Delving into the Mechanisms of Sponge-Associated Enterobacter against Staphylococcal Biofilms
Abstract
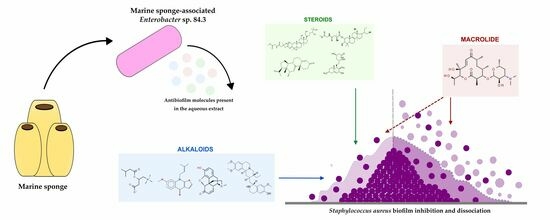
1. Introduction
2. Results
2.1. Antibiofilm Activity of the Aqueous Extract
2.2. Antibiofilm Molecule Identification
2.3. Possible Mode of Action on S. aureus Biofilm
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Culture Conditions
4.2. Fermentation Conditions and Extract Preparation
4.3. Scanning Electron Microscopy Observation of Antibiofilm Activity
4.4. Chemical Analysis, Bioinformatics Screening, and Compound Identification
4.4.1. UHPLC-MS Analysis of the Aqueous Extract
4.4.2. MZmine Parameters
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Balasubramanian, S.; Othman, E.M.; Kampik, D.; Stopper, H.; Hentschel, U.; Ziebuhr, W.; Oelschlaeger, T.A.; Abdelmohsen, U.R. Marine sponge-derived Streptomyces sp. SBT343 extract inhibits staphylococcal biofilm formation. Front. Microbiol. 2017, 8, 236. [Google Scholar] [CrossRef] [PubMed]
- Gajdács, M. The concept of an ideal antibiotic: Implications for drug design. Molecules 2019, 24, 892. [Google Scholar] [CrossRef]
- McCarthy, H.; Rudkin, J.K.; Black, N.S.; Gallagher, L.; O’Neill, E.; O’Gara, J.P. Methicillin resistance and the biofilm phenotype in Staphylococcus aureus. Front. Cell. Infect. Microbiol. 2015, 5, 1. [Google Scholar] [CrossRef]
- Li, X.H.; Lee, J.H. Antibiofilm agents: A new perspective for antimicrobial strategy. J. Microbiol. 2017, 55, 753–766. [Google Scholar] [CrossRef]
- Tuon, F.F.; Suss, P.H.; Telles, J.P.; Dantas, L.R.; Borges, N.H.; Ribeiro, V.S.T. Antimicrobial Treatment of Staphylococcus aureus Biofilms. Antibiotics 2023, 12, 87. [Google Scholar] [CrossRef]
- Geissel, F.J.; Platania, V.; Gogos, A.; Herrmann, I.K.; Belibasakis, G.N.; Chatzinikolaidou, M.; Sotiriou, G.A. Antibiofilm activity of nanosilver coatings against Staphylococcus aureus. J. Colloid. Interface Sci. 2022, 608, 3141–3150. [Google Scholar] [CrossRef] [PubMed]
- Jamal, M.T.; Sathianeson, S. Antibiofilm activity of secondary metabolites of sponge-associated bacterium Alcanivorax sp. from the Red Sea. Front. Mar. Sci. 2022, 9, 980418. [Google Scholar] [CrossRef]
- Raj, V.; Kim, Y.; Kim, Y.G.; Lee, J.H.; Lee, J. Chitosan-gum arabic embedded alizarin nanocarriers inhibit biofilm formation of multispecies microorganisms. Carbohydr. Polym. 2022, 284, 118959. [Google Scholar] [CrossRef]
- Barros, T.F.; Lin, V.; Borges, J.S.; Primon-Barros, M.; Zanuncio, V.S.S.; Silva, D.B.; Trentin, D.S. Staphylococcus aureus antibiofilm agents from Combretum species (Combretaceae) by metabolomics profiling. Ind. Crops Prod. 2023, 193, 116280. [Google Scholar] [CrossRef]
- Abo-Salem, H.M.; Abd El Salam, H.A.; Abdel-Aziem, A.; Abdel-Aziz, M.S.; El-Sawy, E.R. Synthesis, molecular docking, and biofilm formation inhibitory activity of bis (indolyl) pyridines analogues of the marine alkaloid nortopsentin. Molecules 2021, 26, 4112. [Google Scholar] [CrossRef]
- Rajput, A.; Bhamare, K.T.; Thakur, A.; Kumar, M. Anti-Biofilm: Machine Learning Assisted Prediction of IC50 Activity of Chemicals Against Biofilms of Microbes Causing Antimicrobial Resistance and Implications in Drug Repurposing. J. Mol. Biol. 2023, 168115. [Google Scholar] [CrossRef]
- Santos-Gandelman, J.F.; Giambiagi-deMarval, M.; Oelemann, W.M.R.; Laport, M.S. Biotechnological potential of sponge-associated bacteria. Curr. Pharm. Biotechnol. 2014, 15, 143–155. [Google Scholar] [CrossRef] [PubMed]
- Nunes, S.O.; Rosa, H.S.; Canellas, A.L.B.; Romanos, M.T.V.; dos Santos, K.R.; Muricy, G.; Oelemann, W.M.R.; Laport, M.S. High reduction of staphylococcal biofilm by aqueous extract from marine sponge-isolated Enterobacter sp. Res. Microbiol. 2021, 172, 103787. [Google Scholar] [CrossRef]
- Shastry, R.P.; Rai, V.R. Molecular identification of aiiA homologous gene from endophytic Enterobacter species and in silico analysis of putative tertiary structure of AHL-lactonase. Biochem. Biophys. Res. Commun. 2014, 443, 290–295. [Google Scholar] [CrossRef]
- Rajesh, P.S.; Rai, V.R. Purification and antibiofilm activity of AHL-lactonase from endophytic Enterobacter aerogenes VT66. Int. J. Biol. Macromol. 2015, 81, 1046–1052. [Google Scholar] [CrossRef]
- Shastry, R.P.; Dolan, S.K.; Abdelhamid, Y.; Vittal, R.R.; Welch, M. Purification and characterisation of a quorum quenching AHL-lactonase from the endophytic bacterium Enterobacter sp. CS66. FEMS Microbiol. Lett. 2018, 365, fny054. [Google Scholar] [CrossRef]
- Shastry, R.P.; Rekha, P.D.; Rai, V.R. Biofilm inhibitory activity of metallo-protein AHL-lactonase from cell-free lysate of endophytic Enterobacter species isolated from Coscinium fenestratum Gaertn. Biocatal. Agric. Biotechnol. 2019, 18, 101009. [Google Scholar] [CrossRef]
- Guimarães, L.C.; de Souza, B.M.; de Oliveira Whitaker, C.; Abreu, F.; Ferreira, R.B.R.; Dos Santos, K.R.N. Increased biofilm formation by Staphylococcus aureus clinical isolates on surfaces covered with plasma proteins. J. Med. Microbiol. 2021, 70, 001389. [Google Scholar] [CrossRef]
- Boles, B.R.; Thoendel, M.; Roth, A.J.; Horswill, A.R. Identification of genes involved in polysaccharide-independent Staphylococcus aureus biofilm formation. PLoS ONE 2010, 5, e10146. [Google Scholar] [CrossRef]
- Idrees, M.; Sawant, S.; Karodia, N.; Rahman, A. Staphylococcus aureus Biofilm: Morphology, Genetics, Pathogenesis and Treatment Strategies. Int. J. Environ. Res. Public. Health 2021, 18, 7602. [Google Scholar] [CrossRef] [PubMed]
- Artini, M.; Papa, R.; Vrenna, G.; Lauro, C.; Ricciardelli, A.; Casillo, A.; Corsaro, M.M.; Tutino, M.L.; Parrilli, E.; Selan, L. Cold adapted bacterial extracts as source of anti-infective and antimicrobial compounds against Staphylococcus aureus. Future Microbiol. 2019, 14, 1369e82. [Google Scholar] [CrossRef] [PubMed]
- Laport, M.S. Isolating bacteria from sponges: Why and how? Curr. Pharm. Biotechnol. 2017, 18, 1224–1236. [Google Scholar] [CrossRef] [PubMed]
- Parai, D.; Banerjee, M.; Dey, P.; Mukherjee, S.K. Reserpine attenuates biofilm formation and virulence of Staphylococcus aureus. Microb. Pathog. 2020, 138, 103790. [Google Scholar] [CrossRef] [PubMed]
- Fujimura, S.; Sato, T.; Mikami, T.; Kikuchi, T.; Gomi, K.; Watanabe, A. Combined efficacy of clarithromycin plus cefazolin or vancomycin against Staphylococcus aureus biofilms formed on titanium medical devices. Int. J. Antimicrob. Agents 2008, 32, 481–484. [Google Scholar] [CrossRef]
- Fujimura, S.; Sato, T.; Kikuchi, T.; Zaini, J.; Gomi, K.; Watanabe, A. Efficacy of clarithromycin plus vancomycin in mice with implant-related infection caused by biofilm-forming Staphylococcus aureus. J. Orthop. Sci. 2009, 14, 658–661. [Google Scholar] [CrossRef]
- Arshad, M.S.; Zahra, A.T.; Zafar, S.; Zaman, H.; Akhtar, A.; Ayaz, M.M.; Kucuk, I.; Maniruzzaman, M.; Chang, M.W.; Ahma, Z. Antibiofilm Effects of Macrolide Loaded Microneedle Patches: Prospects in Healing Infected Wounds. Pharm. Res. 2021, 38, 165–177. [Google Scholar] [CrossRef]
- Parra-Ruiz, J.; Vidaillac, C.; Rybak, M.J. Macrolides and staphylococcal biofilms. Rev. Esp. Quimioter. 2012, 25, 10–16. [Google Scholar]
- Gui, Z.; Wang, H.; Ding, T.; Zhu, W.; Zhuang, X.; Chu, W. Azithromycin Reduces the Production of α-hemolysin and Biofilm Formation in Staphylococcus aureus. Indian. J. Microbiol. 2014, 54, 114–117. [Google Scholar] [CrossRef]
- Bode, H.B.; Zeggel, B.; Silakowski, B.; Wenzel, S.C.; Reichenbach, H.; Müller, R. Steroid biosynthesis in prokaryotes: Identification of myxobacterial steroids and cloning of the first bacterial 2,3(S)-oxidosqualene cyclase from the myxobacterium Stigmatella aurantiaca. Mol. Microbiol. 2003, 47, 471–481. [Google Scholar] [CrossRef]
- Melander, R.J.; Basak, A.K.; Melander, C. Natural products as inspiration for the development of bacterial antibiofilm agents. Nat. Prod. Rep. 2020, 37, 1454–1477. [Google Scholar] [CrossRef]
- Goggin, R.; Jardeleza, C.; Wormald, P.J.; Vreugde, S. Corticosteroids directly reduce Staphylococcus aureus biofilm growth: An in vitro study. Laryngoscope 2014, 124, 602–607. [Google Scholar] [CrossRef] [PubMed]
- Vollaro, A.; Esposito, A.; Antonaki, E.; Iula, V.D.; D’Alonzo, D.; Guaragna, A.; De Gregorio, E. Steroid Derivatives as potential antimicrobial agents against Staphylococcus aureus planktonic cells. Microorganisms 2020, 8, 468. [Google Scholar] [CrossRef]
- Resch, A.; Leicht, S.; Saric, M.; Pásztor, L.; Jakob, A.; Götz, F.; Nordheim, A. Comparative proteome analysis of Staphylococcus aureus biofilm and planktonic cells and correlation with transcriptome profiling. Proteomics 2006, 6, 1867–1877. [Google Scholar] [CrossRef] [PubMed]
- Sycz, Z.; Tichaczek-Goska, D.; Wojnicz, D. Anti-Planktonic and Anti-Biofilm Properties of Pentacyclic Triterpenes—Asiatic Acid and Ursolic Acid as Promising Antibacterial Future Pharmaceuticals. Biomolecules 2022, 12, 98. [Google Scholar] [CrossRef] [PubMed]
- Ren, D.; Zuo, R.; González Barrios, A.F.; Bedzyk, L.A.; Eldridge, G.R.; Pasmore, M.E.; Wood, T.K. Differential gene expression for investigation of Escherichia coli biofilm inhibition by plant extract ursolic acid. Appl. Environ. Microbiol. 2005, 71, 4022–4034. [Google Scholar] [CrossRef]
- da Silva, G.N.S.; Primon-Barros, M.; Macedo, A.J.; Gnoatto, S.C.B. Triterpene derivatives as relevant scaffold for new antibiofilm drugs. Biomolecules 2019, 9, 58. [Google Scholar] [CrossRef]
- Kessner, D.; Chambers, M.; Burke, R.; Agus, D.; Mallick, P. ProteoWizard: Open source software for rapid proteomics tools development. Bioinformatics 2008, 24, 2534–2536. [Google Scholar] [CrossRef]
- Katajamaa, M.; Miettinen, J.; Orešič, M. MZmine: Toolbox for processing and visualization of mass spectrometry based molecular profile data. Bioinformatics 2006, 22, 634–636. [Google Scholar] [CrossRef]
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Orešič, M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform. 2010, 11, 395. [Google Scholar] [CrossRef]


| Compounds | Name | Class | RT (min) | Area | Molecular Ion [M − H]− |
|---|---|---|---|---|---|
| 1 | Isovalerylcarnitine | Alkaloid | 1.01 | 5.0 × 105 | 244.1554 |
| 2 | Acrophylline or morphinone | Alkaloid | 1.42 | 1.2 × 106 | 282.1125 |
| 3 | Novamethymycin | Macrolide | 2.74 | 3.3 × 105 | 484.2883 |
| 4 | Fasciculol E or F | Tetracyclic triterpene | 11.37 | 3.2 × 105 | 722.4543 |
| 5 | 12-methylpregna-4,9(11)-diene-3,20-dione | Steroid | 13.51 | 3.1 × 105 | 325.2181 |
| 6 | cephaeline | Alkaloid | 14.73 | 4.6 × 105 | 465.2727 |
| 7 | (3alpha,5beta,11beta,17beta)-9-fluoro-17-methylandrostane-3,11,17-triol | Steroid | 15.54 | 5.7 × 105 | 339.2165 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Canellas, A.L.B.; de Oliveira, B.F.R.; Nunes, S.d.O.; Malafaia, C.A.; Amaral, A.C.F.; Simas, D.L.R.; Leal, I.C.R.; Laport, M.S. Delving into the Mechanisms of Sponge-Associated Enterobacter against Staphylococcal Biofilms. Molecules 2023, 28, 4843. https://doi.org/10.3390/molecules28124843
Canellas ALB, de Oliveira BFR, Nunes SdO, Malafaia CA, Amaral ACF, Simas DLR, Leal ICR, Laport MS. Delving into the Mechanisms of Sponge-Associated Enterobacter against Staphylococcal Biofilms. Molecules. 2023; 28(12):4843. https://doi.org/10.3390/molecules28124843
Chicago/Turabian StyleCanellas, Anna Luiza Bauer, Bruno Francesco Rodrigues de Oliveira, Suzanne de Oliveira Nunes, Camila Adão Malafaia, Ana Claudia F. Amaral, Daniel Luiz Reis Simas, Ivana Correa Ramos Leal, and Marinella Silva Laport. 2023. "Delving into the Mechanisms of Sponge-Associated Enterobacter against Staphylococcal Biofilms" Molecules 28, no. 12: 4843. https://doi.org/10.3390/molecules28124843
APA StyleCanellas, A. L. B., de Oliveira, B. F. R., Nunes, S. d. O., Malafaia, C. A., Amaral, A. C. F., Simas, D. L. R., Leal, I. C. R., & Laport, M. S. (2023). Delving into the Mechanisms of Sponge-Associated Enterobacter against Staphylococcal Biofilms. Molecules, 28(12), 4843. https://doi.org/10.3390/molecules28124843