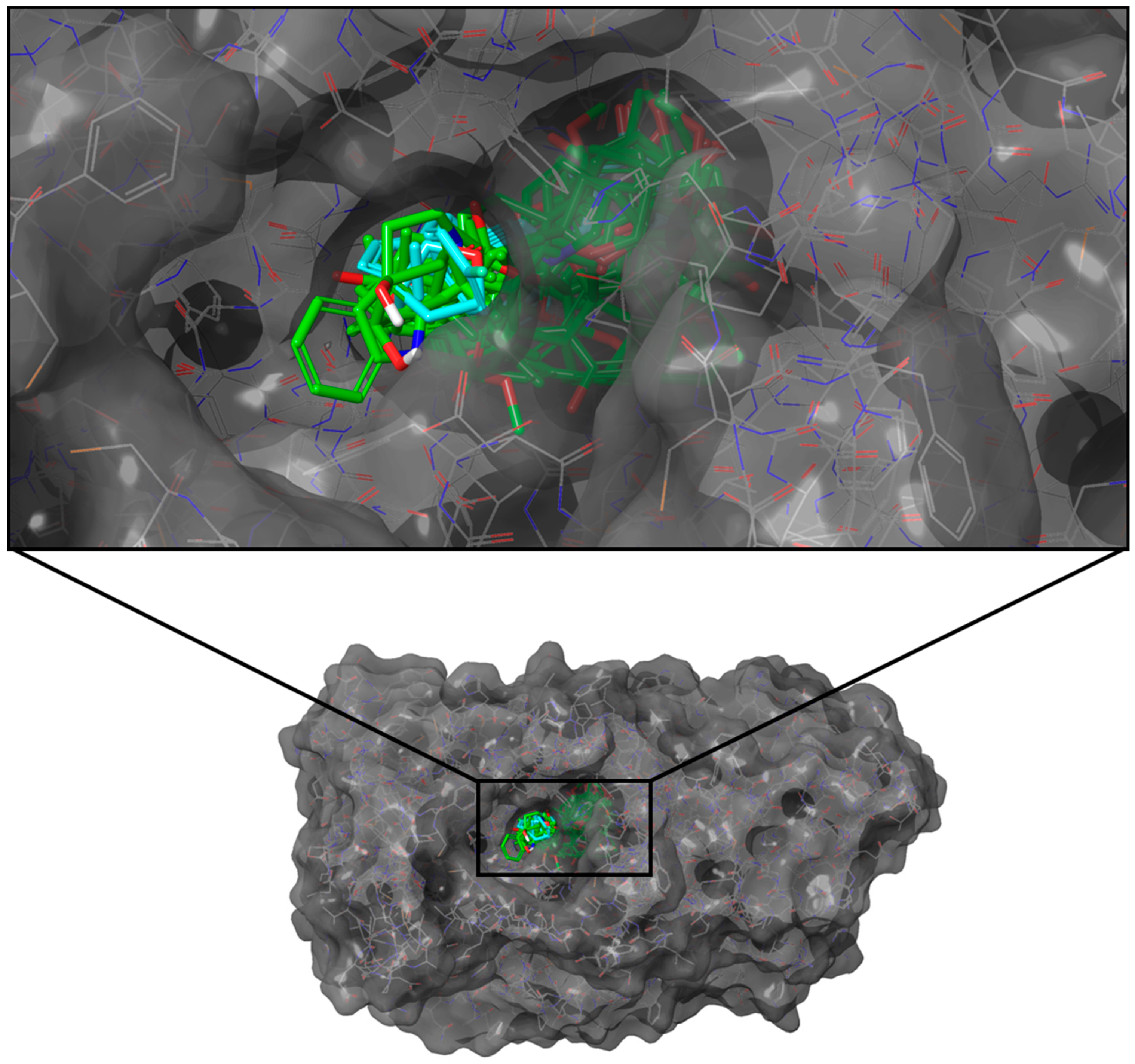
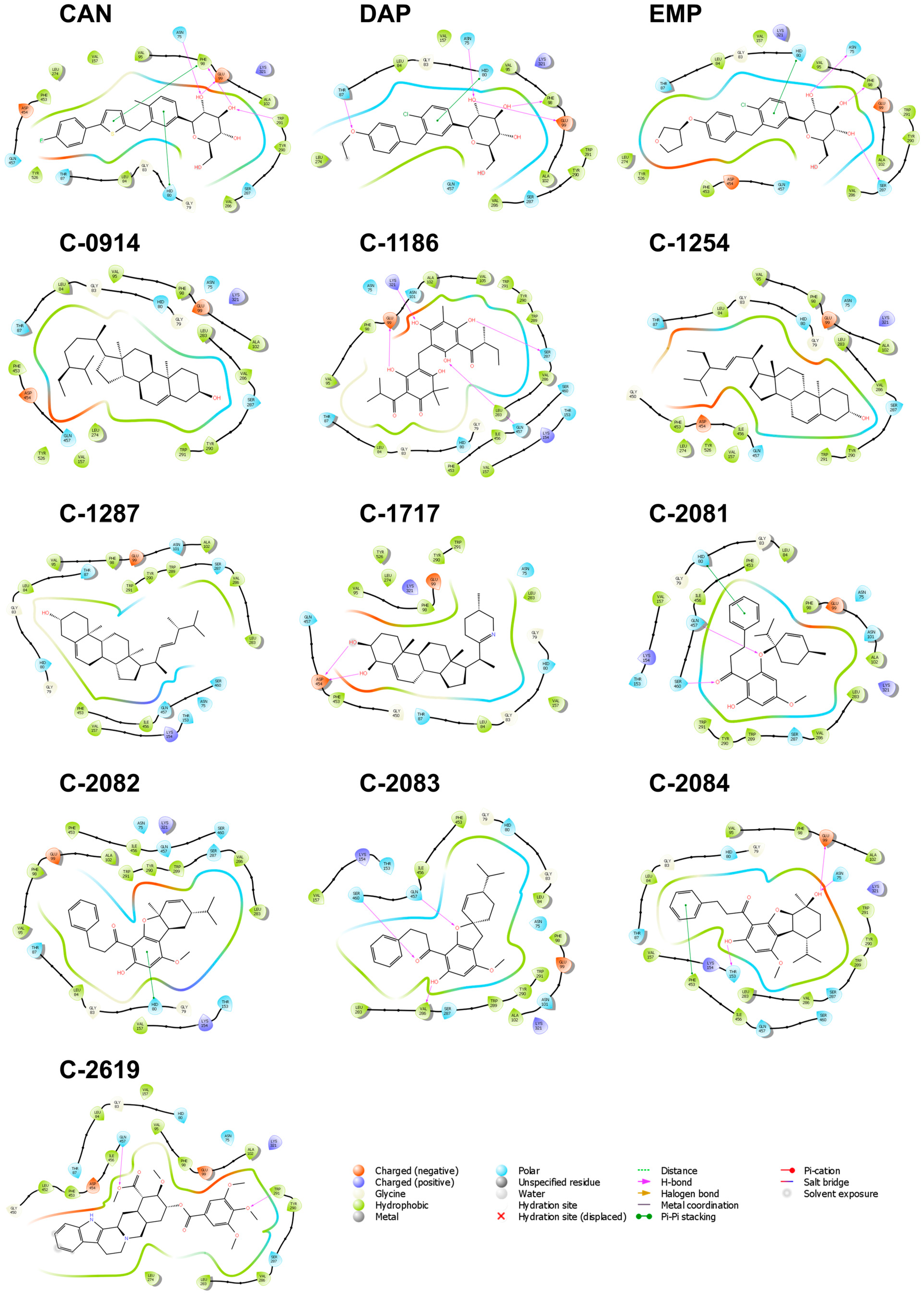
2.3.3. SGLT-2
2.3.4. FBPase
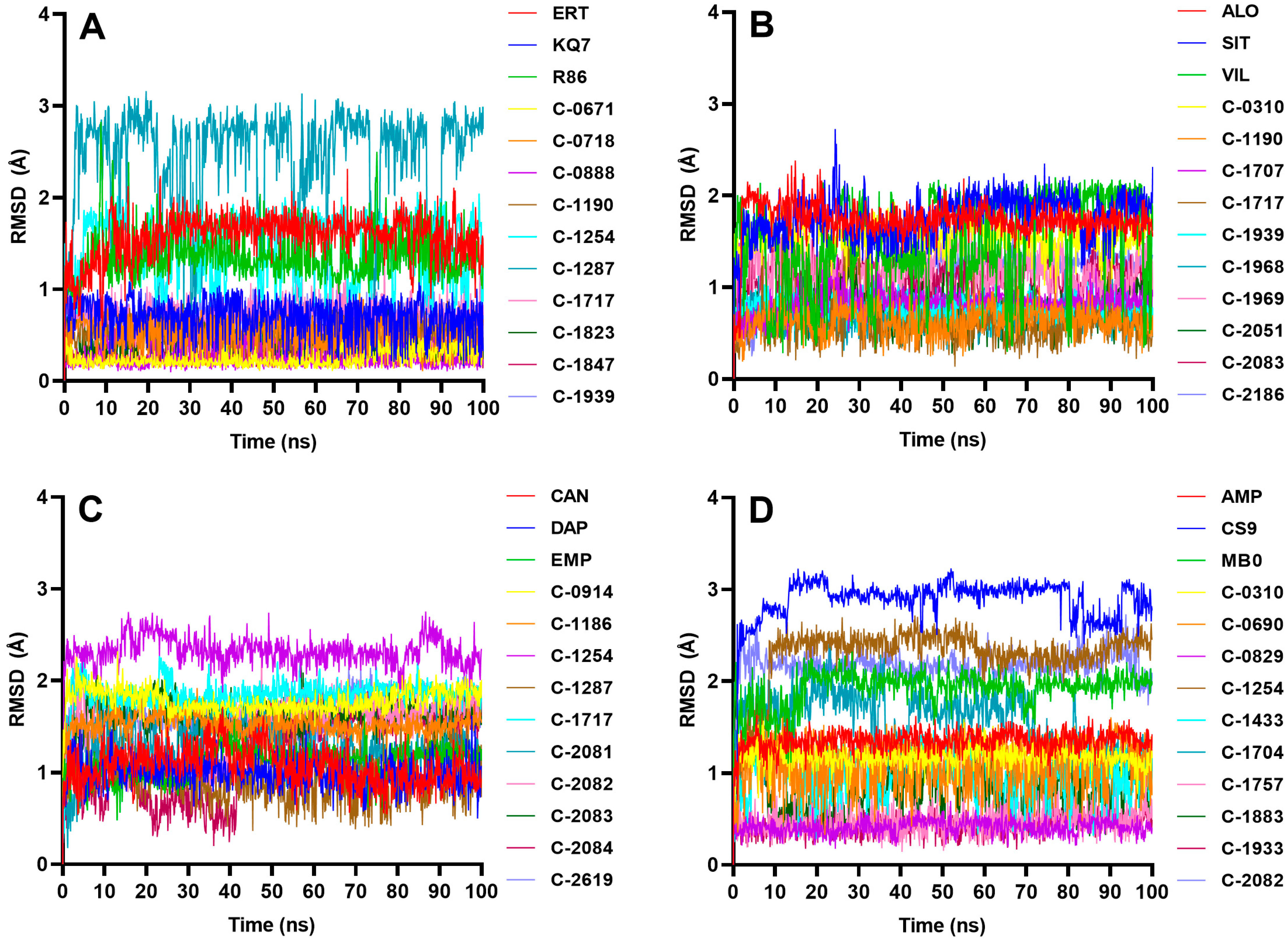
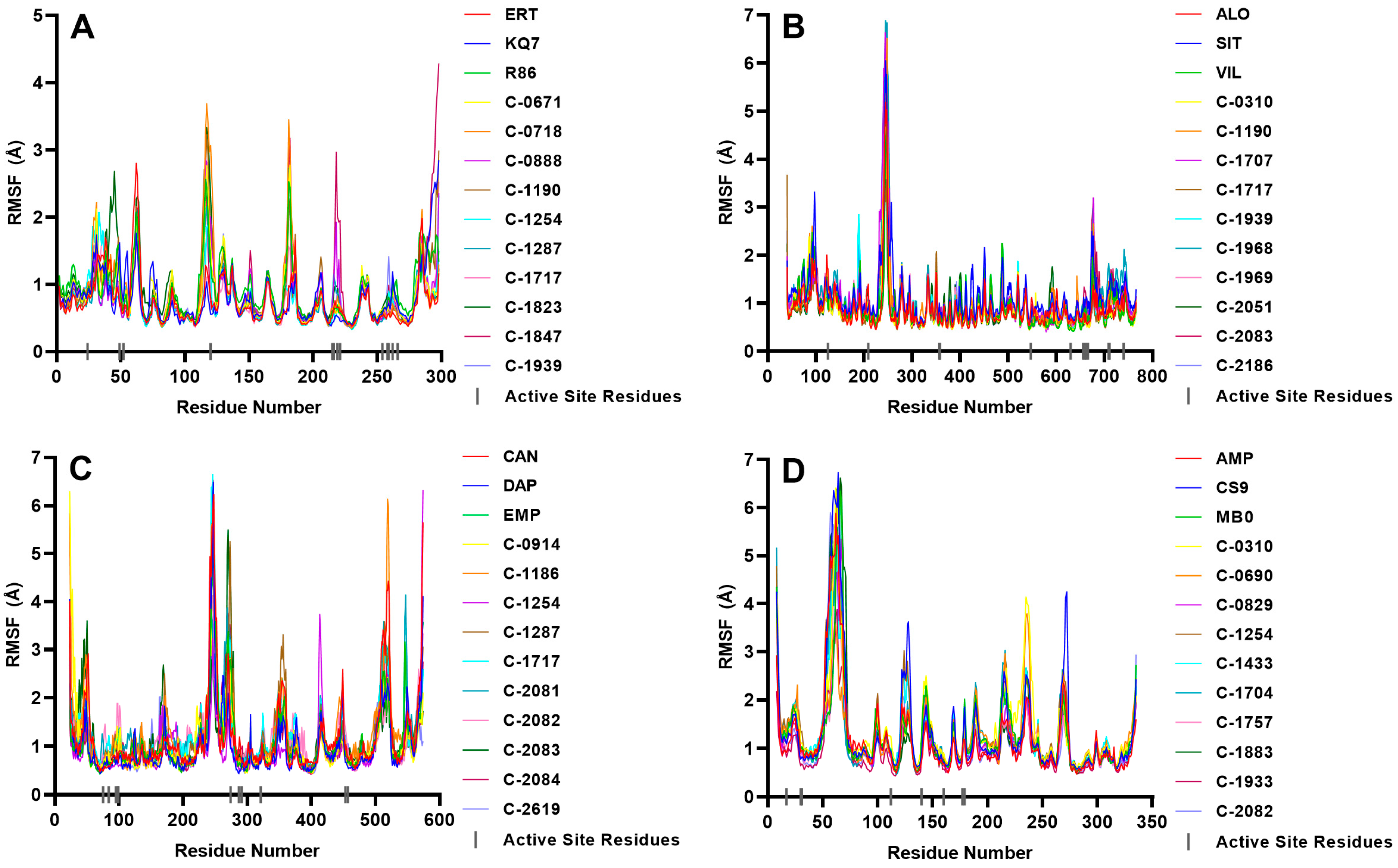
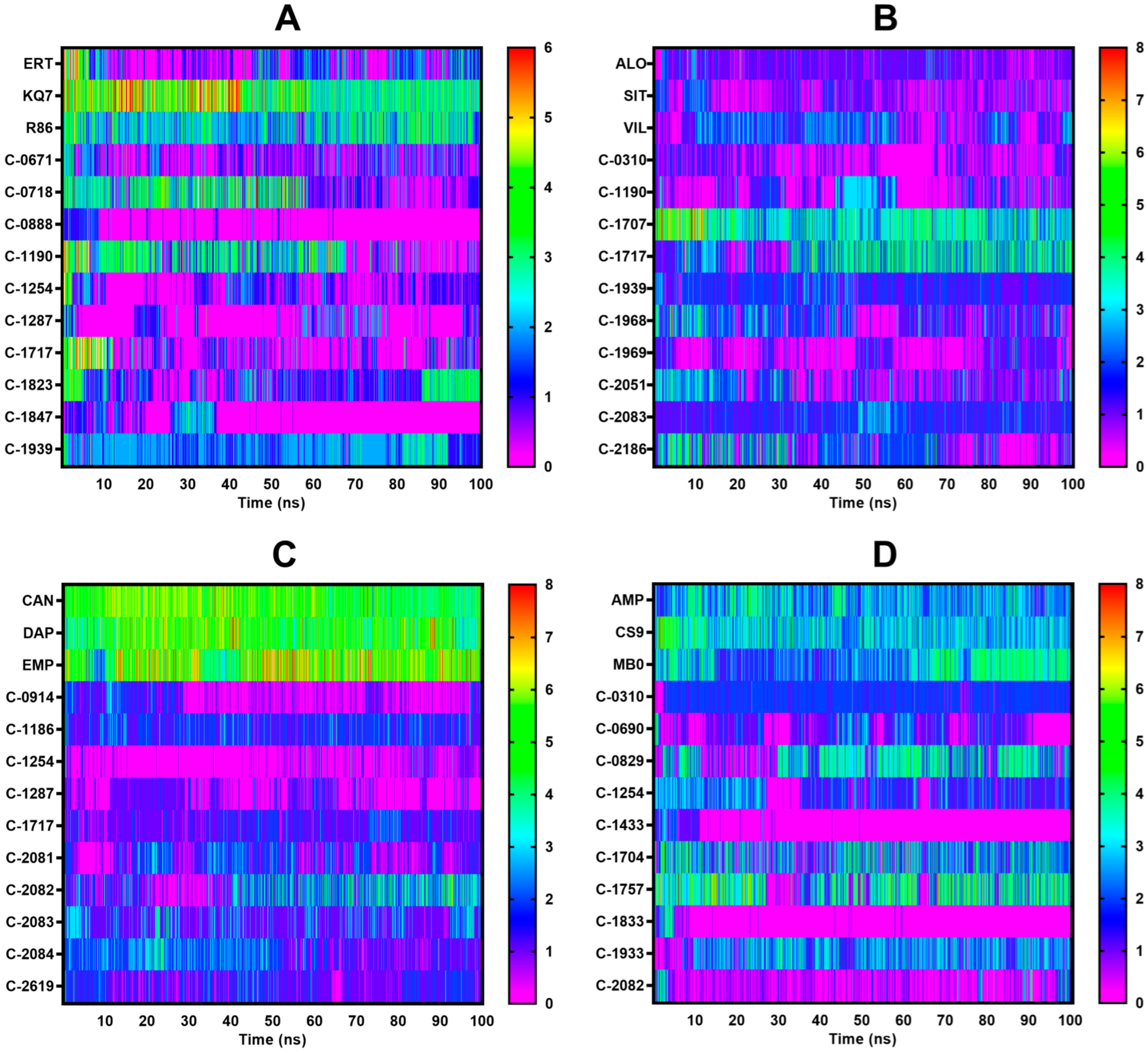
2.4. Molecular Dynamics Simulations
2.4.1. PTP1B
2.4.2. DPP-4
2.4.3. SGLT-2
2.4.4. FBPase
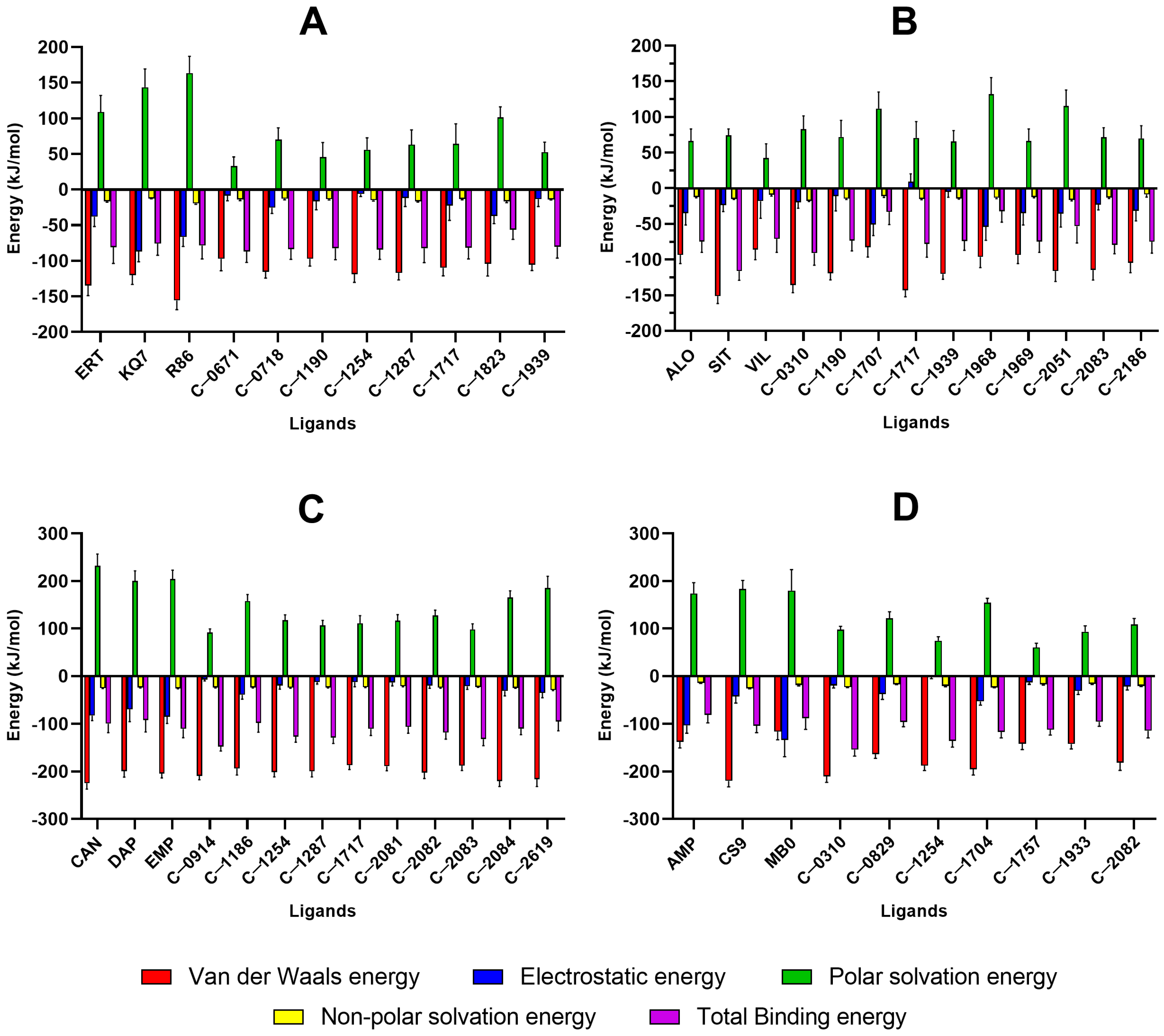
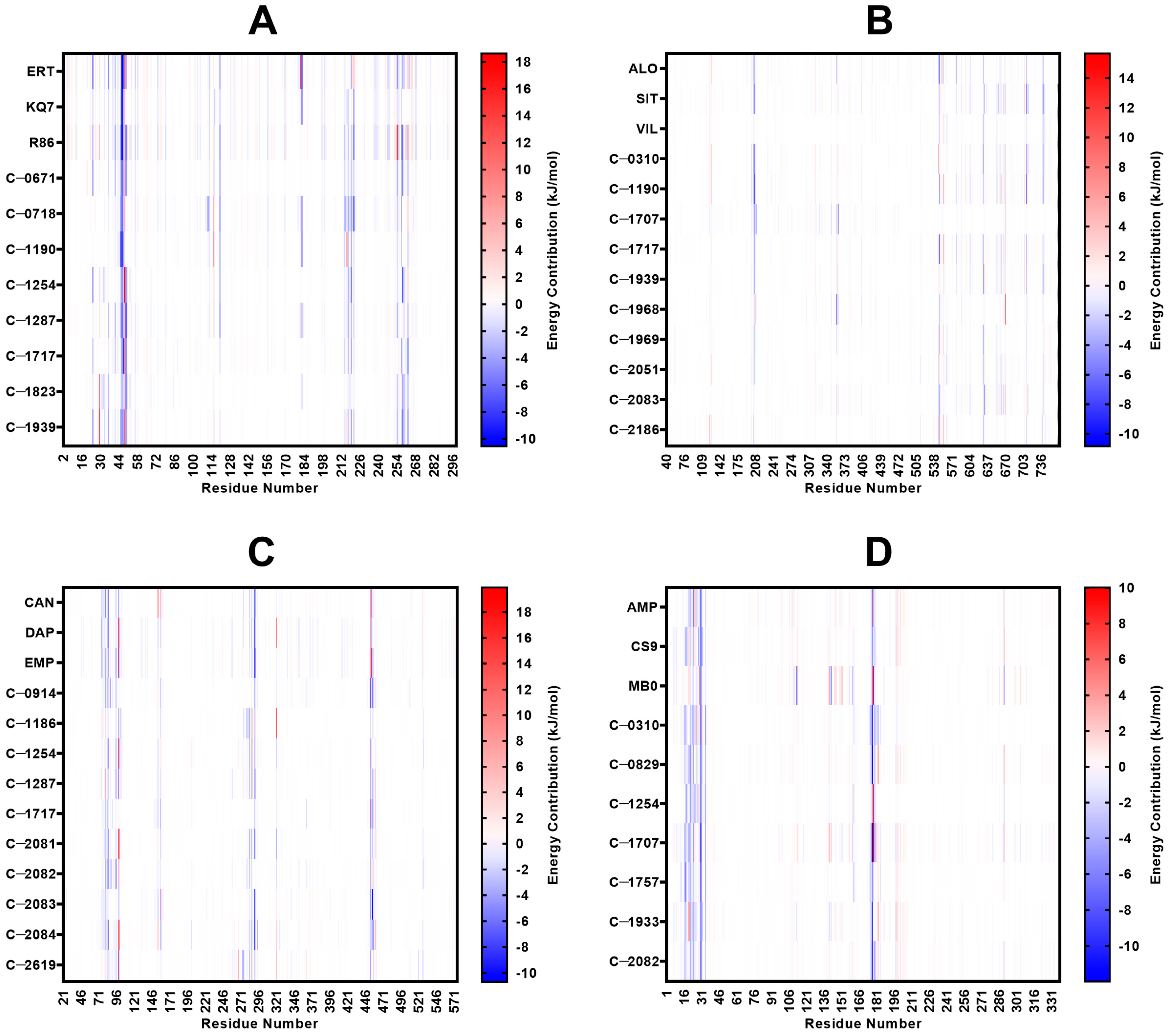
2.5. MM/PBSA Analysis
2.5.1. PTP1B
2.5.2. DPP-4
2.5.3. SGLT-2
2.5.4. FBPase
2.6. Identification of Top Inhibitor
2.7. Plant Identification
3. Materials and Methods
3.1. ADMET Profiling
3.2. Ligand Structure Optimization Using DFT
3.3. Protein Optimization
3.4. Molecular Docking
3.5. Molecular Dynamics (MD) Simulation
3.6. Molecular Mechanics/Poisson–Boltzmann Surface Area (MM/PBSA) Calculation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Forouhi, N.G.; Wareham, N.J. Epidemiology of Diabetes. Medicine 2019, 47, 22–27. [Google Scholar] [CrossRef]
- Walker, K.Z.; O’Dea, K.; Gomez, M.; Girgis, S.; Colagiuri, R. Diet and Exercise in the Prevention of Diabetes. J. Hum. Nutr. Diet. 2010, 23, 344–352. [Google Scholar] [CrossRef] [PubMed]
- Salim, B. Diabetes Mellitus and Its Treatment. Int. J. Diabetes Metab. 2005, 13, 111–134. [Google Scholar]
- Roglic, G. WHO Global Report on Diabetes: A Summary. Int. J. Noncommun. Dis. 2016, 1, 3. [Google Scholar] [CrossRef]
- Ogurtsova, K.; Guariguata, L.; Barengo, N.C.; Ruiz, P.L.-D.; Sacre, J.W.; Karuranga, S.; Sun, H.; Boyko, E.J.; Magliano, D.J. IDF Diabetes Atlas: Global Estimates of Undiagnosed Diabetes in Adults for 2021. Diabetes Res. Clin. Pract. 2022, 183, 109118. [Google Scholar] [CrossRef]
- Belete, T.M. A Recent Achievement In the Discovery and Development of Novel Targets for the Treatment of Type-2 Diabetes Mellitus. J. Exp. Pharmacol. 2020, 12, 1–15. [Google Scholar] [CrossRef]
- Krentz, A.J.; Patel, M.B.; Bailey, C.J. New Drugs for Type 2 Diabetes Mellitus. Drugs 2008, 68, 2131–2162. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, N.; Thakur, A.K.; Kumar, V.; Dey, A.; Kumar, V. Therapeutic Targets for Diabetes Mellitus: An Update. Clin. Pharm. Biopharm. 2014, 3, 1. [Google Scholar] [CrossRef]
- Zhang, Z.-Y.; Lee, S.-Y. PTP1B Inhibitors as Potential Therapeutics in the Treatment of Type 2 Diabetes and Obesity. Expert Opin. Investig. Drugs 2003, 12, 223–233. [Google Scholar] [CrossRef]
- Zhang, S.; Zhang, Z.-Y. PTP1B as a Drug Target: Recent Developments in PTP1B Inhibitor Discovery. Drug Discov. Today 2007, 12, 373–381. [Google Scholar] [CrossRef]
- Bongard, R.D.; Lepley, M.; Thakur, K.; Talipov, M.R.; Nayak, J.; Lipinski, R.A.J.; Bohl, C.; Sweeney, N.; Ramchandran, R.; Rathore, R.; et al. Serendipitous Discovery of Light-Induced (In Situ) Formation of an Azo-Bridged Dimeric Sulfonated Naphthol as a Potent PTP1B Inhibitor. BMC Biochem. 2017, 18, 10. [Google Scholar] [CrossRef] [PubMed]
- Kasina, S.V.S.K.; Baradhi, K.M. Dipeptidyl Peptidase IV (DPP IV) Inhibitors. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2021. [Google Scholar]
- Makrilakis, K. The Role of DPP-4 Inhibitors in the Treatment Algorithm of Type 2 Diabetes Mellitus: When to Select, What to Expect. Int. J. Environ. Res. Public Health 2019, 16, 2720. [Google Scholar] [CrossRef] [PubMed]
- Hsia, D.S.; Grove, O.; Cefalu, W.T. An Update on SGLT2 Inhibitors for the Treatment of Diabetes Mellitus. Curr. Opin. Endocrinol. Diabetes Obes. 2017, 24, 73. [Google Scholar]
- Kalra, S. Sodium Glucose Co-Transporter-2 (SGLT2) Inhibitors: A Review of Their Basic and Clinical Pharmacology. Diabetes Ther. 2014, 5, 355–366. [Google Scholar] [CrossRef] [PubMed]
- Proença, C.; Oliveira, A.; Freitas, M.; Ribeiro, D.; Sousa, J.L.C.; Ramos, M.J.; Silva, A.M.S.; Fernandes, P.A.; Fernandes, E. Structural Specificity of Flavonoids in the Inhibition of Human Fructose 1,6-Bisphosphatase. J. Nat. Prod. 2020, 83, 1541–1552. [Google Scholar] [CrossRef]
- Kaur, R.; Dahiya, L.; Kumar, M. Fructose-1,6-Bisphosphatase Inhibitors: A New Valid Approach for Management of Type 2 Diabetes Mellitus. Eur. J. Med. Chem. 2017, 141, 473–505. [Google Scholar] [CrossRef]
- Veeresham, C. Natural Products Derived from Plants as a Source of Drugs. J. Adv. Pharm. Technol. Res. 2012, 3, 200. [Google Scholar] [CrossRef]
- Wachtel-Galor, S.; Benzie, I.F.F. An Introduction to Its History, Usage, Regulation, Current Trends, and Research Needs in Herbal Medicine: Biomolecular and Clinical Aspects 2011. Available online: https://www.ncbi.nlm.nih.gov/books/NBK92773/ (accessed on 9 November 2021).
- Atanasov, A.G.; Zotchev, S.B.; Dirsch, V.M.; Supuran, C.T. Natural Products in Drug Discovery: Advances and Opportunities. Nat. Rev. Drug Discov. 2021, 20, 200–216. [Google Scholar] [CrossRef]
- Makhoba, X.H.; Viegas, C., Jr.; Mosa, R.A.; Viegas, F.P.; Pooe, O.J. Potential Impact of the Multi-Target Drug Approach in the Treatment of Some Complex Diseases. Drug Des. Dev. Ther. 2020, 14, 3235–3249. [Google Scholar] [CrossRef]
- Lankatillake, C.; Huynh, T.; Dias, D.A. Understanding Glycaemic Control and Current Approaches for Screening Antidiabetic Natural Products from Evidence-Based Medicinal Plants. Plant Methods 2019, 15, 105. [Google Scholar] [CrossRef]
- Jugran, A.K.; Rawat, S.; Devkota, H.P.; Bhatt, I.D.; Rawal, R.S. Diabetes and Plant-Derived Natural Products: From Ethnopharmacological Approaches to Their Potential for Modern Drug Discovery and Development. Phytother. Res. 2021, 35, 223–245. [Google Scholar] [CrossRef] [PubMed]
- Harvey, A.L.; Edrada-Ebel, R.; Quinn, R.J. The Re-Emergence of Natural Products for Drug Discovery in the Genomics Era. Nat. Rev. Drug Discov. 2015, 14, 111–129. [Google Scholar] [CrossRef] [PubMed]
- Maia, E.H.B.; Assis, L.C.; de Oliveira, T.A.; da Silva, A.M.; Taranto, A.G. Structure-Based Virtual Screening: From Classical to Artificial Intelligence. Front. Chem. 2020, 8, 343. [Google Scholar] [CrossRef]
- Bode, A.M.; Dong, Z. Toxic Phytochemicals and Their Potential Risks for Human Cancer. Cancer Prev. Res. 2015, 8, 1–8. [Google Scholar] [CrossRef]
- Jones, R.M. New Therapeutic Strategies for Type 2 Diabetes: Small Molecule Approaches; Royal Society of Chemistry: London, UK, 2012; ISBN 978-1-84973-414-1. [Google Scholar]
- Hähnke, V.D.; Kim, S.; Bolton, E.E. PubChem Chemical Structure Standardization. J. Cheminform. 2018, 10, 36. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Thiessen, P.A.; Bolton, E.E.; Chen, J.; Fu, G.; Gindulyte, A.; Han, L.; He, J.; He, S.; Shoemaker, B.A. PubChem Substance and Compound Databases. Nucleic Acids Res. 2016, 44, D1202–D1213. [Google Scholar] [CrossRef]
- Houston, D.R.; Walkinshaw, M.D. Consensus Docking: Improving the Reliability of Docking in a Virtual Screening Context. J. Chem. Inf. Model. 2013, 53, 384–390. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Liu, R.; Mathieu, C.; Berthelet, J.; Zhang, W.; Dupret, J.-M.; Lima, F.R. Human Protein Tyrosine Phosphatase 1B (PTP1B): From Structure to Clinical Inhibitor Perspectives. Int. J. Mol. Sci. 2022, 23, 7027. [Google Scholar] [CrossRef]
- Arulmozhiraja, S.; Matsuo, N.; Ishitsubo, E.; Okazaki, S.; Shimano, H.; Tokiwa, H. Comparative Binding Analysis of Dipeptidyl Peptidase IV (DPP-4) with Antidiabetic Drugs-An Ab Initio Fragment Molecular Orbital Study. PLoS ONE 2016, 11, e0166275. [Google Scholar] [CrossRef]
- Scapin, G. Structural Chemistry and Molecular Modeling in the Design of DPP4 Inhibitors. In Multifaceted Roles of Crystallography in Modern Drug Discovery; Springer: Berlin/Heidelberg, Germany, 2015; pp. 53–67. [Google Scholar]
- Abbott, C.A.; McCaughan, G.W.; Gorrell, M.D. Two Highly Conserved Glutamic Acid Residues in the Predicted Beta Propeller Domain of Dipeptidyl Peptidase IV Are Required for Its Enzyme Activity. FEBS Lett. 1999, 458, 278–284. [Google Scholar] [CrossRef]
- Bjelke, J.R.; Christensen, J.; Branner, S.; Wagtmann, N.; Olsen, C.; Kanstrup, A.B.; Rasmussen, H.B. Tyrosine 547 Constitutes an Essential Part of the Catalytic Mechanism of Dipeptidyl Peptidase IV. J. Biol. Chem. 2004, 279, 34691–34697. [Google Scholar] [CrossRef]
- Kirby, M.; Yu, D.; O’Connor, S.; Gorrell, M. Inhibitor Selectivity in Clinical Application of DPP-4 Inhibition. Clin. Sci. 2010, 118, 31–41. [Google Scholar] [CrossRef] [PubMed]
- Niu, Y.; Liu, R.; Guan, C.; Zhang, Y.; Chen, Z.; Hoerer, S.; Nar, H.; Chen, L. Structural Basis of Inhibition of the Human SGLT2–MAP17 Glucose Transporter. Nature 2022, 601, 280–284. [Google Scholar] [CrossRef]
- Mutyala, R.; Reddy, R.N.; Sumakanth, M.; Reddanna, P.; Reddy, M.R. Calculation of relative binding affinities of fructose 1,6-bisphosphatase mutants with adenosine monophosphate using free energy perturbation method. J. Comput. Chem. 2007, 28, 932–937. [Google Scholar] [CrossRef]
- Topaz, G.; Epiter-Smith, V.; Robalo, C.; Emad, M.; Ford, V.; Daley, J.; Byron, J.; Stieglitz, K.A. Characterization of Recombinant Fructose-1,6-Bisphosphatase Gene Mutations: Evidence of Inhibition/Activation of FBPase Protein by Gene Mutation. Biosci. Rep. 2019, 39, BSR20180960. [Google Scholar] [CrossRef]
- Zhang, X.; Jiang, H.; Li, W.; Wang, J.; Cheng, M. Computational Insight into Protein Tyrosine Phosphatase 1B Inhibition: A Case Study of the Combined Ligand- and Structure-Based Approach. Comput. Math. Methods Med. 2017, 2017, e4245613. [Google Scholar] [CrossRef]
- Anslyn, E.V.; Dougherty, D.A. Modern Physical Organic Chemistry; University Science Books: Melville, NY, USA, 2006. [Google Scholar]
- Mourad, A.A.E.; Khodir, A.E.; Saber, S.; Mourad, M.A.E. Novel Potent and Selective DPP-4 Inhibitors: Design, Synthesis and Molecular Docking Study of Dihydropyrimidine Phthalimide Hybrids. Pharmaceuticals 2021, 14, 144. [Google Scholar] [CrossRef] [PubMed]
- Barciszewski, J.; Wisniewski, J.; Kolodziejczyk, R.; Jaskolski, M.; Rakus, D.; Dzugaj, A. T-to-R Switch of Muscle Fructose-1,6-Bisphosphatase Involves Fundamental Changes of Secondary and Quaternary Structure. Acta Crystallogr. D Struct. Biol. 2016, 72, 536–550. [Google Scholar] [CrossRef] [PubMed]
- Mohanraj, K.; Karthikeyan, B.S.; Vivek-Ananth, R.P.; Chand, R.P.; Aparna, S.R.; Mangalapandi, P.; Samal, A. IMPPAT: A curated database of Indian Medicinal Plants, Phytochemistry And Therapeutics. Sci. Rep. 2018, 8, 4329. [Google Scholar] [CrossRef]
- Kirtikar, K.; Basu, B. Indian Medicinal Plants, 2nd ed.; M/S Bishen Singh Pal Singh: Delhi, India, 1975; pp. 1465–1472. [Google Scholar]
- Timalsina, D.; Devkota, H.P. Eclipta Prostrata (L.) L. (Asteraceae): Ethnomedicinal Uses, Chemical Constituents, and Biological Activities. Biomolecules 2021, 11, 1738. [Google Scholar] [CrossRef] [PubMed]
- Debnath, M.; Pandey, M.; Sharma, R.; Thankur, G.; Lal, P. Biotechnological Intervention of Agave Sisalana: A Unique Fibe Yielding Plant with Medicinal Property. J. Med. Plant Res. 2010, 4, 177–187. [Google Scholar]
- Gutiérrez, A.; Rodríguez, I.M.; del Río, J.C. Chemical Composition of Lipophilic Extractives from Sisal (Agave Sisalana) Fibers. Ind. Crops Prod. 2008, 28, 81–87. [Google Scholar] [CrossRef]
- Duke, J.; Bogenschutz, M.J. Dr. Duke’s Phytochemical and Ethnobotanical Databases; Agricultural Research Service, USDA: Washington, DC, USA, 1994.
- Moreira, D.D.L.; Guimarães, E.F.; Kaplan, M.A.C. A Chromene from Piper Aduncum. Phytochemistry 1998, 48, 1075–1077. [Google Scholar] [CrossRef]
- Orjala, J.; Wright, A.D.; Erdelmeier, C.A.; Sticher, O.; Rali, T. New Monoterpene-Substituted Dihydrochalcones from Piper Aduncum. Helv. Chim. Acta 1993, 76, 1481–1488. [Google Scholar] [CrossRef]
- Orjala, J.; Wright, A.D.; Behrends, H.; Folkers, G.; Sticher, O.; Rüegger, H.; Rali, T. Cytotoxic and Antibacterial Dihydrochalcones from Piper Aduncum. J. Nat. Prod. 1994, 57, 18–26. [Google Scholar] [CrossRef]
- Wang, N.; Zhao, G.; Zhang, Y.; Wang, X.; Zhao, L.; Xu, P.; Shou, D. A Network Pharmacology Approach to Determine the Active Components and Potential Targets of Curculigo Orchioides in the Treatment of Osteoporosis. Med. Sci. Monit. 2017, 23, 5113–5122. [Google Scholar] [CrossRef][Green Version]
- Akinwumi, K.A.; Eleyowo, O.O.; Oladipo, O.O.; Akinwumi, K.A.; Eleyowo, O.O.; Oladipo, O.O. A Review on the Ethnobotanical Uses, Phytochemistry and Pharmacological Effect of Luffa Cylindrinca; IntechOpen: London, UK, 2021; ISBN 978-1-80356-021-2. [Google Scholar]
- Maamoun, A.A.; El-akkad, R.H.; Farag, M.A. Mapping Metabolome Changes in Luffa Aegyptiaca Mill Fruits at Different Maturation Stages via MS-Based Metabolomics and Chemometrics. J. Adv. Res. 2021, 29, 179–189. [Google Scholar] [CrossRef] [PubMed]
- Adewuyi, A.; Oderinde, R.A. Analysis of the Lipids and Molecular Speciation of the Triacylglycerol of the Oils of Luffa Cylindrica and Adenopus Breviflorus. CyTA-J. Food 2012, 10, 313–320. [Google Scholar] [CrossRef]
- Rastogi, R.P.; Mehrotra, B.N. Compendium of Indian Medicinal Plants; Central Drug Research Institute: Lucknow, India, 1990. [Google Scholar]
- Leone, A.; Spada, A.; Battezzati, A.; Schiraldi, A.; Aristil, J.; Bertoli, S. Moringa Oleifera Seeds and Oil: Characteristics and Uses for Human Health. Int. J. Mol. Sci. 2016, 17, 2141. [Google Scholar] [CrossRef] [PubMed]
- Özcan, M.M. Moringa Spp: Composition and Bioactive Properties. S. Afr. J. Bot. 2020, 129, 25–31. [Google Scholar] [CrossRef]
- Piironen, V.; Toivo, J.; Puupponen-Pimiä, R.; Lampi, A.-M. Plant Sterols in Vegetables, Fruits and Berries. J. Sci. Food Agric. 2003, 83, 330–337. [Google Scholar] [CrossRef]
- Fernández-Cuesta, A.; Velasco, L.; Fernández-Martínez, J.M. Phytosterols in the Seeds of Wild Sunflower Species/Fitoesteroles En Las Semillas de Especies Silvestres de Girasol. Helia 2011, 34, 31–38. [Google Scholar] [CrossRef]
- Jaiswal, N.; Bhatia, V.; Srivastava, S.P.; Srivastava, A.K.; Tamrakar, A.K. Antidiabetic Effect of Eclipta Alba Associated with the Inhibition of Alpha-Glucosidase and Aldose Reductase. Nat. Prod. Res. 2012, 26, 2363–2367. [Google Scholar] [CrossRef] [PubMed]
- Ananthi, J.; Prakasam, A.; Pugalendi, K.V. Antihyperglycemic Activity of Eclipta Alba Leaf on Alloxan-Induced Diabetic Rats. Yale J. Biol. Med. 2003, 76, 97–102. [Google Scholar] [PubMed]
- Chege, B.M.; Nyaga, N.M.; Kaur, P.S.; Misigo, W.O.; Khan, N.; Wanyonyi, W.C.; Mwangi, P.W. The Significant Antidyslipidemic, Hypoglycemic, Antihyperglycemic, and Antiobesity Activities of the Aqueous Extracts of Agave Sisalana Juice Are Partly Mediated via Modulation of Calcium Signaling Pathways. Heliyon 2023, 9, e12400. [Google Scholar] [CrossRef]
- Marques, A.M.; Pereira, S.L.; Paiva, R.A.; Cavalcante, C.V.; Sudo, S.Z.; Tinoco, L.W.; Moreira, D.L.; Guimaraes, E.F.; Sudo, R.T.; Kaplan, M.A.C. Hypoglycemic Effect of the Methanol Flower Extract of Piper Claussenianum and the Major Constituent 2′, 6′-Dihydroxy-4′-Methoxychalcone in Streptozotocin Diabetic Rats. Indian J. Pharm. Sci. 2015, 77, 237. [Google Scholar]
- Madhavan, V.; Joshi, R.; Murali, A.; Yoganarasimhan, S.N. Antidiabetic Activity of Curculigo Orchioides. Root Tuber. Pharm. Biol. 2007, 45, 18–21. [Google Scholar] [CrossRef]
- Azmi, M.B.; Qureshi, S.A. Methanolic Root Extract of Rauwolfia Serpentina Benth Improves the Glycemic, Antiatherogenic, and Cardioprotective Indices in Alloxan-Induced Diabetic Mice. Adv. Pharmacol. Pharm. Sci. 2012, 2012, e376429. [Google Scholar] [CrossRef]
- Hazra, M.; KunduSen, S.; Bhattacharya, S.; Haldar, P.K.; Gupta, M.; Mazumder, U.K. Evaluation of Hypoglycemic and Antihyperglycemic Effects of Luffa Cylindrica Fruit Extract in Rats. J. Adv. Pharm. Educ. Res 2011, 2, 138–146. [Google Scholar]
- Babu, S.; Jayaraman, S. An Update on β-Sitosterol: A Potential Herbal Nutraceutical for Diabetic Management. Biomed. Pharmacother. 2020, 131, 110702. [Google Scholar] [CrossRef]
- Baker, W.L.; Baker, E.L.; Coleman, C.I. The Effect of Plant Sterols or Stanols on Lipid Parameters in Patients with Type 2 Diabetes: A Meta-Analysis. Diabetes Res. Clin. Pract. 2009, 84, e33–e37. [Google Scholar] [CrossRef] [PubMed]
- Jayaraman, S.; Roy, A.; Vengadassalapathy, S.; Sekar, R.; Veeraraghavan, V.P.; Rajagopal, P.; Rengasamy, G.; Mukherjee, R.; Sekar, D.; Manjunathan, R. An Overview on the Therapeutic Function of Foods Enriched with Plant Sterols in Diabetes Management. Antioxidants 2021, 10, 1903. [Google Scholar] [CrossRef]
- Wilson, D.P.; Wan, Z.-K.; Xu, W.-X.; Kirincich, S.J.; Follows, B.C.; Joseph-McCarthy, D.; Foreman, K.; Moretto, A.; Wu, J.; Zhu, M.; et al. Structure-Based Optimization of Protein Tyrosine Phosphatase 1B Inhibitors: From the Active Site to the Second Phosphotyrosine Binding Site. J. Med. Chem. 2007, 50, 4681–4698. [Google Scholar] [CrossRef]
- Xiong, G.; Wu, Z.; Yi, J.; Fu, L.; Yang, Z.; Hsieh, C.; Yin, M.; Zeng, X.; Wu, C.; Lu, A. ADMETlab 2.0: An Integrated Online Platform for Accurate and Comprehensive Predictions of ADMET Properties. Nucleic Acids Res. 2021, 49, W5–W14. [Google Scholar] [CrossRef]
- Yan, A.; Wang, Z.; Cai, Z. Prediction of Human Intestinal Absorption by GA Feature Selection and Support Vector Machine Regression. Int. J. Mol. Sci. 2008, 9, 1961–1976. [Google Scholar] [CrossRef]
- Giannozzi, P.; Baroni, S.; Bonini, N.; Calandra, M.; Car, R.; Cavazzoni, C.; Ceresoli, D.; Chiarotti, G.L.; Cococcioni, M.; Dabo, I. QUANTUM ESPRESSO: A Modular and Open-Source Software Project for Quantum Simulations of Materials. J. Phys. Condens. Matter 2009, 21, 395502. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M.; Battistuz, T.; Bhat, T.N.; Bluhm, W.F.; Bourne, P.E.; Burkhardt, K.; Feng, Z.; Gilliland, G.L.; Iype, L.; Jain, S. The Protein Data Bank. Acta Crystallogr. Sect. D Biol. Crystallogr. 2002, 58, 899–907. [Google Scholar] [CrossRef] [PubMed]
- Schrödinger Release 2023-2: Maestro; Schrödinger, LLC: New York, NY, USA, 2023.
- Eswar, N.; Eramian, D.; Webb, B.; Shen, M.-Y.; Sali, A. Protein Structure Modeling with MODELLER. Struct. Proteom. High-Throughput Methods 2008, 426, 145–159. [Google Scholar]
- Deshpande, T.A.; Isshak, M.; Priefer, R. PTP1B Inhibitors as Potential Target for Type II Diabetes. Curr. Res. Diabetes Obes. J. 2020, 14, 555876. [Google Scholar] [CrossRef]
- Singh, U.C.; Kollman, P.A. An Approach to Computing Electrostatic Charges for Molecules. J. Comput. Chem. 1984, 5, 129–145. [Google Scholar] [CrossRef]
- Quiroga, R.; Villarreal, M.A. Vinardo: A Scoring Function Based on Autodock Vina Improves Scoring, Docking, and Virtual Screening. PLoS ONE 2016, 11, e0155183. [Google Scholar] [CrossRef]
- Berendsen, H.J.; van der Spoel, D.; van Drunen, R. GROMACS: A Message-Passing Parallel Molecular Dynamics Implementation. Comput. Phys. Commun. 1995, 91, 43–56. [Google Scholar] [CrossRef]
- Brooks, B.R.; Bruccoleri, R.E.; Olafson, B.D.; States, D.J.; Swaminathan, S.A.; Karplus, M. CHARMM: A Program for Macromolecular Energy, Minimization, and Dynamics Calculations. J. Comput. Chem. 1983, 4, 187–217. [Google Scholar] [CrossRef]
- Vanommeslaeghe, K.; Hatcher, E.; Acharya, C.; Kundu, S.; Zhong, S.; Shim, J.; Darian, E.; Guvench, O.; Lopes, P.; Vorobyov, I. CHARMM General Force Field: A Force Field for Drug-like Molecules Compatible with the CHARMM All-Atom Additive Biological Force Fields. J. Comput. Chem. 2010, 31, 671–690. [Google Scholar] [CrossRef]
- Kumari, R.; Kumar, R.; Consortium, O.S.D.D.; Lynn, A. G_mmpbsa A GROMACS Tool for High-Throughput MM-PBSA Calculations. J. Chem. Inf. Model. 2014, 54, 1951–1962. [Google Scholar] [CrossRef]
- Case, D.A.; Aktulga, H.M.; Belfon, K.; Ben-Shalom, I.; Brozell, S.R.; Cerutti, D.S.; Cheatham, T.E., III; Cruzeiro, V.W.D.; Darden, T.A.; Duke, R.E. Amber 2021; University of California: San Francisco, CA, USA, 2021. [Google Scholar]
- Genheden, S.; Ryde, U. The MM/PBSA and MM/GBSA Methods to Estimate Ligand-Binding Affinities. Expert Opin. Drug Discov. 2015, 10, 449–461. [Google Scholar] [CrossRef] [PubMed]





| ADMET Property | Passed | Failed |
|---|---|---|
| Human Intestinal Absorption | 1994 | 663 |
| Human Oral Bioavailability | 984 | 1673 |
| Carcinogenicity | 1962 | 695 |
| Hepatoxicity | 1641 | 1016 |
| Acute Toxicity Rule | 2648 | 9 |
| Lipinski’s Rule of Five | 2187 | 470 |
| All six parameters | 373 | 2284 |
| PTP1B | DPP-4 | SGLT-2 | FBPase | |||||
|---|---|---|---|---|---|---|---|---|
| Rank | Compound | Consensus Docking Score (kJ/mol) | Compound | Consensus Docking Score (kJ/mol) | Compound | Consensus Docking Score (kJ/mol) | Compound | Consensus Docking Score (kJ/mol) |
| ERT * | −79.08 | ALO | −80.79 | CAN | −96.06 | CS9 * | −60.46 | |
| KQ7 * | −75.77 | SIT | −75.44 | DAP | −87.11 | MB0 * | −58.87 | |
| R86 * | −96.61 | VIL | −70.33 | EMP | −92.17 | AMP * | −59.96 | |
| 1 | C-1823 | −81.96 | C-1939 | −83.85 | C-1254 | −94.43 | C-0829 | −67.53 |
| 2 | C-0671 | −81.50 | C-1968 | −82.30 | C-2084 | −94.22 | C-1757 | −66.23 |
| 3 | C-0718 | −79.08 | C-2186 | −81.88 | C-2083 | −91.46 | C-1433 | −64.35 |
| 4 | C-1254 | −77.49 | C-1190 | −80.42 | C-1186 | −91.21 | C-1254 | −64.27 |
| 5 | C-1847 | −77.40 | C-0310 | −78.91 | C-1287 | −89.24 | C-2082 | −64.14 |
| 6 | C-0888 | −77.24 | C-1717 | −78.70 | C-0914 | −89.16 | C-0310 | −63.93 |
| 7 | C-1287 | −76.02 | C-1707 | −78.49 | C-2082 | −88.37 | C-1704 | −63.51 |
| 8 | C-1190 | −75.56 | C-2083 | −76.27 | C-2619 | −87.82 | C-0690 | −63.47 |
| 9 | C-1939 | −74.77 | C-2051 | −76.23 | C-2081 | −87.70 | C-1933 | −63.47 |
| 10 | C-1717 | −74.39 | C-1969 | −75.94 | C-1717 | −87.40 | C-1883 | −62.97 |
| PTP1B | DPP-4 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Compound | Average Complex–RMSD | Stability Rank | Binding Energy (kJ/mol) | Binding Affinity Rank | MD Consensus Score | Overall Rank | Compound | Average Complex–RMSD | Stability Rank | Binding Energy (kJ/mol) | Binding Affinity Rank | MD Consensus Score | Overall Rank |
| C-0671 | 2.230 | 1 | −19.279 | 1 | −17.049 | 1 | C-2083 | 2.508 | 4 | −22.289 | 1 | −19.781 | 1 |
| C-1287 | 2.319 | 5 | −19.163 | 2 | −16.843 | 2 | C-1969 | 2.395 | 2 | −19.235 | 2 | −16.841 | 2 |
| C-0718 | 2.304 | 3 | −18.641 | 4 | −16.337 | 3 | C-0310 | 2.132 | 1 | −18.867 | 3 | −16.735 | 3 |
| C-1939 | 2.371 | 6 | −18.666 | 3 | −16.295 | 4 | C-2186 | 2.711 | 6 | −17.731 | 4 | −15.020 | 4 |
| C-1254 | 2.275 | 2 | −18.533 | 5 | −16.258 | 5 | C-1717 | 2.464 | 3 | −17.185 | 7 | −14.722 | 5 |
| C-1717 | 2.315 | 4 | −18.229 | 6 | −15.915 | 6 | C-1939 | 2.999 | 7 | −17.551 | 5 | −14.552 | 6 |
| C-1190 | 2.638 | 7 | −17.872 | 7 | −15.234 | 7 | C-1190 | 2.685 | 5 | −17.204 | 6 | −14.519 | 7 |
| SGLT-2 | FBPase | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Compound | Average Complex–RMSD | Stability Rank | Binding Energy (kJ/mol) | Binding Affinity Rank | MD Consensus Score | Overall Rank | Compound | Average Complex–RMSD | Stability Rank | Binding Energy (kJ/mol) | Binding Affinity Rank | MD Consensus Score | Overall Rank |
| C-0914 | 3.113 | 8 | −34.815 | 1 | −31.702 | 1 | C-1254 | 2.415 | 6 | −31.798 | 1 | −29.384 | 1 |
| C-2083 | 2.977 | 2 | −30.268 | 2 | −27.291 | 2 | C-0310 | 2.499 | 7 | −28.676 | 2 | −26.177 | 2 |
| C-1287 | 3.008 | 4 | −29.796 | 3 | −26.788 | 3 | C-1757 | 2.262 | 2 | −25.427 | 3 | −23.165 | 3 |
| C-2082 | 3.215 | 10 | −27.630 | 4 | −24.415 | 4 | C-2082 | 2.401 | 4 | −25.289 | 4 | −22.888 | 4 |
| C-1254 | 3.064 | 6 | −27.183 | 5 | −24.119 | 5 | C-1704 | 2.410 | 5 | −22.591 | 5 | −20.181 | 5 |
| C-2081 | 3.050 | 5 | −25.333 | 6 | −22.282 | 6 | C-0829 | 2.236 | 1 | −22.380 | 6 | −20.144 | 6 |
| C-1717 | 3.070 | 7 | −24.787 | 7 | −21.717 | 7 | C-1933 | 2.339 | 3 | −17.946 | 7 | −15.607 | 7 |
| C-1186 | 2.981 | 3 | −24.065 | 8 | −21.084 | 8 | |||||||
| C-2084 | 3.118 | 9 | −23.333 | 9 | −20.215 | 9 | |||||||
| C-2619 | 2.920 | 1 | −22.378 | 10 | −19.458 | 10 | |||||||
| PTP1B | DPP-4 | SGLT-2 | FBPase | ||||
|---|---|---|---|---|---|---|---|
| C-0671 | Gypsogenin | C-0310 | Campesterol | C-0914 | Sitosterol | C-0310 | Campesterol |
| C-0718 | Quillaic acid | C-1190 | Yuccagenin | C-1186 | Saroaspidin B | C-0829 | Actinodaphnine |
| C-1190 | Yuccagenin | C-1717 | 4beta-Hydroxyverazine | C-1254 | Stigmasterol | C-1254 | Stigmasterol |
| C-1254 | Stigmasterol | C-1939 | 9-Dehydrohecogenin | C-1287 | Brassicasterol | C-1704 | Piperaduncin A |
| C-1287 | Brassicasterol | C-1969 | Veramiline | C-1717 | 4beta-Hydroxyverazine | C-1757 | Copalic Acid |
| C-1717 | 4beta-Hydroxyverazine | C-2083 | Adunctin C | C-2081 | Adunctin A | C-1933 | Norisoboldine |
| C-1939 | 9-Dehydrohecogenin | C-2186 | Hongguanggenin | C-2082 | Adunctin B | C-2082 | Adunctin B |
| C-2083 | Adunctin C | ||||||
| C-2084 | Adunctin E | ||||||
| C-2619 | Deserpidine | ||||||
| Plant | Part | Protein Target | Phytochemical | Reference |
|---|---|---|---|---|
| Eclipta prostata | Leaves | PTP1B | 4beta-Hydroxyverazine | [47] |
| Stigmasterol | [48] | |||
| DPP-4 | 4beta-Hydroxyverazine | [47] | ||
| Veramiline | [47] | |||
| SGLT-2 | 4beta-Hydroxyverazine | [47] | ||
| Stigmasterol | [48] | |||
| Sitosterol | [48] | |||
| FBPase | Stigmasterol | [48] | ||
| Agave sisalana | Leaves | PTP1B | 9-Dehydrohecogenin | [49] |
| Stigmasterol | [50] | |||
| DPP-4 | 9-Dehydrohecogenin | [49] | ||
| Hongguanggenin | [51] | |||
| SGLT-2 | Stigmasterol | [50] | ||
| Sitosterol | [50] | |||
| FBPase | Stigmasterol | [50] | ||
| Campesterol | [50] | |||
| Piper aduncum | Leaves | PTP1B | Stigmasterol | [52] |
| DPP-4 | Adunctin C | [53] | ||
| SGLT-2 | Adunctin A | [53] | ||
| Adunctin B | [53] | |||
| Adunctin C | [53] | |||
| Adunctin E | [53] | |||
| Stigmasterol | [52] | |||
| FBPase | Adunctin B | [53] | ||
| Stigmasterol | [52] | |||
| Piperaduncin A | [54] | |||
| Curculigo orchioides | Rhizomes | PTP1B | Yuccagenin | [55] |
| Stigmasterol | [55] | |||
| DPP-4 | Yuccagenin | [55] | ||
| SGLT-2 | Stigmasterol | [55] | ||
| Sitosterol | [55] | |||
| FBPase | Stigmasterol | [55] | ||
| Luffa cylindrica | Seeds | PTP1B | Gypsogenin | [56] |
| Quillaic Acid | [57] | |||
| Stigmasterol | [58] | |||
| DPP-4 | Campesterol | [59] | ||
| Luffa cylindrica | Seeds | SGLT-2 | Stigmasterol | [58] |
| Sitosterol | [58] | |||
| FBPase | Stigmasterol | [58] | ||
| Campesterol | [59] | |||
| Moringa oleifera | Seeds | PTP1B | Brassicasterol | [60,61] |
| Stigmasterol | [60,61] | |||
| DPP-4 | Campesterol | [60,61] | ||
| SGLT-2 | Brassicasterol | [60,61] | ||
| Stigmasterol | [60,61] | |||
| FBPase | Stigmasterol | [60,61] | ||
| Campesterol | [60,61] | |||
| Alium cepa | Bulb | PTP1B | Brassicasterol | [62] |
| Stigmasterol | [62] | |||
| DPP-4 | Campesterol | [62] | ||
| SGLT-2 | Brassicasterol | [62] | ||
| Stigmasterol | [62] | |||
| Sitosterol | [62] | |||
| FBPase | Stigmasterol | [62] | ||
| Campesterol | [62] | |||
| Stigmasterol | [62] | |||
| Helianthus annuus | Seeds | PTP1B | Stigmasterol | [63] |
| DPP-4 | Campesterol | [63] | ||
| SGLT-2 | Stigmasterol | [63] | ||
| Sitosterol | [63] | |||
| FBPase | Stigmasterol | [63] | ||
| Campesterol | [63] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Macalalad, M.A.B.; Gonzales, A.A., III. In Silico Screening and Identification of Antidiabetic Inhibitors Sourced from Phytochemicals of Philippine Plants against Four Protein Targets of Diabetes (PTP1B, DPP-4, SGLT-2, and FBPase). Molecules 2023, 28, 5301. https://doi.org/10.3390/molecules28145301
Macalalad MAB, Gonzales AA III. In Silico Screening and Identification of Antidiabetic Inhibitors Sourced from Phytochemicals of Philippine Plants against Four Protein Targets of Diabetes (PTP1B, DPP-4, SGLT-2, and FBPase). Molecules. 2023; 28(14):5301. https://doi.org/10.3390/molecules28145301
Chicago/Turabian StyleMacalalad, Mark Andrian B., and Arthur A. Gonzales, III. 2023. "In Silico Screening and Identification of Antidiabetic Inhibitors Sourced from Phytochemicals of Philippine Plants against Four Protein Targets of Diabetes (PTP1B, DPP-4, SGLT-2, and FBPase)" Molecules 28, no. 14: 5301. https://doi.org/10.3390/molecules28145301
APA StyleMacalalad, M. A. B., & Gonzales, A. A., III. (2023). In Silico Screening and Identification of Antidiabetic Inhibitors Sourced from Phytochemicals of Philippine Plants against Four Protein Targets of Diabetes (PTP1B, DPP-4, SGLT-2, and FBPase). Molecules, 28(14), 5301. https://doi.org/10.3390/molecules28145301







