Identification of Gentian-Related Species Based on Two-Dimensional Correlation Spectroscopy (2D-COS) Combined with Residual Neural Network (ResNet)
Abstract
1. Introduction
2. Results
2.1. Data Pre-Processing
2.2. Results of Feature Variable Selection
2.3. Two-Dimensional Correlation Spectroscopy (2D-COS) Image Acquisition
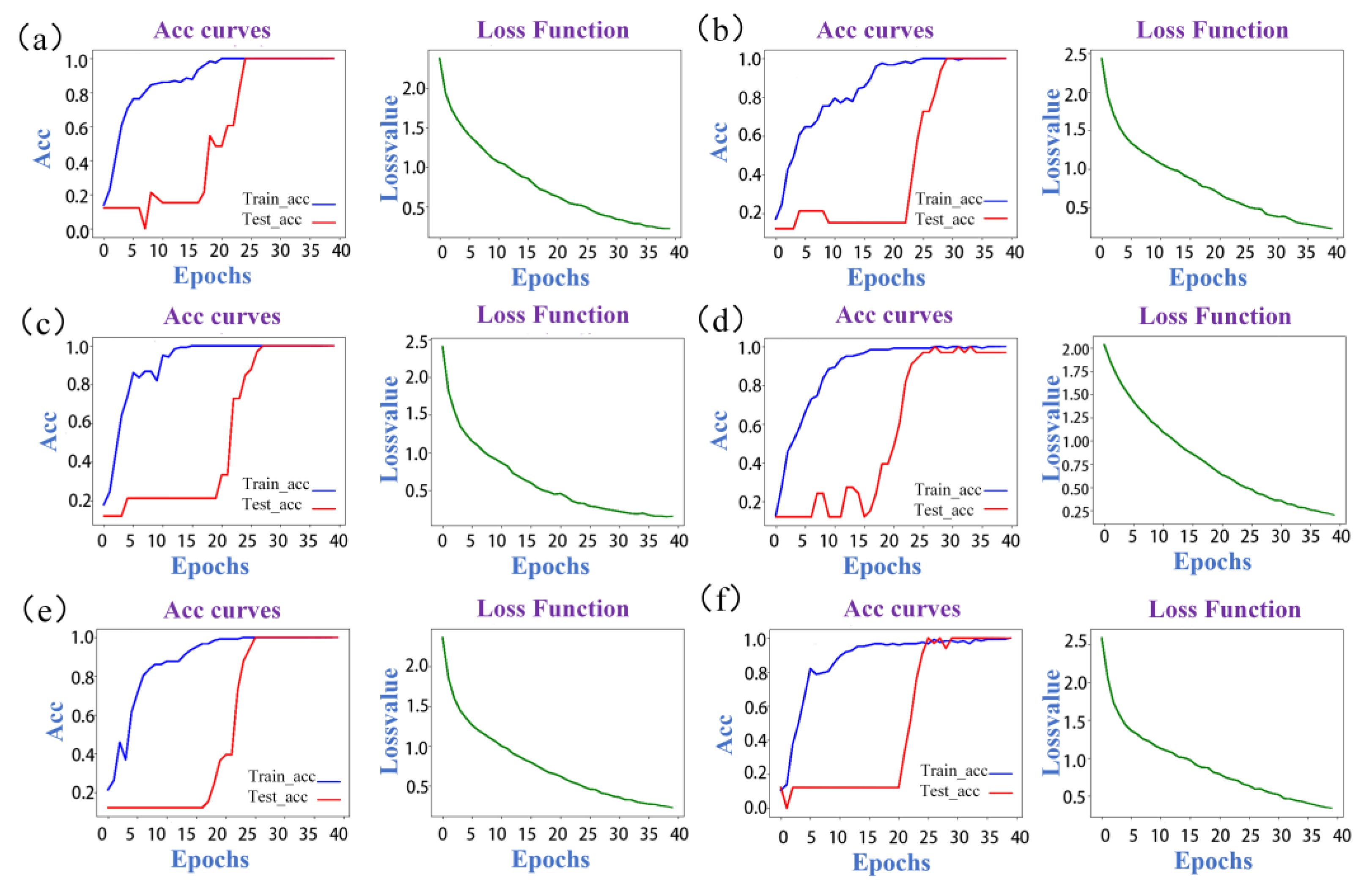
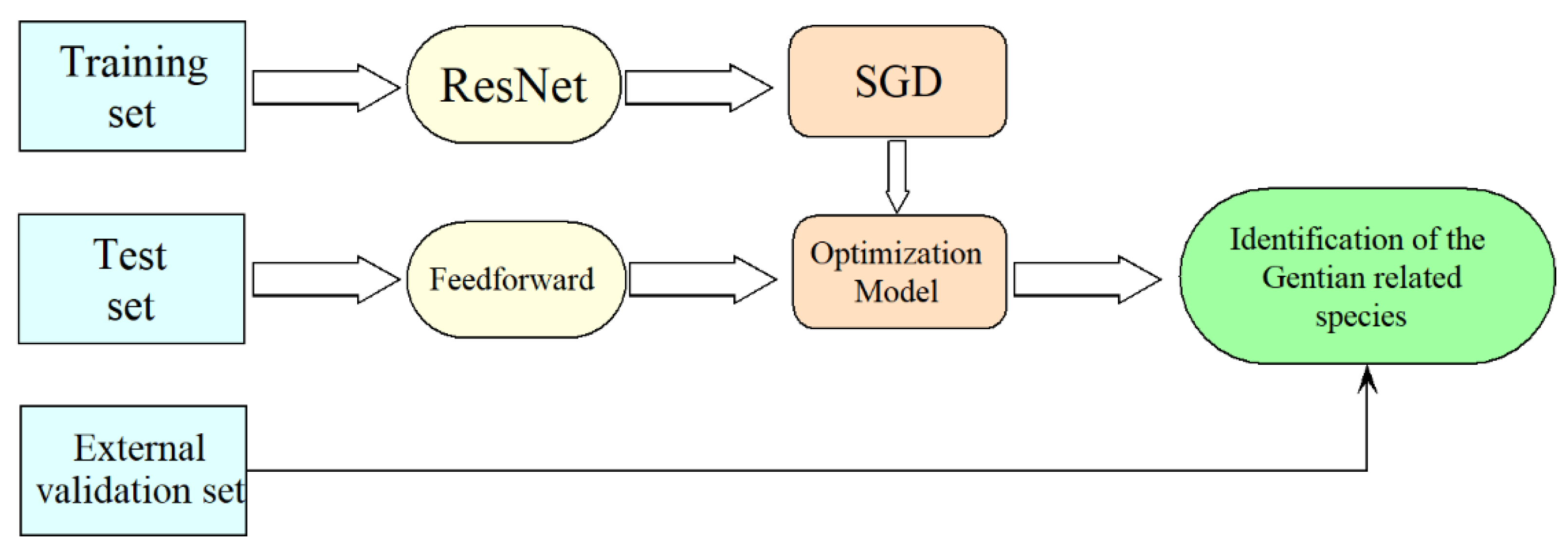
2.4. Discrimination Results of ResNet
3. Discussion
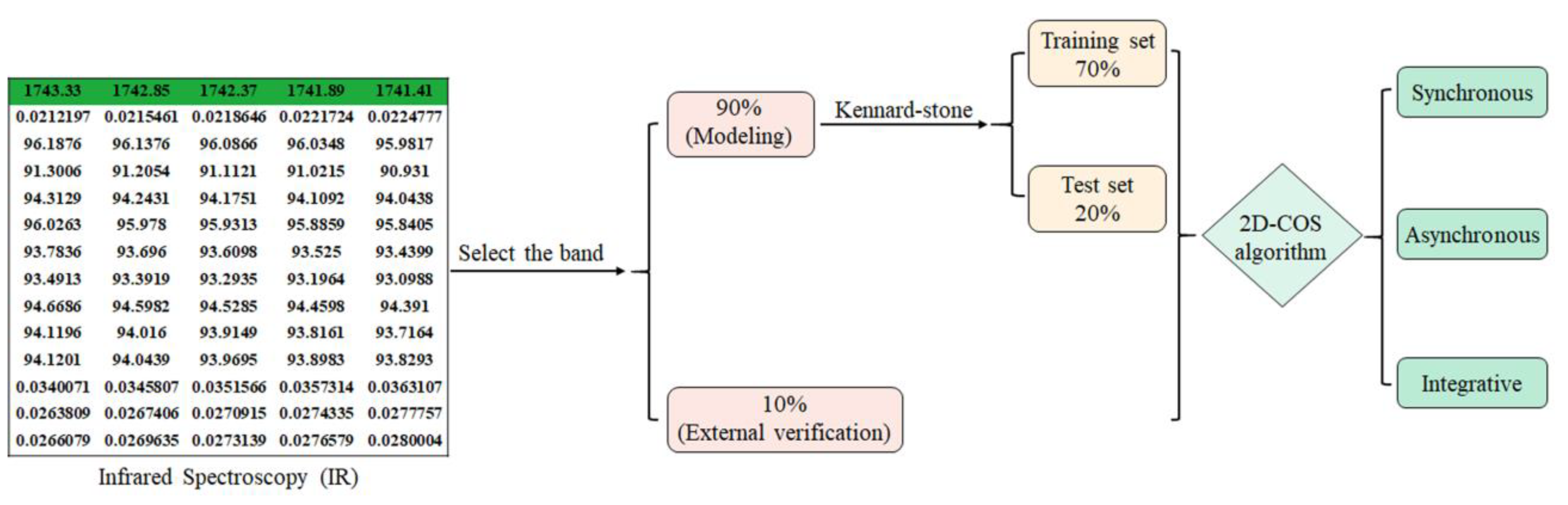
4. Materials and Methods
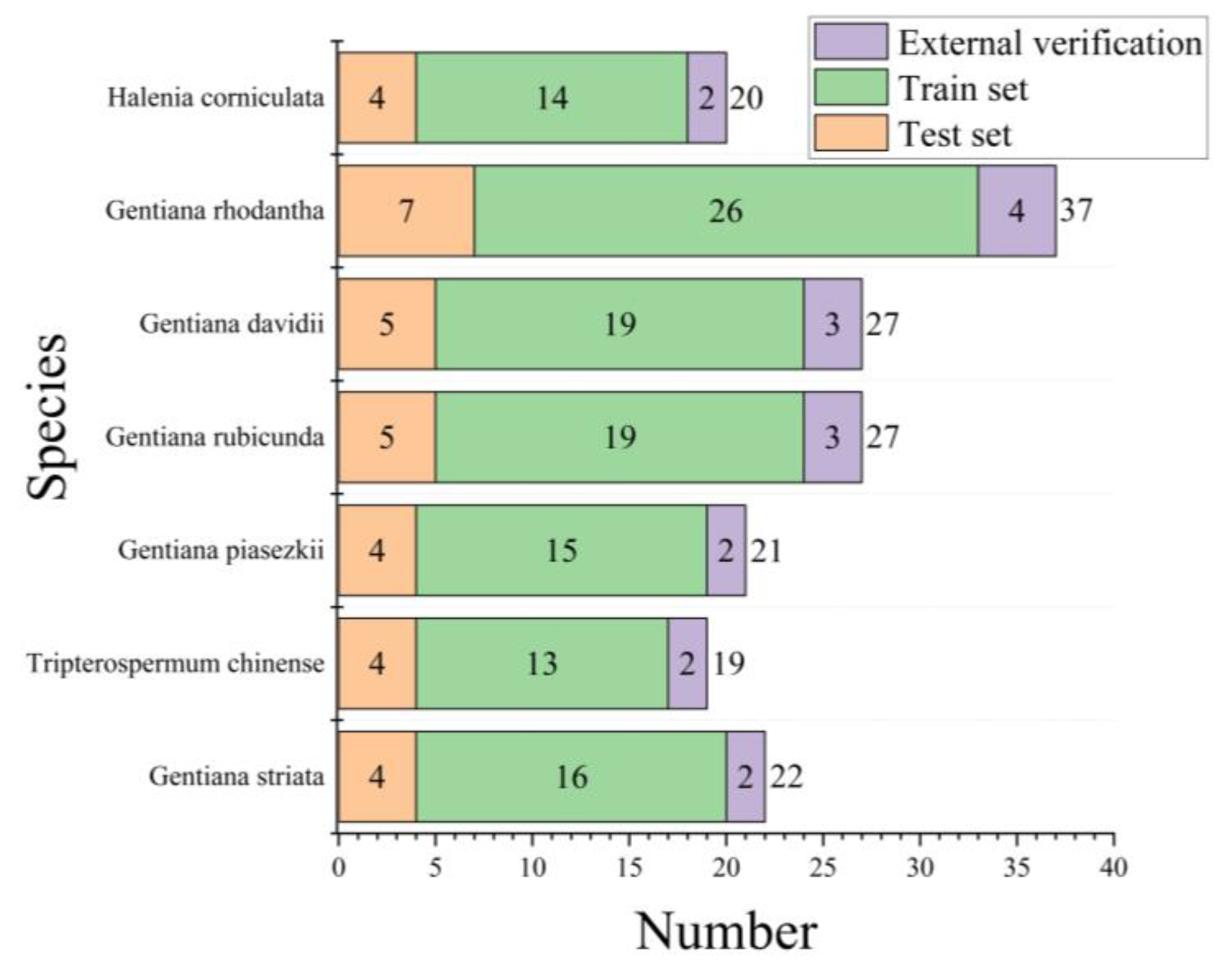
4.1. Sample Information
4.2. IR Acquisition
4.3. Two-Dimensional Correlation Spectroscopy (2D-COS) and Integrated 2D-COS Spectra Image Acquisition
4.3.1. Introduction of 2D-COS
4.3.2. The 2D-COS Algorithm
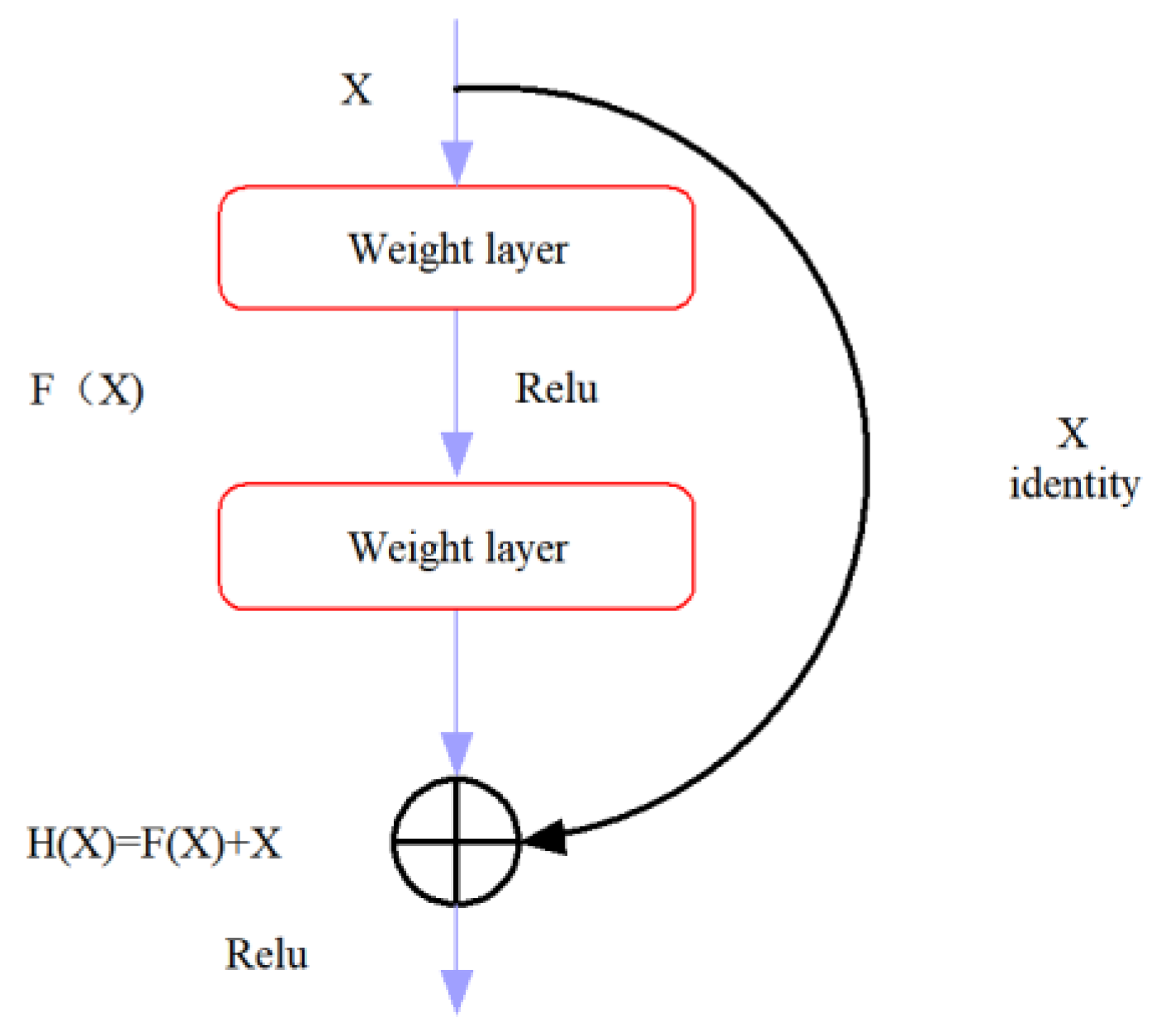
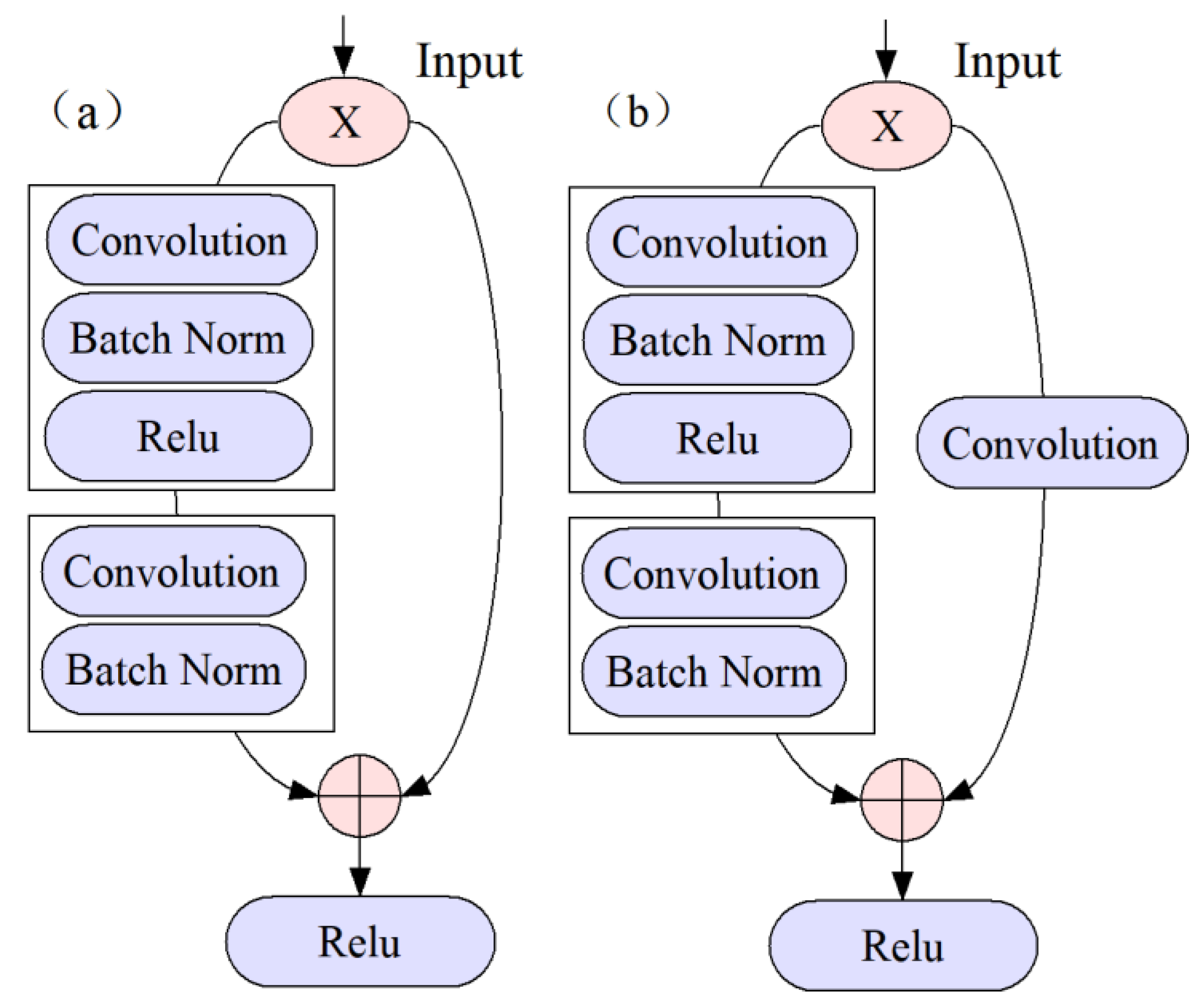
4.4. CNN
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Zheng, Y.Y.; Fang, D.; Huang, C.Y.; Zhao, L.N.; Gan, L.M.; Chen, Y.L.; Liu, F.B. Gentiana scabra Restrains Hepatic Pro-Inflammatory Macrophages to Ameliorate Non-Alcoholic Fatty Liver Disease. Front. Pharmacol. 2021, 12, 816032. [Google Scholar] [CrossRef]
- Jiang, M.; Cui, B.W.; Wu, Y.L.; Nan, J.X.; Lian, L.H. Genus Gentiana: A review on phytochemistry, pharmacology and molecular mechanism. J. Ethnopharmacol. 2021, 264, 113391. [Google Scholar] [CrossRef]
- Huang, S.H.; Chen, E.H.; Wu, C.T.; Kuo, C.L.; Tsay, H.S. Comparative analysis among three Taiwan-specific Gentiana species and Chinese medicinal plant Gentiana scabra. Bot. Stud. 2013, 54, 54. [Google Scholar] [CrossRef]
- Qu, J.B.; Zhang, T.D.; Liu, J.X.; Su, Y.Y.; Wang, H.N. Considerations for the Quality Control of Newly Registered Traditional Chinese Medicine in China: A Review. J. AOAC Int. 2018, 102, 689–694. [Google Scholar] [CrossRef]
- Mlynárik, V. Introduction to nuclear magnetic resonance. Anal. Biochem. 2017, 529, 4–9. [Google Scholar] [CrossRef]
- Bārzdiņa, A.; Paulausks, A.; Bandere, D.; Brangule, A. The Potential Use of Herbal Fingerprints by Means of HPLC and TLC for Characterization and Identification of Herbal Extracts and the Distinction of Latvian Native Medicinal Plants. Molecules 2022, 27, 2555. [Google Scholar] [CrossRef]
- Saparbaev, E.; Yamaletdinov, R.; Boyarkin, O.V. Identification of Isomeric Lipids by UV Spectroscopy of Noncovalent Complexes with Aromatic Molecules. Anal. Chem. 2021, 93, 12822–12826. [Google Scholar] [CrossRef]
- Zhang, Y.Y.; Shen, T.; Zuo, Z.T.; Wang, Y.Z. ResNet and MaxEnt modeling for quality assessment of Wolfiporia cocos based on FT-NIR fingerprints. Front. Plant Sci. 2022, 13, 996069. [Google Scholar] [CrossRef]
- Zeng, P.; Li, X.K.; Wu, X.X.; Diao, Y.; Liu, Y.; Liu, P.Z. Rapid Identification of Wild Gentiana Genus in Different Geographical Locations Based on FT-IR and an Improved Neural Network Structure Double-Net. Molecules 2022, 27, 5979. [Google Scholar] [CrossRef]
- Yan, Z.Y.; Liu, H.G.; Li, T.; Li, J.Q.; Wang, Y.Z. Two dimensional correlation spectroscopy combined with ResNet: Efficient method to identify bolete species compared to traditional machine learning. Lwt-Food Sci. Technol. 2022, 162, 113490. [Google Scholar] [CrossRef]
- Chen, J.B.; Wang, Y.; Rong, L.X.; Wang, J.J. Integrative two-dimensional correlation spectroscopy (i2DCOS) for the intuitive identification of adulterated herbal materials. J. Mol. Struct. 2018, 1163, 327–335. [Google Scholar] [CrossRef]
- Nodo, I. Generalized Two-Dimensional Correlation Method Applicable to Infrared, Raman, and Other Types of Spectroscopy. Appl. Spectrosc. 1993, 47, 1329–1336. [Google Scholar] [CrossRef]
- Gawehn, E.; Hiss, J.A.; Schneider, G. Deep Learning in Drug Discovery. Mol. Inform. 2016, 35, 3–14. [Google Scholar] [CrossRef]
- Vogt, M. An overview of deep learning techniques. At-Autom. 2018, 66, 690–703. [Google Scholar] [CrossRef]
- Geert, L.; Thijs, K.; Babak, E.B.; Arnaud, A.A.A.; Francesco, C.; Mohsen, G. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar]
- Lee, H.; Kwon, H. Going Deeper with Contextual CNN for Hyperspectral Image Classification. IEEE Trans. Image Process. 2017, 26, 4843–4855. [Google Scholar] [CrossRef]
- Wang, L.B.; Liu, J.Y.; Zhang, J.; Wang, J.; Fan, X.F. Corn Seed Defect Detection Based on Watershed Algorithm and Two-Pathway Convolutional Neural Networks. Front. Plant Sci. 2022, 13, 730190. [Google Scholar] [CrossRef]
- Anwar, S.M.; Majid, M.; Qayyum, A.; Awais, M.; Alnowami, M.; Khan, M.K. Medical Image Analysis using Convolutional Neural Networks: A Review. J. Med. Syst. 2018, 42, 226. [Google Scholar] [CrossRef]
- He, K.M.; Zhang, X.Y.; Ren, S.Q.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Li, J.; Cheng, K.W.; Wang, S.H.; Morstatter, F.; Trevino, R.P.; Tang, J.; Liu, H. Feature Selection: A Data Perspective. ACM 2016, 50, 1–45. [Google Scholar] [CrossRef]
- Wang, K.; Coyle, M.E.; Mansu, S.; Zhang, A.L.; Xue, C.C. Gentiana scabra Bunge. Formula for Herpes Zoster: Biological Actions of Key Herbs and Systematic Review of Efficacy and Safety. Phytother. Res. 2017, 31, 375–386. [Google Scholar] [CrossRef]
- Zhao, R.; Yin, S.Y.; Xue, J.N.; Liu, C.; Xing, Y.P.; Yin, H.B.; Ren, X.; Chen, J.X.; Jia, D.D. Sequencing and comparative analysis of chloroplast genomes of three medicinal plants: Gentiana manshurica, G. scabra and G. triflora. Physiol. Mol. Biol. Plants 2022, 28, 1421–1435. [Google Scholar] [CrossRef]
- Paiva, D.N.A.; Perdiz, R.O.; Almeida, T.E. Using near-infrared spectroscopy to discriminate closely related species: A case study of neotropical ferns. J. Plant Res. 2021, 134, 509–520. [Google Scholar] [CrossRef]
- Yao, S.; Li, T.; Li, J.; Liu, H.; Wang, Y. Geographic identifification of Boletus mushrooms by data fusion of FT-IR and UV spectroscopies combined with multivariate statistical analysis. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2018, 198, 257–263. [Google Scholar] [CrossRef]
- Wu, Z.; Zhao, Y.; Zhang, J.; Wang, Y. Quality Assessment of Gentiana rigescens from Different Geographical Origins Using FT-IR Spectroscopy Combined with HPLC. Molecules 2017, 22, 1238. [Google Scholar] [CrossRef]
- Shen, T.; Yu, H.; Wang, Y.Z. Discrimination of Gentiana and Its Related Species Using IR Spectroscopy Combined with Feature Selection and Stacked Generalization. Molecules 2020, 25, 1442. [Google Scholar] [CrossRef]
- Devos, O.; Downey, G.; Duponchel, L. Simultaneous data pre-processing and SVM classification model selection based on a parallel genetic algorithm applied to spectroscopic data of olive oils. Food Chem. 2014, 148, 124–130. [Google Scholar] [CrossRef]
- Bazgir, O.; Zhang, R.; Dhruba, S.R.; Rahman, R.; Ghosh, S.; Pal, R. Representation of features as images with neighborhood dependencies for compatibility with convolutional neural networks. Nat. Commun. 2020, 11, 4391. [Google Scholar] [CrossRef]
- Shinzawa, H.; Hashimoto, K.; Sato, H.; Kanematsu, W.; Noda, I. Multiple-perturbation two-dimensional (2d) correlation analysis for spectroscopic imaging data. J. Mol. Struct. 2014, 1069, 176–182. [Google Scholar] [CrossRef]
- Lindsay, G.W. Convolutional Neural Networks as a Model of the Visual System: Past, Present, and Future. J. Cogn. Neurosci. 2021, 33, 2017–2031. [Google Scholar] [CrossRef]
- Itoh, H.; Hanajima, N.; Muraoka, Y.; Ohata, M.; Fujihira, Y. Exercise classification using CNN with image frames produced from time-series motion data. J. Robot. Netw. Artif. Life 2017, 4, 18. [Google Scholar] [CrossRef]
- Kai, Z.; Zuo, W.M.; Chen, Y.J.; Meng, D.Y.; Lei, Z. Beyond a Gaussian Denoiser: Residual Learning of Deep CNN for Image Denoising. IEEE Press. 2017, 26, 3142–3155. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Szegedy, C.; Liu, W.; Jia, Y.Q.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar]
- Lu, Z.; Bai, Y.; Chen, Y.; Su, C.; Wang, S. The Classification of Gliomas Based on a Pyramid Dilated Convolution ResNet Model. Pattern Recognit. Lett. 2020, 133, 173–179. [Google Scholar] [CrossRef]
- Wang, P.; Luo, F.; Wang, L.H.; Li, C.S.; Niu, Q.; LI, H. S-ResNet: An improved ResNet neural model capable of the identification of small insects. Front. Plant Sci. 2022, 13, 1066115. [Google Scholar] [CrossRef]
- Wu, Z.; Shen, C.; Hengel, A. Wider or Deeper: Revisiting the ResNet Model for Visual Recognition. Pattern Recogn. 2019, 90, 119–133. [Google Scholar] [CrossRef]
- Qian, Q.; Jin, R.; Yi, J.F.; Zhang, L.J.; Zhu, S.G. Efficient distance metric learning by adaptive sampling and mini-batch stochastic gradient descent (SGD). Mach. Learn. 2015, 99, 353–372. [Google Scholar] [CrossRef]
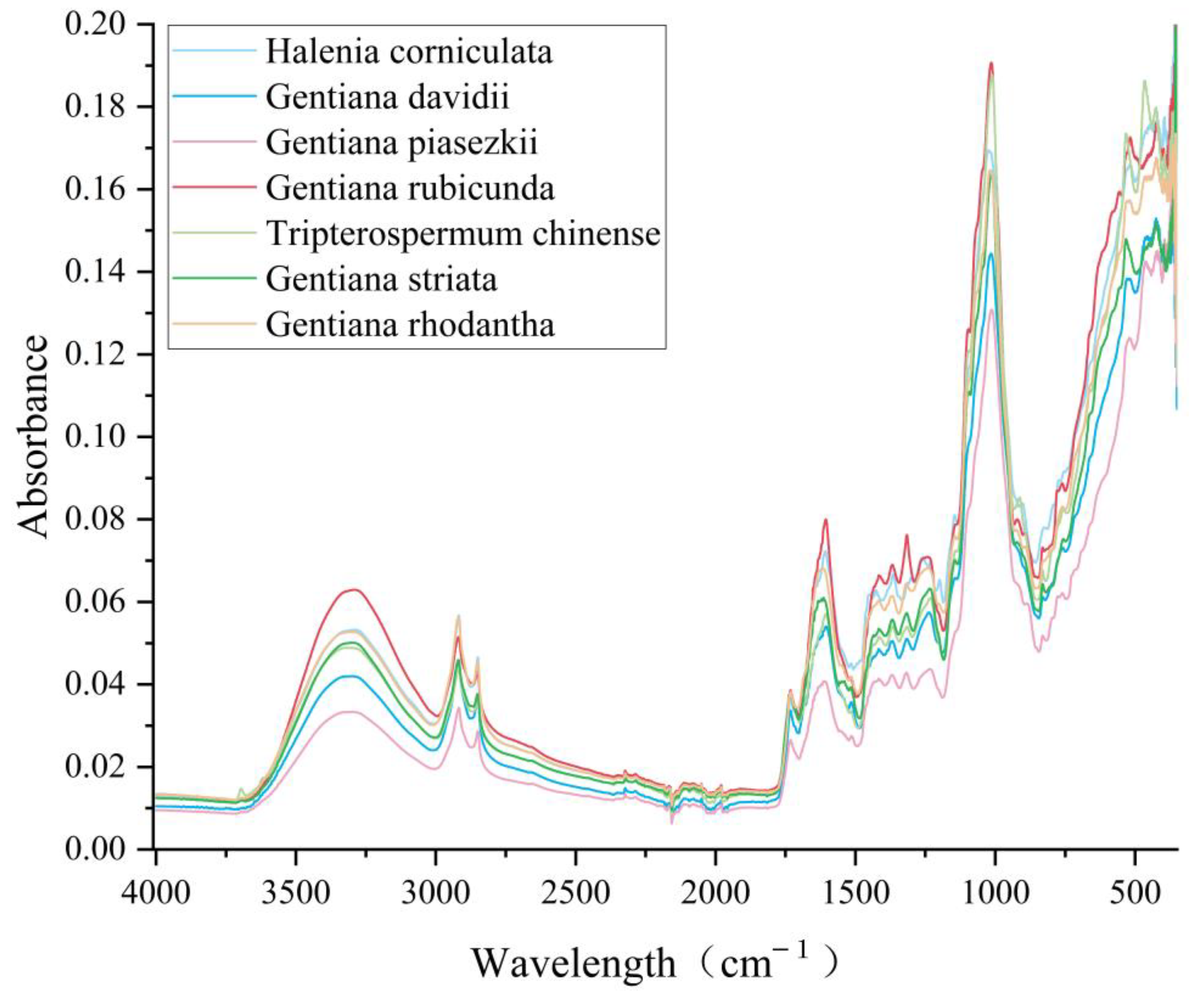
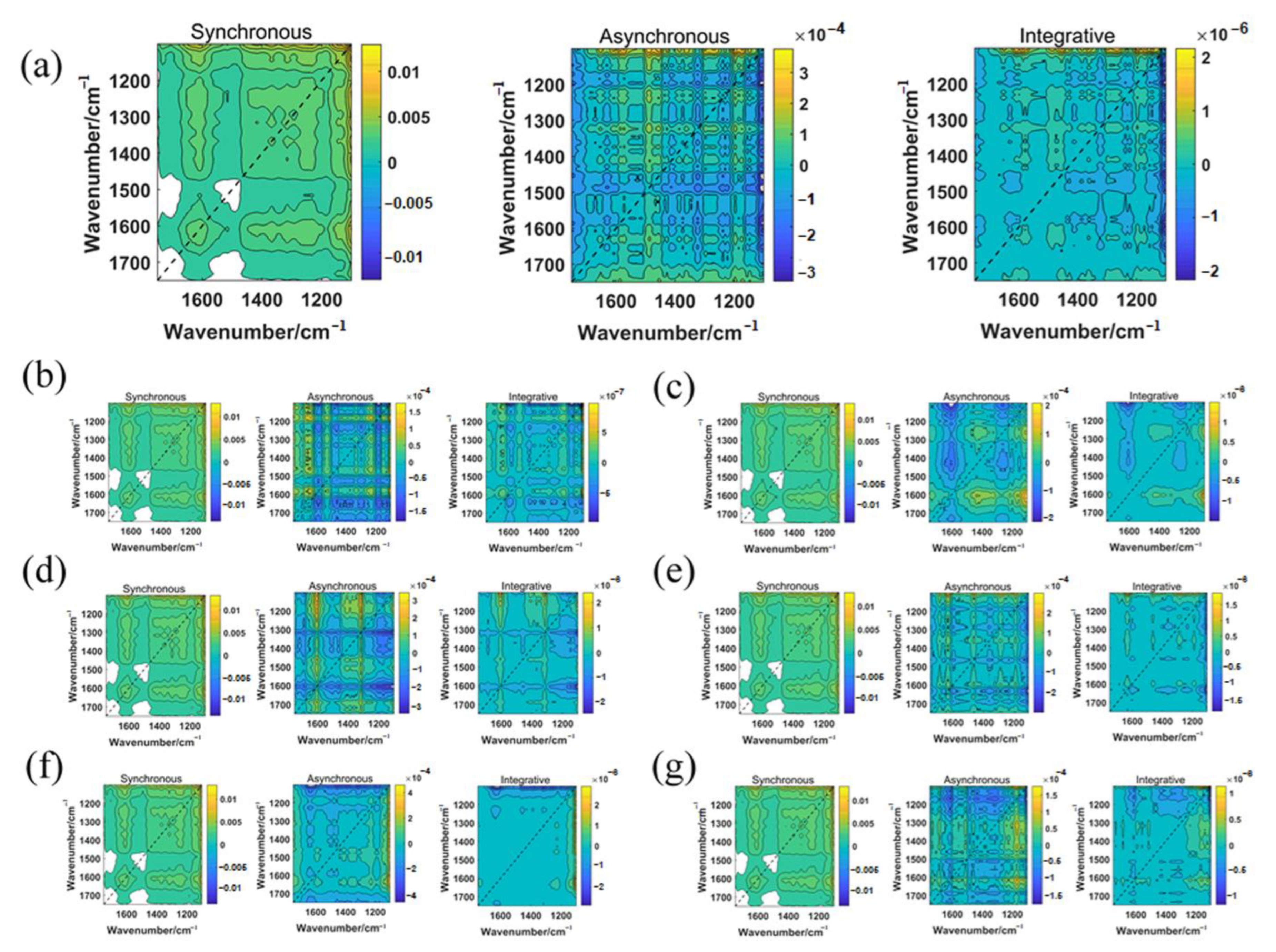

| Feature Bands (cm−1) | Wavenumber (cm−1) | Vibration Mode |
|---|---|---|
| 3500–3000 | 3292 | O-H stretching vibration |
| 3000–2750 | 2922 | Methylene asymmetrical stretching vibration |
| 2849 | Methylene symmetrical stretching vibration | |
| 1750–1100 | 1732 | Ester substance C=O stretching vibration |
| 1610 | Terpenoid C-C asymmetrical stretching vibration | |
| 1510, 1422 | Lignin benzene ring skeleton vibration | |
| 1371 | Methylene deformation vibration | |
| 1100–400 | 1030 | Saccharide C-OH stretching vibration |
| 920 | C-H bending vibration |
| Model Number | Band (cm−1) | The Type of 2D-COS | Loss Value | Train Set Acc | Test Set Acc | Validation Set Acc |
|---|---|---|---|---|---|---|
| A | 3500–3000 | Syn- | 0.222 | 100.00% | 100.00% | 100.00% |
| Asy- | 1.277 | 59.84% | 36.36% | 44.40% | ||
| Int- | 1.187 | 67.21% | 30.30% | 50.00% | ||
| B | 3000–2750 | Syn- | 0.227 | 100.00% | 100.00% | 100.00% |
| Asy- | 1.483 | 68.85% | 36.36% | 27.78% | ||
| Int- | 1.31 | 64.75% | 45.45% | 44.44% | ||
| C | 1750–1100 | Syn- | 0.155 | 100.00% | 100.00% | 100.00% |
| Asy- | 1.346 | 67.21% | 54.55% | 38.89% | ||
| Int- | 1.153 | 63.93% | 63.64% | 66.67% | ||
| D | 1100–400 | Syn- | 0.202 | 100.00% | 96.97% | 100.00% |
| Asy- | 1.304 | 63.11% | 45.45% | 27.78% | ||
| Int- | 1.265 | 59.02% | 51.52% | 44.44% | ||
| E | A + B + C + D | Syn- | 0.223 | 100.00% | 100.00% | 100.00% |
| F | Full spectra | Syn- | 0.339 | 100.00% | 100.00% | 100.00% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, X.; Yang, X.; Cheng, Z.; Li, S.; Li, X.; Zhang, H.; Diao, Y. Identification of Gentian-Related Species Based on Two-Dimensional Correlation Spectroscopy (2D-COS) Combined with Residual Neural Network (ResNet). Molecules 2023, 28, 5000. https://doi.org/10.3390/molecules28135000
Wu X, Yang X, Cheng Z, Li S, Li X, Zhang H, Diao Y. Identification of Gentian-Related Species Based on Two-Dimensional Correlation Spectroscopy (2D-COS) Combined with Residual Neural Network (ResNet). Molecules. 2023; 28(13):5000. https://doi.org/10.3390/molecules28135000
Chicago/Turabian StyleWu, Xunxun, Xintong Yang, Zhiyun Cheng, Suyun Li, Xiaokun Li, Haiyun Zhang, and Yong Diao. 2023. "Identification of Gentian-Related Species Based on Two-Dimensional Correlation Spectroscopy (2D-COS) Combined with Residual Neural Network (ResNet)" Molecules 28, no. 13: 5000. https://doi.org/10.3390/molecules28135000
APA StyleWu, X., Yang, X., Cheng, Z., Li, S., Li, X., Zhang, H., & Diao, Y. (2023). Identification of Gentian-Related Species Based on Two-Dimensional Correlation Spectroscopy (2D-COS) Combined with Residual Neural Network (ResNet). Molecules, 28(13), 5000. https://doi.org/10.3390/molecules28135000







