The Effect of Ethanol on Lipid Nanoparticle Stabilization from a Molecular Dynamics Simulation Perspective
Abstract
1. Introduction
2. Result
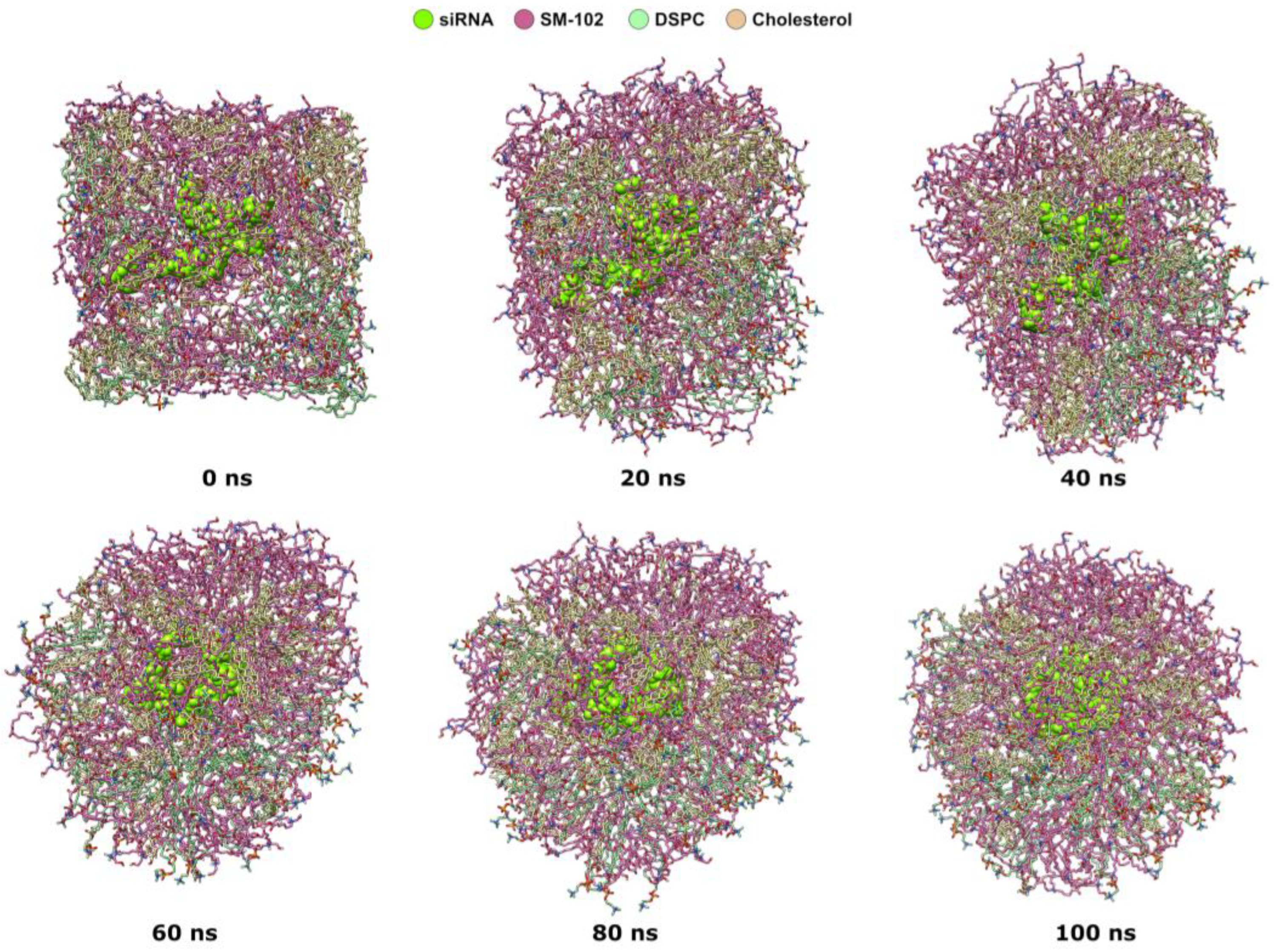
2.1. Molecular Dynamics Simulations of Lipid Nanoparticle Self-Assembly
2.2. Molecular Dynamics Simulations of A Lipid Nanoparticle in an Aqueous Ethanol Environment
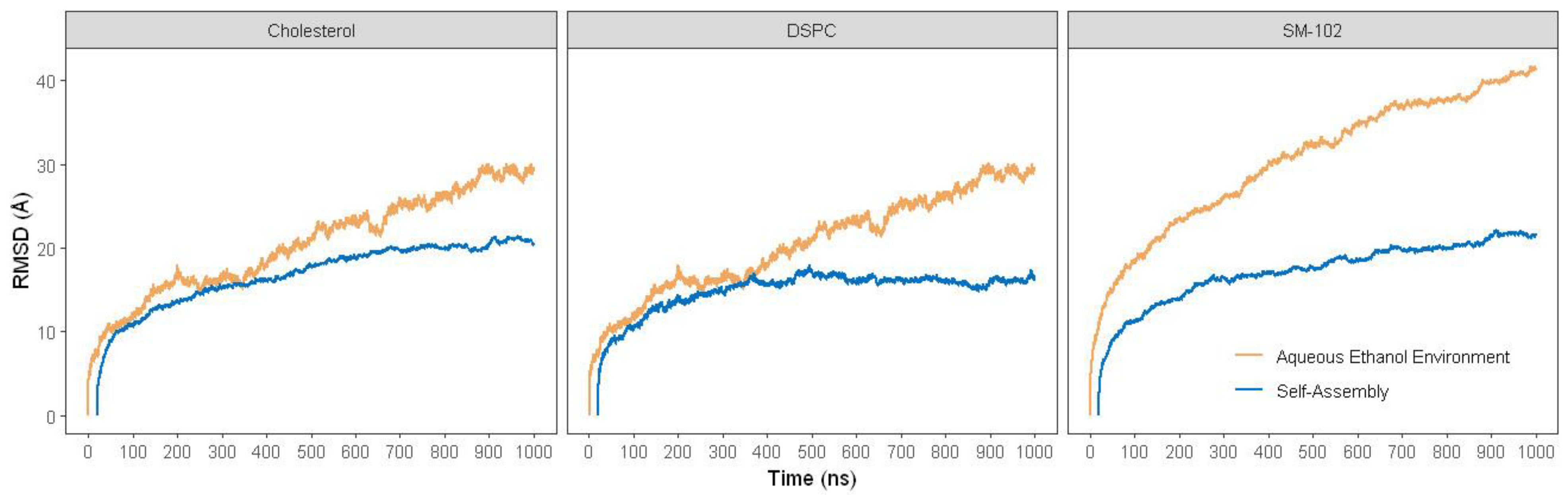
2.2.1. RMSD
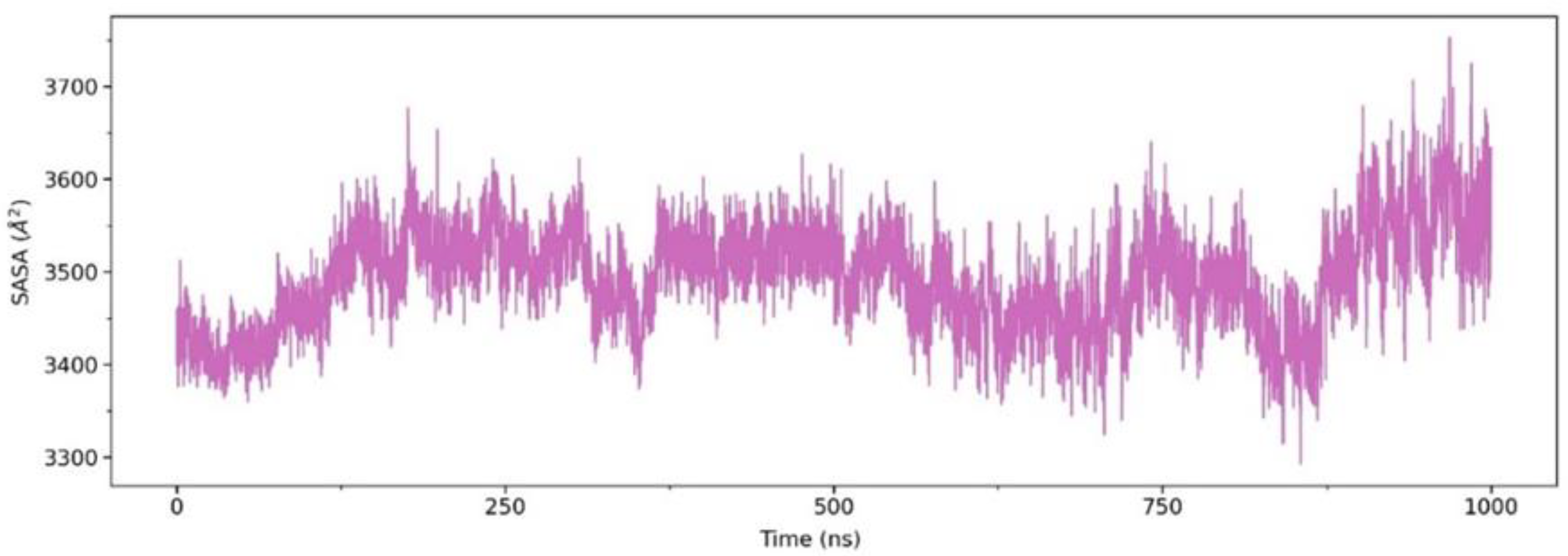
2.2.2. Solvent Accessible Surface Area (SASA)
2.2.3. Electron Density Profiles (EDPs)
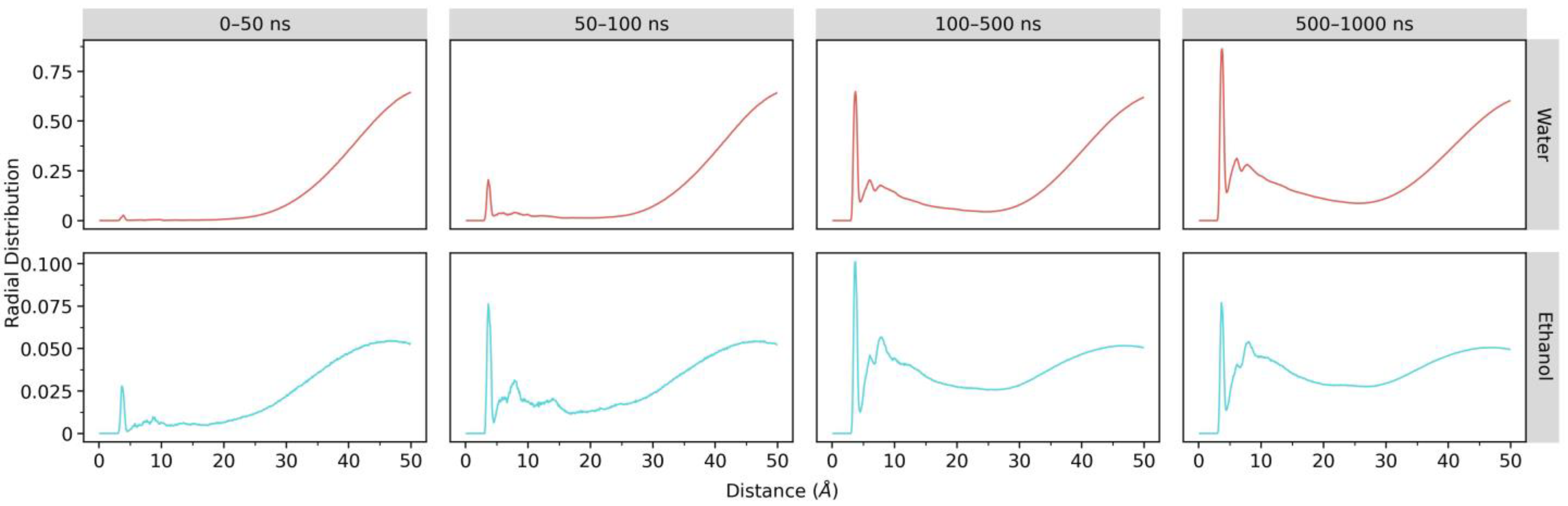
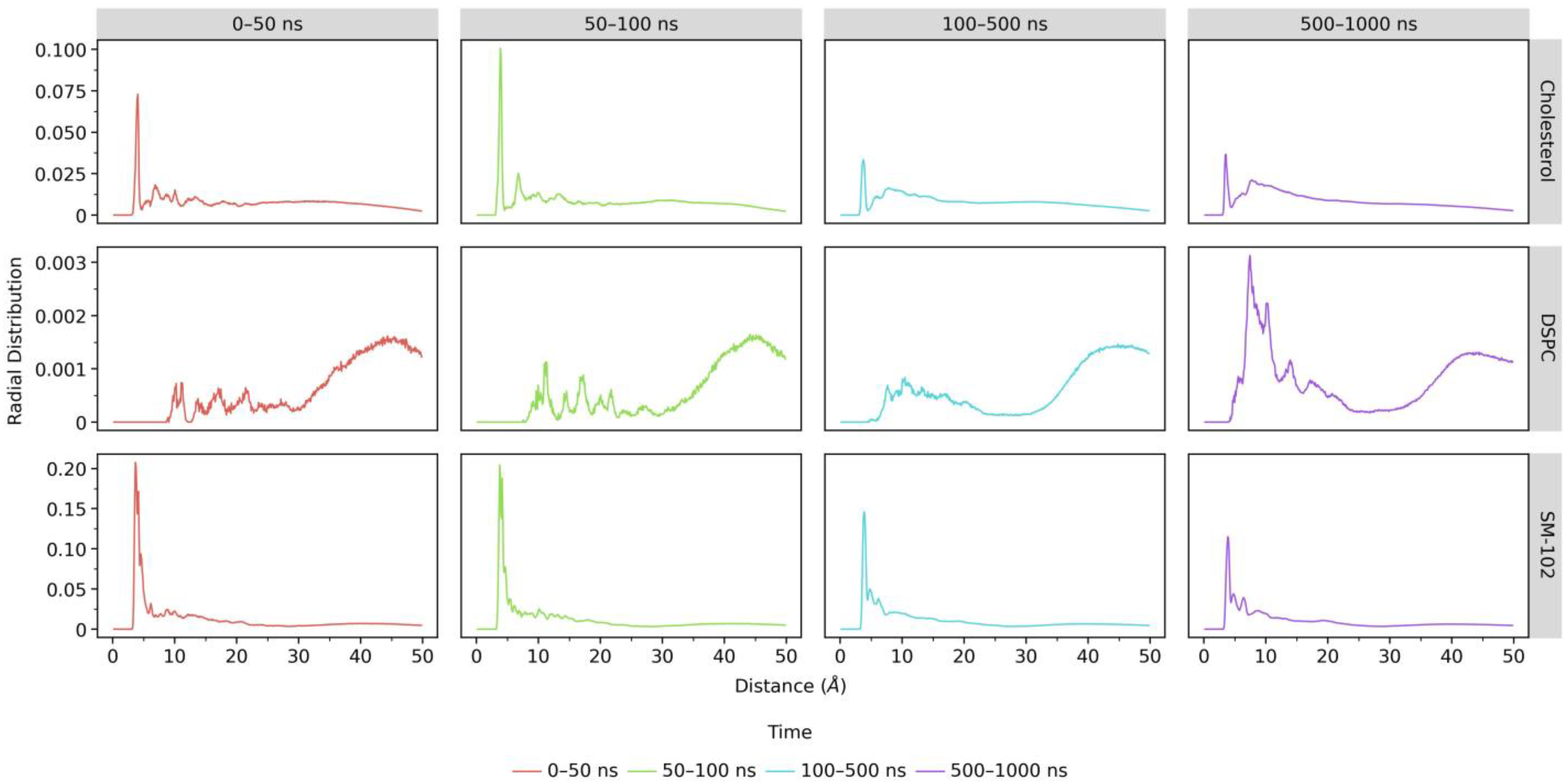
2.2.4. Radial Distribution Function (RDF)
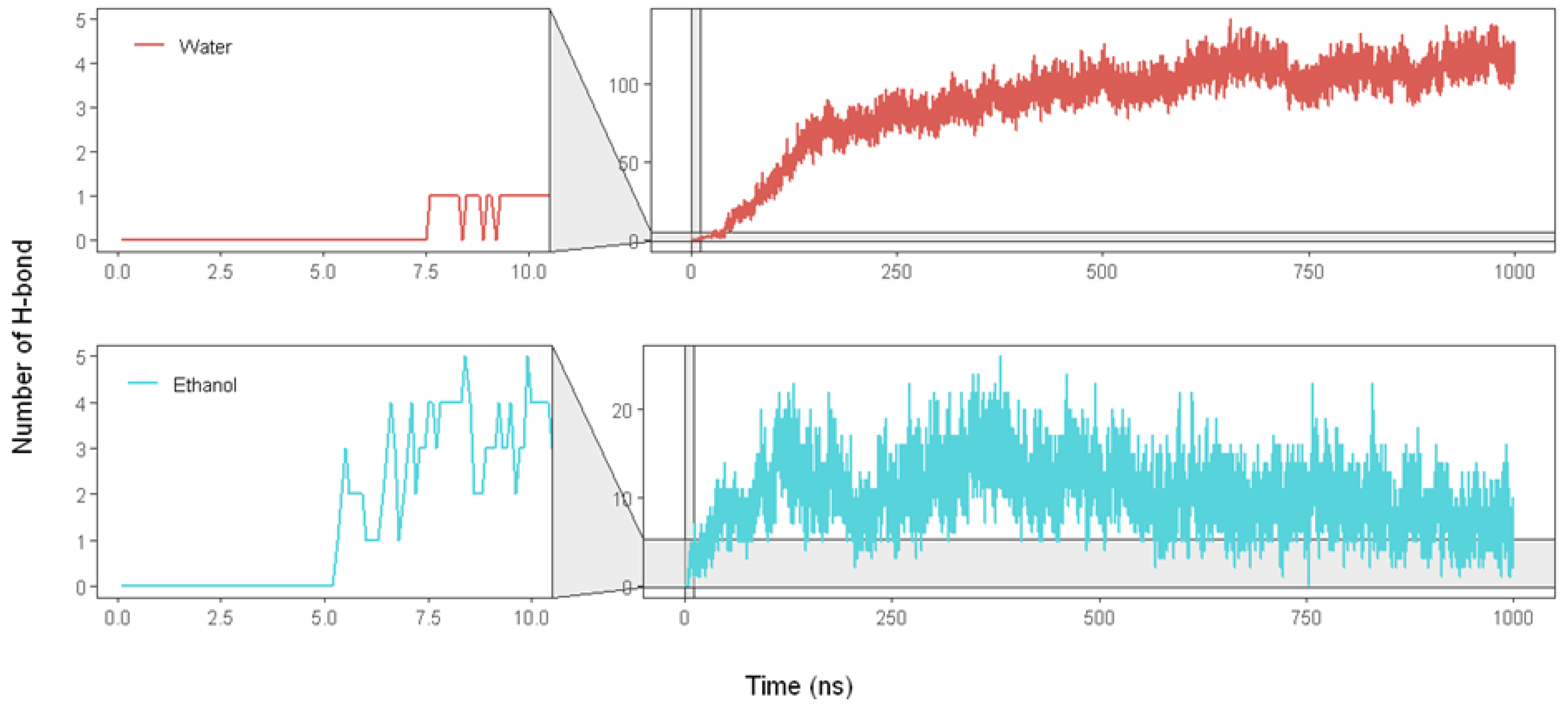
2.2.5. Hydrogen Bonding (H-Bond)
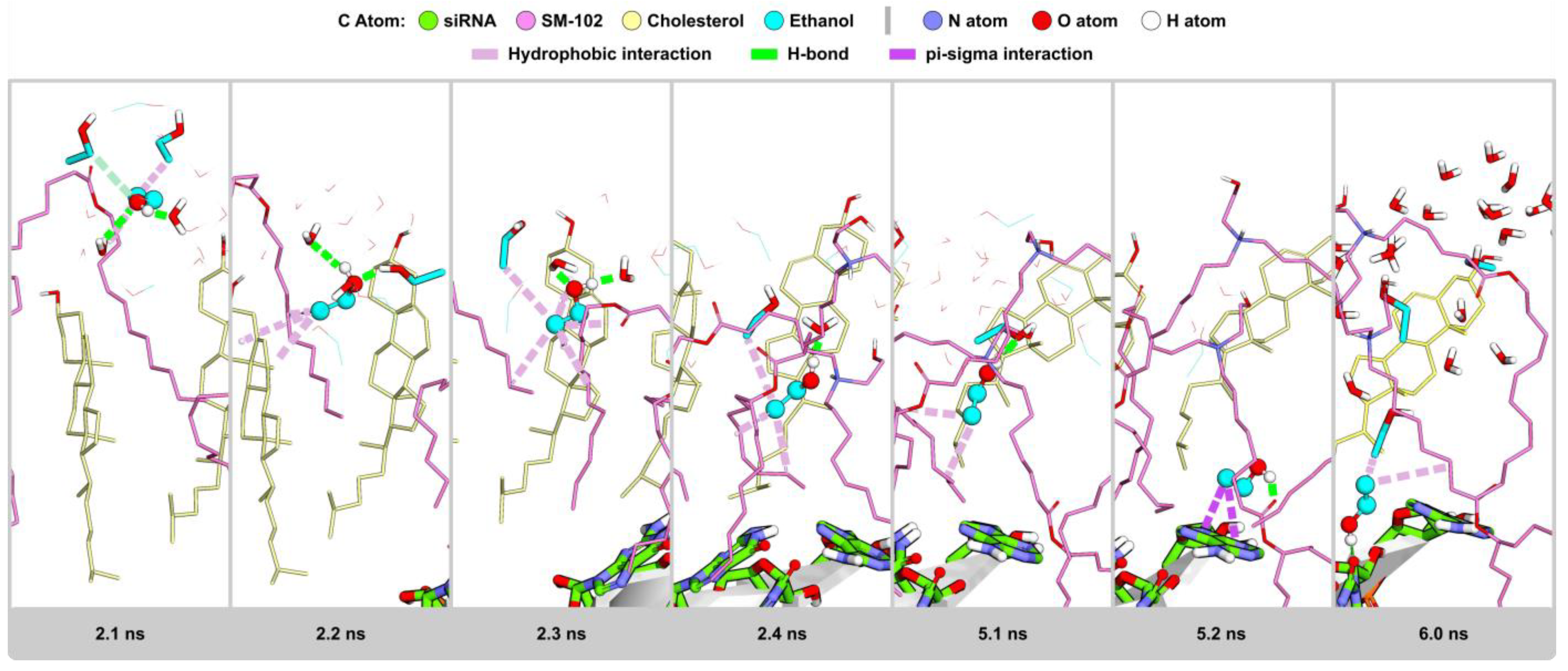
2.2.6. Visualization Analysis
3. Materials and Methods
3.1. Force Fields
3.2. Molecular Dynamics Simulations
3.3. Simulation Analyses
3.4. Molecular Visualization and Graph Generations
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Chen, X.; Mangala, L.S.; Rodriguez-Aguayo, C.; Kong, X.; Lopez-Berestein, G.; Sood, A.K. RNA Interference-Based Therapy and Its Delivery Systems. Cancer Metastasis Rev. 2018, 37, 107–124. [Google Scholar] [CrossRef]
- Cullis, P.R.; Hope, M.J. Lipid Nanoparticle Systems for Enabling Gene Therapies. Mol. Ther. 2017, 25, 1467–1475. [Google Scholar] [CrossRef] [PubMed]
- Brader, M.L.; Williams, S.J.; Banks, J.M.; Hui, W.H.; Zhou, Z.H.; Jin, L. Encapsulation State of Messenger RNA inside Lipid Nanoparticles. Biophys. J. 2021, 120, 2766–2770. [Google Scholar] [CrossRef] [PubMed]
- Anderson, E.J.; Rouphael, N.G.; Widge, A.T.; Jackson, L.A.; Roberts, P.C.; Makhene, M.; Chappell, J.D.; Denison, M.R.; Stevens, L.J.; Pruijssers, A.J.; et al. Safety and Immunogenicity of SARS-CoV-2 MRNA-1273 Vaccine in Older Adults. N. Engl. J. Med. 2020, 383, 2427–2438. [Google Scholar] [CrossRef]
- Labouta, H.I.; Langer, R.; Cullis, P.R.; Merkel, O.M.; Prausnitz, M.R.; Gomaa, Y.; Nogueira, S.S.; Kumeria, T. Role of Drug Delivery Technologies in the Success of COVID-19 Vaccines: A Perspective. Drug Deliv. Transl. Res. 2022, 12, 2581–2588. [Google Scholar] [CrossRef] [PubMed]
- Hald Albertsen, C.; Kulkarni, J.A.; Witzigmann, D.; Lind, M.; Petersson, K.; Simonsen, J.B. The Role of Lipid Components in Lipid Nanoparticles for Vaccines and Gene Therapy. Adv. Drug Deliv. Rev. 2022, 188, 114416. [Google Scholar] [CrossRef] [PubMed]
- Kularatne, R.N.; Crist, R.M.; Stern, S.T. The Future of Tissue-Targeted Lipid Nanoparticle-Mediated Nucleic Acid Delivery. Pharmaceuticals 2022, 15, 897. [Google Scholar] [CrossRef]
- Schoenmaker, L.; Witzigmann, D.; Kulkarni, J.A.; Verbeke, R.; Kersten, G.; Jiskoot, W.; Crommelin, D.J. mRNA-lipid nanoparticle COVID-19 vaccines: Structure and stability. Int. J. Pharm. 2021, 601, 120586. [Google Scholar] [CrossRef]
- European Medicines Agency. ICH Guideline Q3C (R6) on Impurities: Guideline for Residual Solvents Step 5; European Medicines Agency: Amsterdam, The Netherlands, 2015. [Google Scholar]
- Kimura, N.; Maeki, M.; Sato, Y.; Note, Y.; Ishida, A.; Tani, H.; Harashima, H.; Tokeshi, M. Development of the ILiNP Device: Fine Tuning the Lipid Nanoparticle Size within 10 Nm for Drug Delivery. ACS Omega 2018, 3, 5044–5051. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Luengo, X.F.; Camacho, J.; Faraudo, J. Computer Simulations of Lipid Nanoparticles. Nanomaterials 2017, 7, 461. [Google Scholar] [CrossRef]
- Jing, H.; Wang, Y.; Desai, P.R.; Ramamurthi, K.S.; Das, S. Formation and Properties of a Self-Assembled Nanoparticle-Supported Lipid Bilayer Probed through Molecular Dynamics Simulations. Langmuir 2020, 36, 5524–5533. [Google Scholar] [CrossRef] [PubMed]
- Settanni, G.; Brill, W.; Haas, H.; Schmid, F. PH-Dependent Behavior of Ionizable Cationic Lipids in MRNA-Carrying Lipoplexes Investigated by Molecular Dynamics Simulations. Macromol. Rapid. Commun. 2022, 43, 2100683. [Google Scholar] [CrossRef]
- Bailey-Hytholt, C.M.; Ghosh, P.; Dugas, J.; Zarraga, I.E.; Bandekar, A. Formulating and Characterizing Lipid Nanoparticles for Gene Delivery Using a Microfluidic Mixing Platform. J Vis Exp 2021, 2021. [Google Scholar] [CrossRef]
- Jeffs, L.B.; Palmer, L.R.; Ambegia, E.G.; Giesbrecht, C.; Ewanick, S.; MacLachlan, I. A Scalable, Extrusion-Free Method for Efficient Liposomal Encapsulation of Plasmid DNA. Pharm. Res. 2005, 22, 362–372. [Google Scholar] [CrossRef] [PubMed]
- Rissanou, A.N.; Ouranidis, A.; Karatasos, K. Complexation of Single Stranded RNA with an Ionizable Lipid: An All-Atom Molecular Dynamics Simulation Study. Soft Matter 2020, 16, 6993–7005. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Feng, S.; Ye, Z.; Gao, H.; Lin, J.; Ouyang, D. Prediction of Lipid Nanoparticles for MRNA Vaccines by the Machine Learning Algorithm. Acta Pharm. Sin. B 2022, 12, 2950–2962. [Google Scholar] [CrossRef]
- Nakamura, K.; Aihara, K.; Ishida, T. Importance of Process Parameters Influencing the Mean Diameters of SiRNA-Containing Lipid Nanoparticles (LNPs) on the in Vitro Activity of Prepared LNPs. Biol. Pharm. Bull. 2022, 45, 497–507. [Google Scholar] [CrossRef]
- Ding, A.; Han, B.; Zhang, S.; Huang, Q.; Wang, J.; Wei, C.; Du, Y.; Seifert, H.J. Ab Initio Molecular Dynamics Study on the Disordered Li–Ga–Sn System. Phys. Chem. Chem. Phys. 2022, 24, 10537–10547. [Google Scholar] [CrossRef]
- Sun, G.Y.; Sun, A.Y. Ethanol and Membrane Lipids. Alcohol. Clin. Exp. Res. 1985, 9, 164–180. [Google Scholar] [CrossRef]
- Lundberg, S.; Karlsson, E.; Dahlberg, H.; Glansk, M.; Larsson, S.; Larsson, S.; Carlsson, K. Exosomes and Lipid Nanoparticles—The Future of Targeted Drug Delivery; Uppsala Universitet: Uppsala, Sweden, 2020. [Google Scholar]
- Jia, N.; Lin, S.; Yu, Y.; Zhang, G.; Li, L.; Zheng, D.; Liu, D. The Effects of Ethanol and Rutin on the Structure and Gel Properties of Whey Protein Isolate and Related Mechanisms. Foods 2022, 11, 3480. [Google Scholar] [CrossRef]
- Daragan, V.A.; Voloshin, A.M.; Chochina, S.V.; Khazanovich, T.N.; Wood, W.G.; Avdulov, N.A.; Mayo, K.H. Specific Binding of Ethanol to Cholesterol in Organic Solvents. Biophys. J. 2000, 79, 406–415. [Google Scholar] [CrossRef] [PubMed]
- Dickson, C.J.; Walker, R.C.; Gould, I.R. Lipid21: Complex Lipid Membrane Simulations with AMBER. J. Chem. Theory Comput. 2022, 18, 1726–1736. [Google Scholar] [CrossRef] [PubMed]
- Jakalian, A.; Jack, D.B.; Bayly, C.I. Fast, Efficient Generation of High-Quality Atomic Charges. AM1-BCC Model: II. Parameterization and Validation. J. Comput. Chem. 2002, 23, 1623–1641. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Man, V.H.; Yang, W.; Lee, T.S.; Wang, J. A Fast and High-Quality Charge Model for the next Generation General AMBER Force Field. J. Chem. Phys. 2020, 153, 114502. [Google Scholar] [CrossRef] [PubMed]
- Case Ross, C.; Walker, D.A.; Wang, J.; Robert Duke, T.E. Amber 2019 Reference Manual Principal Contributors to the Current Codes; University of California: San Francisco, CA, USA, 2019. [Google Scholar]
- Malde, A.K.; Zuo, L.; Breeze, M.; Stroet, M.; Poger, D.; Nair, P.C.; Oostenbrink, C.; Mark, A.E. An Automated Force Field Topology Builder (ATB) and Repository: Version 1.0. J. Chem. Theory Comput. 2011, 7, 4026–4037. [Google Scholar] [CrossRef]
- Zgarbová, M.; Otyepka, M.; Šponer, J.; Mládek, A.; Banáš, P.; Cheatham, T.E.; Jurečka, P. Refinement of the Cornell et al. Nucleic Acids Force Field Based on Reference Quantum Chemical Calculations of Glycosidic Torsion Profiles. J. Chem. Theory Comput. 2011, 7, 2886–2902. [Google Scholar] [CrossRef]
- Putral, L.N.; Bywater, M.J.; Gu, W.; Saunders, N.A.; Gabrielli, B.G.; Leggatt, G.R.; McMillan, N.A.J. RNA Interference against Human Papillomavirus Oncogenes in Cervical Cancer Cells Results in Increased Sensitivity to Cisplatin. Mol. Pharm. 2005, 68, 1311–1319. [Google Scholar] [CrossRef]
- Martinez, L.; Andrade, R.; Birgin, E.G.; Martínez, J.M. PACKMOL: A Package for Building Initial Configurations for Molecular Dynamics Simulations. J. Comput. Chem. 2009, 30, 2157–2164. [Google Scholar] [CrossRef]
- Case, D.A.; Belfon, K.; Ben-Shalom, I.Y.; Brozell, S.R.; Cerutti, D.S.; Cheatham, T.E., III; Cruzeiro, V.W.D.; Darden, T.A.; Duke, R.E.; Giambasu, G.; et al. AMBER 2020 2020; University of California: San Francisco, CA, USA, 2019. [Google Scholar]
- Gary, D.J.; Min, J.; Kim, Y.; Park, K.; Won, Y.Y. The Effect of N/P Ratio on the in Vitro and in Vivo Interaction Properties of PEGylated Poly[2-(Dimethylamino)Ethyl Methacrylate]-Based SiRNA Complexes. Macromol. Biosci. 2013, 13, 1059–1071. [Google Scholar] [CrossRef]
- Kubota, K.; Onishi, K.; Sawaki, K.; Li, T.; Mitsuoka, K.; Sato, T.; Takeoka, S. Effect of the Nanoformulation of SiRNA-Lipid Assemblies on Their Cellular Uptake and Immune Stimulation. Int. J. Nanomed. 2017, 12, 5121–5133. [Google Scholar] [CrossRef]
- Weerapol, Y.; Manmuan, S.; Chaothanaphat, N.; Limmatvapirat, S.; Sirirak, J.; Tamdee, P.; Tubtimsri, S. New Approach for Preparing Solid Lipid Nanoparticles with Volatile Oil-Loaded Quercetin Using the Phase-Inversion Temperature Method. Pharmaceutics 2022, 14, 1984. [Google Scholar] [CrossRef] [PubMed]
- Connolly, M.L. Analytical Molecular Surface Calculation. J. Appl. Cryst. 1983, 16, 548–558. [Google Scholar] [CrossRef]
- Dickson, C.J.; Madej, B.D.; Skjevik, Å.A.; Betz, R.M.; Teigen, K.; Gould, I.R.; Walker, R.C. Lipid14: The Amber Lipid Force Field. J. Chem. Theory Comput. 2014, 10, 865–879. [Google Scholar] [CrossRef] [PubMed]
- Ayoub, A.T.; Craddock, T.J.A.; Klobukowski, M.; Tuszynski, J. Analysis of the Strength of Interfacial Hydrogen Bonds between Tubulin Dimers Using Quantum Theory of Atoms in Molecules. Biophys. J. 2014, 107, 740–750. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera--a Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Wickham, H.; Vaughan, D. Girlich M Tidyr: Tidy Messy Data. 2023. Available online: https://tidyr.tidyverse.org (accessed on 1 May 2023).
- Pedersen, T.L. “ggforce”: Accelerating “Ggplot2”. 2021. Available online: https://ggforce.data-imaginist.com (accessed on 1 May 2023).
- Auguie, B. GridExtra: Miscellaneous Functions for Grid Graphics. 2017. Available online: https://cran.r-project.org/web/packages/gridExtra/index.html (accessed on 1 May 2023).
- Kassambara, A. “ggpubr”: “Ggplot2” Based Publication Ready Plots. 2022. Available online: https://cran.r-project.org/web/packages/ggpubr/index.html (accessed on 1 May 2023).
- Pedersen, T.L. Patchwork: The Composer of Plots. 2022. Available online: https://patchwork.data-imaginist.com (accessed on 1 May 2023).
- Granger, B.; Pérez, F. Jupyter: Thinking and Storytelling with Code and Data. Comput. Sci. Eng. 2021, 23, 7–14. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing 2019; R Core Team: Vienna, Austria, 2019. [Google Scholar]
- Project, 2020. Inkscape. Available online: https://inkscape.org (accessed on 1 May 2023).



| Parameter | N/P (5:2) | N/P (10:2) | N/P (15:2) |
|---|---|---|---|
| siRNA | 1 | 1 | 1 |
| SM-102 | 50 | 100 | 150 |
| Cholesterol | 39 | 80 | 120 |
| DSPC | 10 | 20 | 30 |
| Water | 11,352 | 67,358 | 75,800 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hardianto, A.; Muscifa, Z.S.; Widayat, W.; Yusuf, M.; Subroto, T. The Effect of Ethanol on Lipid Nanoparticle Stabilization from a Molecular Dynamics Simulation Perspective. Molecules 2023, 28, 4836. https://doi.org/10.3390/molecules28124836
Hardianto A, Muscifa ZS, Widayat W, Yusuf M, Subroto T. The Effect of Ethanol on Lipid Nanoparticle Stabilization from a Molecular Dynamics Simulation Perspective. Molecules. 2023; 28(12):4836. https://doi.org/10.3390/molecules28124836
Chicago/Turabian StyleHardianto, Ari, Zahra Silmi Muscifa, Wahyu Widayat, Muhammad Yusuf, and Toto Subroto. 2023. "The Effect of Ethanol on Lipid Nanoparticle Stabilization from a Molecular Dynamics Simulation Perspective" Molecules 28, no. 12: 4836. https://doi.org/10.3390/molecules28124836
APA StyleHardianto, A., Muscifa, Z. S., Widayat, W., Yusuf, M., & Subroto, T. (2023). The Effect of Ethanol on Lipid Nanoparticle Stabilization from a Molecular Dynamics Simulation Perspective. Molecules, 28(12), 4836. https://doi.org/10.3390/molecules28124836







