Antibacterial and Cytocompatible pH-Responsive Peptide Hydrogel
Abstract
1. Introduction
2. Results
3. Discussion
4. Materials and Methods
Solubilization of Hydrogel
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Schneider, L.A.; Korber, A.; Grabbe, S.; Dissemond, J. Influence of pH on Wound-healing: A New Perspective for Wound-therapy? Arch. Dermatol. Res. 2007, 298, 413–420. [Google Scholar] [CrossRef]
- Petrof, G.; Abdul-Wahab, A.; McGrath, J.A. Cell therapy in dermatology. Cold Spring Harb. Perspect. Med. 2014, 4, a015156. [Google Scholar] [CrossRef]
- Huang, G.; Li, F.; Zhao, X.; Ma, Y.; Li, Y.; Lin, M.; Jin, G.; Lu, T.J.; Genin, G.M.; Xu, F. Functional and Biomimetic Materials for Engineering of the Three-Dimensional Cell Microenvironment. Chem. Rev. 2017, 117, 12764–12850. [Google Scholar] [CrossRef]
- Nicodemus, G.D.; Bryant, S.J. Cell encapsulation in biodegradable hydrogels for tissue engineering applications. Tissue Eng. Part B. Rev. 2008, 14, 149–165. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Heilshorn, S.C. Adaptable hydrogel networks with reversible linkages for tissue engineering. Adv. Mater. 2015, 27, 3717–3736. [Google Scholar] [CrossRef] [PubMed]
- Guvendiren, M.; Lu, H.D.; Burdick, J.A. Shear-thinning hydrogels for biomedical applications. Soft Matter 2012, 8, 260–272. [Google Scholar] [CrossRef]
- Yan, C.; Pochan, D.J. Rheological properties of peptide-based hydrogels for biomedical and other applications. Chem. Soc. Rev. 2010, 39, 3528–3540. [Google Scholar] [CrossRef]
- Gonzalez-Henriquez, C.M.; Sarabia-Vallejos, M.A.; Rodriguez-Hernandez, J. Advances in the Fabrication of Antimicrobial Hydrogels for Biomedical Applications. Materials 2017, 10, 232. [Google Scholar] [CrossRef]
- Veiga, A.S.; Schneider, J.P. Antimicrobial Hydrogels for the Treatment of Infection. Biopolymers 2013, 100, 637–644. [Google Scholar] [CrossRef]
- Field, F.K.; Kerstein, M.D. Overview of wound healing in a moist environment. Am. J. Surg. 1994, 167, 2S–6S. [Google Scholar] [CrossRef] [PubMed]
- Junker, J.P.; Kamel, R.A.; Caterson, E.J.; Eriksson, E. Clinical Impact Upon Wound Healing and Inflammation in Moist, Wet, and Dry Environments. Adv. Wound Care 2013, 2, 348–356. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Xing, R.; Bai, S.; Yan, X. Recent advances of self-assembling peptide-based hydrogels for biomedical applications. Soft Matter 2019, 15, 1704–1715. [Google Scholar] [CrossRef] [PubMed]
- Ahn, W.; Lee, J.-H.; Kim, S.R.; Lee, J.; Lee, E.J. Designed protein- and peptide-based hydrogels for biomedical sciences. J. Mater. Chem. B. 2021, 9, 1919–1940. [Google Scholar] [CrossRef] [PubMed]
- Jonker, A.M.; Löwik, D.W.P.M.; van Hest, J.C.M. Peptide- and Protein-Based Hydrogels. Chem. Mater. 2012, 24, 759–773. [Google Scholar] [CrossRef]
- Mukherjee, N.; Adak, A.; Ghosh, S. Recent trends in the development of peptide and protein-based hydrogel therapeutics for the healing of CNS injury. Soft Matter 2020, 16, 10046–10064. [Google Scholar] [CrossRef]
- Wang, X.J.; Cheng, J.; Zhang, L.Y.; Zhang, J.G. Self-assembling peptides-based nano-cargos for targeted chemotherapy and immunotherapy of tumors: Recent developments, challenges, and future perspectives. Drug. Deliv. 2022, 1, 1184–1200. [Google Scholar] [CrossRef]
- Mahmood, A.; Patel, D.; Hickson, B.; DesRochers, J.; Hu, X. Recent Progress in Biopolymer-Based Hydrogel Materials for Biomedical Applications. Int. J. Mol. Sci. 2022, 23, 1415. [Google Scholar] [CrossRef]
- Zhang, M.; Zhao, X. Alginate hydrogel dressings for advanced wound management. Int. J. Biol. Macromol. 2020, 162, 1414–1428. [Google Scholar] [CrossRef] [PubMed]
- Gray, V.P.; Amelung, C.D.; Duti, I.J.; Laudermilch, E.G.; Letteri, R.A.; Lampe, K.J. Biomaterials via peptide assembly: Design, characterization, and application in tissue engineering. Acta Biomater. 2022, 140, 43–75. [Google Scholar] [CrossRef]
- Hersel, U.; Dahmen, C.; Kessler, H. RGD modified polymers: Biomaterials for stimulated cell adhesion and beyond. Biomaterials 2003, 24, 4385–4415. [Google Scholar] [CrossRef] [PubMed]
- Datta, D.; Chaudhary, N. Chapter 8—Peptide-based hydrogels for biomedical applications. In Translational Biotechnology; Hasija, Y., Ed.; Academic Press: Cambridge, MA, USA, 2021; pp. 203–232. [Google Scholar] [CrossRef]
- D’Souza, A.; Marshall, L.R.; Yoon, J.; Kulesha, A.; Edirisinghe, D.I.U.; Chandrasekaran, S.; Rathee, P.; Prabhakar, R.; Makhlynets, O.V. Peptide hydrogel with self-healing and redox-responsive properties. Nano Converg. 2022, 9, 18. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Han, Q.; Chen, B.; Zheng, Y.; Zhang, K.; Li, Q.; Wang, J. Antimicrobial hydrogels: Promising materials for medical application. Int. J. Nanomed. 2018, 13, 2217–2263. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.S.; Khademhosseini, A. Advances in engineering hydrogels. Science 2017, 356, eaaf3627. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Zhang, Q.; Zhu, S.; Liu, H.; Chen, J. Preparation and applications of peptide-based injectable hydrogels. RSC Adv. 2019, 9, 28299–28311. [Google Scholar] [CrossRef]
- Makhlynets, O.V.; Caputo, G.A. Characteristics and therapeutic applications of antimicrobial peptides. Biophys. Rev. 2021, 2, 011301. [Google Scholar] [CrossRef]
- Talebian, S.; Mehrali, M.; Taebnia, N.; Pennisi, C.P.; Kadumudi, F.B.; Foroughi, J.; Hasany, M.; Nikkhah, M.; Akbari, M.; Orive, G.; et al. Self-Healing Hydrogels: The Next Paradigm Shift in Tissue Engineering? Adv. Sci. 2019, 6, 1801664. [Google Scholar] [CrossRef] [PubMed]
- Francesko, A.; Petkova, P.; Tzanov, T. Hydrogel Dressings for Advanced Wound Management. Curr. Med. Chem. 2018, 25, 5782–5797. [Google Scholar] [CrossRef]
- Saunders, L.; Ma, P.X. Self-Healing Supramolecular Hydrogels for Tissue Engineering Applications. Macromol. Biosci. 2019, 19, e1800313. [Google Scholar] [CrossRef]
- Cavanagh, P.R.; Lipsky, B.A.; Bradbury, A.W.; Botek, G. Treatment for diabetic foot ulcers. Lancet 2005, 366, 1725–1735. [Google Scholar] [CrossRef]
- Jeffcoate, W.J.; Vileikyte, L.; Boyko, E.J.; Armstrong, D.G.; Boulton, A.J.M. Current Challenges and Opportunities in the Prevention and Management of Diabetic Foot Ulcers. Diabetes Care 2018, 41, 645–652. [Google Scholar] [CrossRef]
- Lindholm, C.; Searle, R. Wound management for the 21st century: Combining effectiveness and efficiency. Int. Wound J. 2016, 13 (Suppl. 2), 5–15. [Google Scholar] [CrossRef] [PubMed]
- Cutrona, K.J.; Kaufman, B.A.; Figueroa, D.M.; Elmore, D.E. Role of arginine and lysine in the antimicrobial mechanism of histone-derived antimicrobial peptides. FEBS Lett. 2015, 589, 3915–3920. [Google Scholar] [CrossRef]
- Kohn, E.M.; Shirley, D.J.; Arotsky, L.; Picciano, A.M.; Ridgway, Z.; Urban, M.W.; Carone, B.R.; Caputo, G.A. Role of Cationic Side Chains in the Antimicrobial Activity of C18G. Molecules 2018, 23, 329. [Google Scholar] [CrossRef]
- Yang, C.H.; Chen, Y.C.; Peng, S.Y.; Tsai, A.P.; Lee, T.J.; Yen, J.H.; Liou, J.W. An engineered arginine-rich alpha-helical antimicrobial peptide exhibits broad-spectrum bactericidal activity against pathogenic bacteria and reduces bacterial infections in mice. Sci. Rep. 2018, 8, 14602. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Zeng, X.; Wen, X. Design and Synthesis of New Cationic Antimicrobial Peptides with Low Cytotoxicity. Int. J. Pept. Res. Ther. 2021, 27, 831–840. [Google Scholar] [CrossRef]
- Tomar, D.; Chaudhary, S.; Jena, K.C. Self-assembly of l-phenylalanine amino acid: Electrostatic induced hindrance of fibril formation. RSC Adv. 2019, 9, 12596–12605. [Google Scholar] [CrossRef] [PubMed]
- Rajagopal, K.; Lamm, M.S.; Haines-Butterick, L.A.; Pochan, D.J.; Schneider, J.P. Tuning the pH Responsiveness of β-Hairpin Peptide Folding, Self-Assembly, and Hydrogel Material Formation. Biomacromolecules 2009, 10, 2619–2625. [Google Scholar] [CrossRef]
- Andersson, D.I.; Hughes, D.; Kubicek-Sutherland, J.Z. Mechanisms and consequences of bacterial resistance to antimicrobial peptides. Drug. Resist. Updat. 2016, 26, 43–57. [Google Scholar] [CrossRef]
- Exner, M.; Bhattacharya, S.; Christiansen, B.; Gebel, J.; Goroncy-Bermes, P.; Hartemann, P.; Heeg, P.; Ilschner, C.; Kramer, A.; Larson, E.; et al. Antibiotic resistance: What is so special about multidrug-resistant Gram-negative bacteria? GMS Hyg. Infect. Control. 2017, 12, Doc05. [Google Scholar] [CrossRef]
- Blair, J.M.; Webber, M.A.; Baylay, A.J.; Ogbolu, D.O.; Piddock, L.J. Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 2015, 13, 42–51. [Google Scholar] [CrossRef]
- Epand, R.M.; Walker, C.; Epand, R.F.; Magarvey, N.A. Molecular mechanisms of membrane targeting antibiotics. Biochim. Et. Biophys. Acta -Biomembr. 2016, 1858, 980–987. [Google Scholar] [CrossRef] [PubMed]
- Chan, D.I.; Prenner, E.J.; Vogel, H.J. Tryptophan- and arginine-rich antimicrobial peptides: Structures and mechanisms of action. Biochim. Biophys. Acta 2006, 1758, 1184–1202. [Google Scholar] [CrossRef]
- Jing, W.; Demcoe, A.R.; Vogel, H.J. Conformation of a bactericidal domain of puroindoline a: Structure and mechanism of action of a 13-residue antimicrobial peptide. J. Bacteriol. 2003, 185, 4938–4947. [Google Scholar] [CrossRef] [PubMed]
- Larsen, T.J.; Salick, D.A.; Nagarkar, R.; Schneider, J. Inherently Antibacterial Hydrogels: Altering Activity via Tryptophan/Arginine Interactions. FASEB J. 2009, 23, 863-14. [Google Scholar] [CrossRef]
- Moroz, Y.S.; Binder, W.; Nygren, P.; Caputo, G.A.; Korendovych, I.V. Painting proteins blue: Beta-(1-azulenyl)-L-alanine as a probe for studying protein-protein interactions. Chem. Commun. 2013, 49, 490–492. [Google Scholar] [CrossRef]
- D’Souza, A.R.; Necelis, M.R.; Kulesha, A.; Caputo, G.A.; Makhlynets, O.V. Beneficial Impacts of Incorporating the Non-Natural Amino Acid Azulenyl-Alanine into the Trp-Rich Antimicrobial Peptide buCATHL4B. Biomolecules 2021, 11, 421. [Google Scholar] [CrossRef]
- Pubell, B. Wound healing and the role of fibroblasts. J. Wound Care 2013, 22, 407–412. [Google Scholar] [CrossRef]
- Bellis, S.L. Advantages of RGD peptides for directing cell association with biomaterials. Biomaterials 2011, 32, 4205–4210. [Google Scholar] [CrossRef]
- Pierschbacher, M.D.; Ruoslahti, E. Cell attachment activity of fibronectin can be duplicated by small synthetic fragments of the molecule. Nature 1984, 309, 30–33. [Google Scholar] [CrossRef]
- Ruoslahti, E. RGD and other recognition sequences for integrins. Annu. Rev. Cell. Dev. Biol. 1996, 12, 697–715. [Google Scholar] [CrossRef] [PubMed]
- Hern, D.L.; Hubbell, J.A. Incorporation of adhesion peptides into nonadhesive hydrogels useful for tissue resurfacing. J. Biomed. Mater. Res. 1998, 39, 266–276. [Google Scholar] [CrossRef]
- Zhou, M.; Ulijn, R.V.; Gough, J.E. Extracellular matrix formation in self-assembled minimalistic bioactive hydrogels based on aromatic peptide amphiphiles. J. Tissue Eng. 2014, 5, 1593. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Smith, A.M.; Das, A.K.; Hodson, N.W.; Collins, R.F.; Ulijn, R.V.; Gough, J.E. Self-assembled peptide-based hydrogels as scaffolds for anchorage-dependent cells. Biomaterials 2009, 30, 2523–2530. [Google Scholar] [CrossRef] [PubMed]
- Storrie, H.; Guler, M.O.; Abu-Amara, S.N.; Volberg, T.; Rao, M.; Geiger, B.; Stupp, S.I. Supramolecular crafting of cell adhesion. Biomaterials 2007, 28, 4608–4618. [Google Scholar] [CrossRef] [PubMed]
- Guler, M.O.; Hsu, L.; Soukasene, S.; Harrington, D.A.; Hulvat, J.F.; Stupp, S.I. Presentation of RGDS epitopes on self-assembled nanofibers of branched peptide amphiphiles. Biomacromolecules 2006, 7, 1855–1863. [Google Scholar] [CrossRef]
- Li, Y.; Rodrigues, J.; Tomas, H. Injectable and biodegradable hydrogels: Gelation, biodegradation and biomedical applications. Chem. Soc. Rev. 2012, 41, 2193–2221. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.W.; Park, Y.J.; Lee, S.J.; Lee, S.K.; Lee, K.Y. The effect of spacer arm length of an adhesion ligand coupled to an alginate gel on the control of fibroblast phenotype. Biomaterials 2010, 31, 5545–5551. [Google Scholar] [CrossRef] [PubMed]
- Menger, F.M.; Caran, K.L. Anatomy of a Gel. Amino Acid Derivatives that Rigidify Water at Submillimolar Concentration. J. Am. Chem. Soc. 2000, 122, 11679–11691. [Google Scholar] [CrossRef]
- Lansdown, A.B. Calcium: A potential central regulator in wound healing in the skin. Wound Repair. Regen. 2002, 10, 271–285. [Google Scholar] [CrossRef]
- Lange, T.S.; Bielinsky, A.K.; Kirchberg, K.; Bank, I.; Herrmann, K.; Krieg, T.; Scharffetter-Kochanek, K. Mg2+ and Ca2+ differentially regulate beta 1 integrin-mediated adhesion of dermal fibroblasts and keratinocytes to various extracellular matrix proteins. Exp. Cell. Res. 1994, 214, 381–388. [Google Scholar] [CrossRef]
- Grzesiak, J.J.; Pierschbacher, M.D. Shifts in the concentrations of magnesium and calcium in early porcine and rat wound fluids activate the cell migratory response. J. Clin. Investig. 1995, 95, 227–233. [Google Scholar] [CrossRef]
- Lin, B.F.; Megley, K.A.; Viswanathan, N.; Krogstad, D.V.; Drews, L.B.; Kade, M.J.; Qian, Y.; Tirrell, M.V. pH-responsive branched peptide amphiphile hydrogel designed for applications in regenerative medicine with potential as injectable tissue scaffolds. J. Mater. Chem. 2012, 22, 19447–19454. [Google Scholar] [CrossRef]
- Hatip Koc, M.; Cinar Ciftci, G.; Baday, S.; Castelletto, V.; Hamley, I.W.; Guler, M.O. Hierarchical Self-Assembly of Histidine-Functionalized Peptide Amphiphiles into Supramolecular Chiral Nanostructures. Langmuir 2017, 33, 7947–7956. [Google Scholar] [CrossRef]
- Aguado, B.A.; Mulyasasmita, W.; Su, J.; Lampe, K.J.; Heilshorn, S.C. Improving viability of stem cells during syringe needle flow through the design of hydrogel cell carriers. Tissue Eng. Part A 2012, 18, 806–815. [Google Scholar] [CrossRef]
- Wang, Y.; He, X.; Bruggeman, K.F.; Gayen, B.; Tricoli, A.; Lee, W.M.; Williams, R.J.; Nisbet, D.R. Peptide programmed hydrogels as safe sanctuary microenvironments for cell transplantation. Adv. Funct. Mater. 2020, 30, 1900390. [Google Scholar] [CrossRef]
- Madl, A.C.; Madl, C.M.; Myung, D. Injectable Cucurbit[8]uril-Based Supramolecular Gelatin Hydrogels for Cell Encapsulation. ACS Macro Lett. 2020, 9, 619–626. [Google Scholar] [CrossRef]
- Wei, Z.; Yang, J.H.; Liu, Z.; Xu, F.; Zhou, J.X.; Zrínyi, M.; Osada, Y.; Chen, Y.M. Novel Biocompatible Polysaccharide-Based Self-Healing Hydrogel. Adv. Funct. Mater. 2015, 25, 421. [Google Scholar] [CrossRef]
- Tang, J.D.; Roloson, E.B.; Amelung, C.D.; Lampe, K.J. Rapidly Assembling Pentapeptides for Injectable Delivery (RAPID) Hydrogels as Cytoprotective Cell Carriers. ACS Biomater. Sci. Eng. 2019, 5, 2117–2121. [Google Scholar] [CrossRef] [PubMed]
- Amer, M.H.; Felicity, R.A.J.R.; Shakesheff, K.M.; Modo, M.; White, L.J. Translational considerations in injectable cell-based therapeutics for neurological applications: Concepts, progress and challenges. NPJ Regen. Med. 2017, 2, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Kuipers, B.J.H.; Gruppen, H. Prediction of Molar Extinction Coefficients of Proteins and Peptides Using UV Absorption of the Constituent Amino Acids at 214 nm To Enable Quantitative Reverse Phase High-Performance Liquid Chromatography−Mass Spectrometry Analysis. J. Agric. Food Chem. 2007, 55, 5445–5451. [Google Scholar] [CrossRef]
- Bowerman, C.J.; Liyanage, W.; Federation, A.J.; Nilsson, B.L. Tuning Beta-sheet Peptide Self-assembly and Hydrogelation Behavior by Modification of Sequence Hydrophobicity and Aromaticity. Biomacromolecules 2011, 12, 2735–2745. [Google Scholar] [CrossRef] [PubMed]
- Ozbas, B.; Kretsinger, J.; Rajagopal, K.; Schneider, J.P.; Pochan, D.J. Salt-triggered Peptide Folding and Consequent Self-assembbly into Hydrogels with Tunable Modulus. Macromolecules 2004, 37, 7331–7337. [Google Scholar] [CrossRef]
- Caplan, M.R.; Schwartzfarb, E.M.; Zhang, S.; Kamm, R.D.; Lauffenburger, D.A. Control of Self-assembling Oligopeptide Matrix Formation Through Systematic Variation of Amino Acid Sequence. Biomaterials 2002, 23, 219–227. [Google Scholar] [CrossRef]
- D’Souza, A.; Yoon, J.H.; Beaman, H.; Gosavi, P.; Lengyel-Zhand, Z.; Sternisha, A.; Centola, G.; Marshall, L.R.; Wehrman, M.D.; Schultz, K.M.; et al. Nine-Residue Peptide Self-Assembles in the Presence of Silver to Produce a Self-Healing, Cytocompatible, Antimicrobial Hydrogel. ACS Appl. Mater. Interfaces 2020, 12, 17091–17099. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Wang, L.; Li, Y.; Huang, Z.; Luo, S.; He, Y.; Han, H.; Raza, F.; Wu, J.; Ge, L. Injectable pH and redox dual responsive hydrogels based on self-assembled peptides for anti-tumor drug delivery. Biomater Sci 2020, 8, 5415–5426. [Google Scholar] [CrossRef]
- Ghosh, G.; Barman, R.; Sarkar, J.; Ghosh, S. pH-Responsive Biocompatible Supramolecular Peptide Hydrogel. J. Phys. Chem. B 2019, 123, 5909–5915. [Google Scholar] [CrossRef]
- Raza, F.; Zhu, Y.; Chen, L.; You, X.; Zhang, J.; Khan, A.; Khan, M.W.; Hasnat, M.; Zafar, H.; Wu, J.; et al. Paclitaxel-loaded pH responsive hydrogel based on self-assembled peptides for tumor targeting. Biomater. Sci. 2019, 7, 2023–2036. [Google Scholar] [CrossRef]
- Kaur, H.; Sharma, P.; Patel, N.; Pal, V.K.; Roy, S. Accessing Highly Tunable Nanostructured Hydrogels in a Short Ionic Complementary Peptide Sequence via pH Trigger. Langmuir 2020, 36, 12107–12120. [Google Scholar] [CrossRef]
- Liu, Y.; Ran, Y.; Ge, Y.; Raza, F.; Li, S.; Zafar, H.; Wu, Y.; Paiva-Santos, A.C.; Yu, C.; Sun, M.; et al. pH-Sensitive Peptide Hydrogels as a Combination Drug Delivery System for Cancer Treatment. Pharmaceutics 2022, 14. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, S.; Nishimura, K.; Morita, K.; Kanemitsu, S.; Nishida, Y.; Morimoto, T.; Aoi, T.; Tamura, A.; Maruyama, T. Microenvironment pH-Induced Selective Cell Death for Potential Cancer Therapy Using Nanofibrous Self-Assembly of a Peptide Amphiphile. Biomacromolecules 2021, 22, 2524–2531. [Google Scholar] [CrossRef]
- Schneider, J.P.; Pochan, D.J.; Ozbas, B.; Rajagopal, K.; Pakstis, L.; Kretsinger, J. Responsive Hydrogels from the Intramolecular Folding and Self-Assembly of a Designed Peptide. J. Am. Chem. Soc. 2002, 124, 15030–15037. [Google Scholar] [CrossRef] [PubMed]
- Dexter, A.F.; Fletcher, N.L.; Creasey, R.G.; Filardo, F.; Boehm, M.W.; Jack, K.S. Fabrication and characterization of hydrogels formed from designer coiled-coil fibril-forming peptides. RSC Advances 2017, 7, 27260–27271. [Google Scholar] [CrossRef]
- Liu, Y.; Yang, Y.; Wang, C.; Zhao, X. Stimuli-responsive self-assembling peptides made from antibacterial peptides. Nanoscale 2013, 5, 6413–6421. [Google Scholar]
- Bortolini, C.; Liu, L.; Hoffmann, S.V.; Jones, N.C.; Knowles, T.P.J.; Dong, M. Exciton Coupling of Phenylalanine Reveals Conformational Changes of Cationic Peptides. ChemistrySelect 2017, 2, 2476–2479. [Google Scholar] [CrossRef]
- Decandio, C.C.; Silva, E.R.; Hamley, I.W.; Castelletto, V.; Liberato, M.S.; Oliveira, V.X.; Oliveira, C.L.P.; Alves, W.A. Self-Assembly of a Designed Alternating Arginine/Phenylalanine Oligopeptide. Langmuir 2015, 31, 4513–4523. [Google Scholar] [CrossRef]
- Perween, S.; Chandanshive, B.; Kotamarthi, H.C.; Khushalani, D. Single amino acid based self-assembled structure. Soft Matter 2013, 9, 10141–10145. [Google Scholar] [CrossRef]
- Krysmann, M.J.; Castelletto, V.; Kelarakis, A.; Hamley, I.W.; Hule, R.A.; Pochan, D.J. Self-Assembly and Hydrogelation of an Amyloid Peptide Fragment. Biochemistry 2008, 47, 4597–4605. [Google Scholar] [CrossRef] [PubMed]
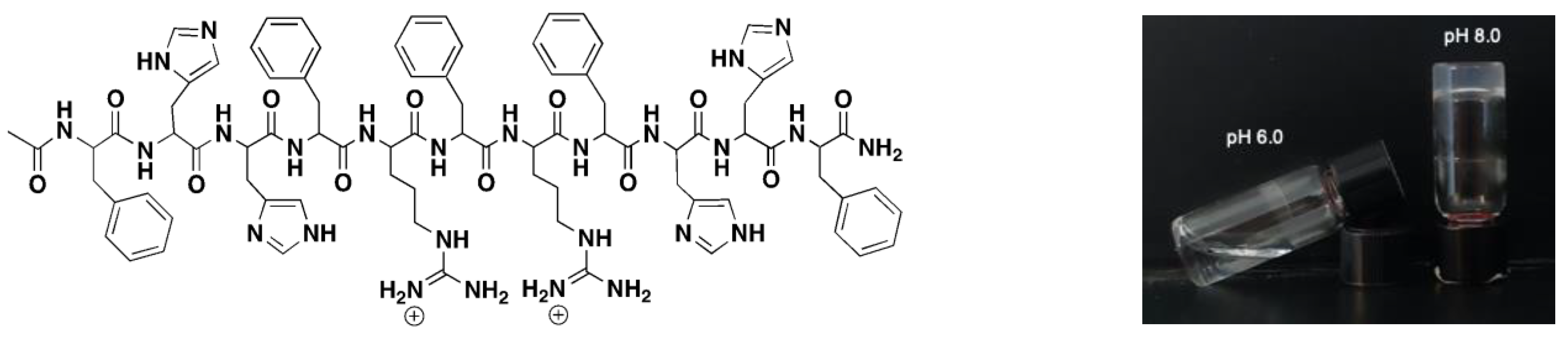
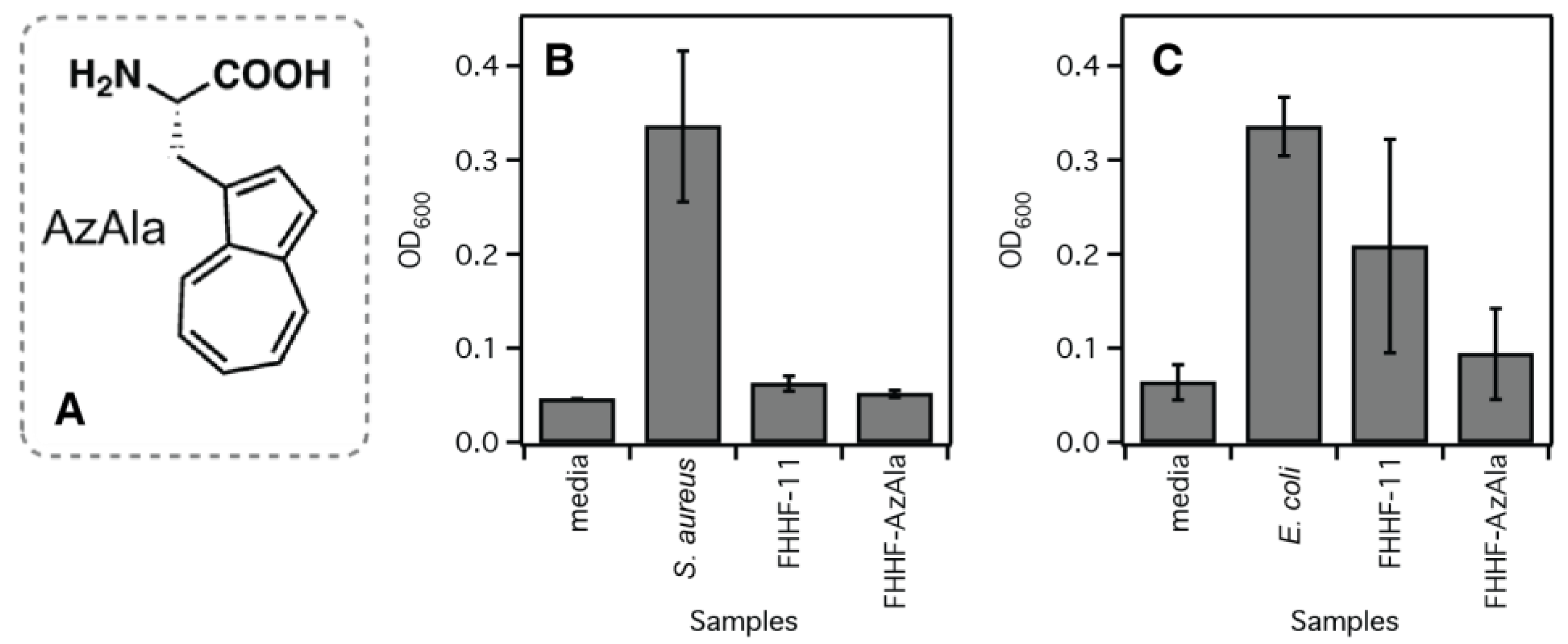
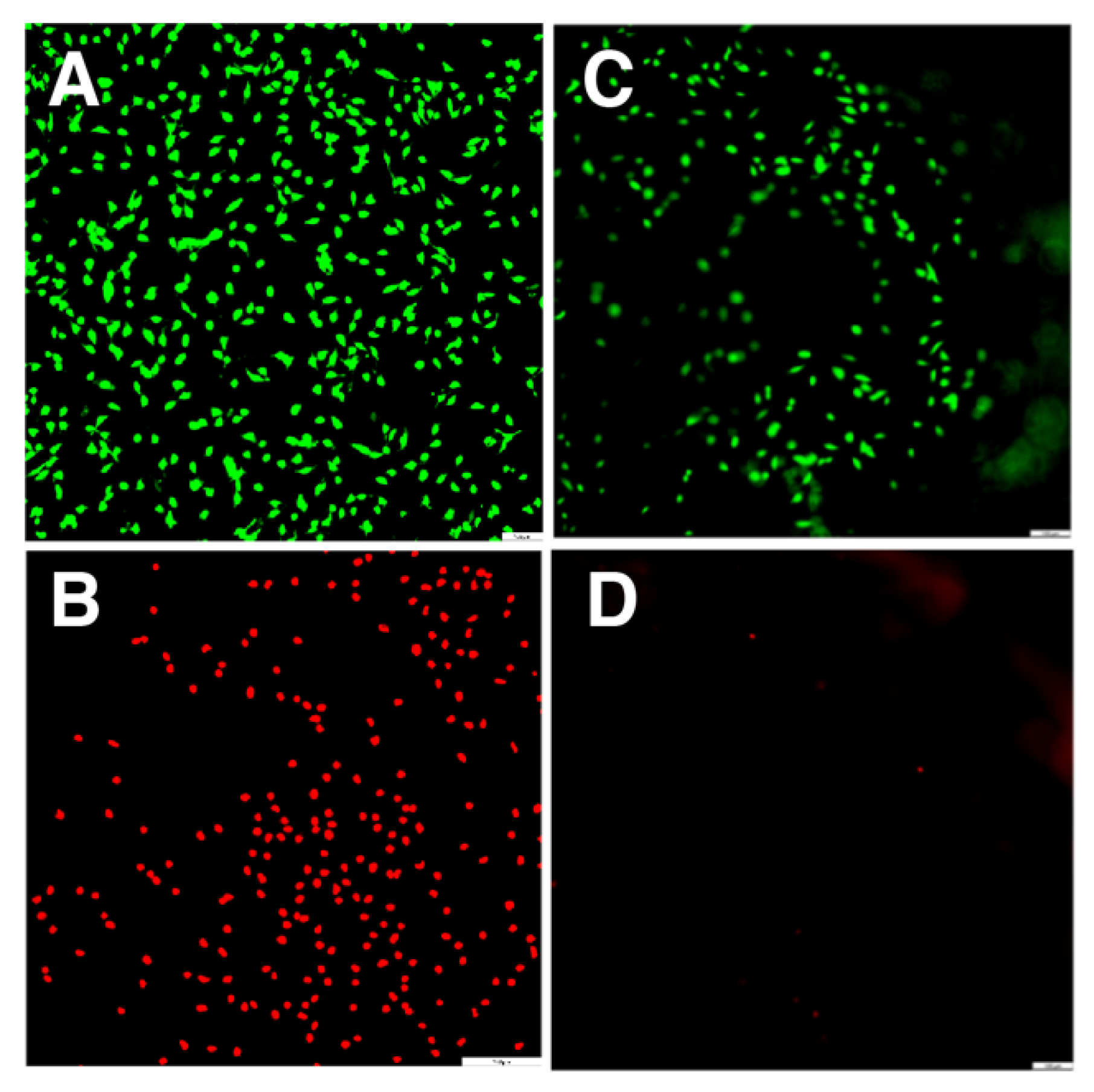
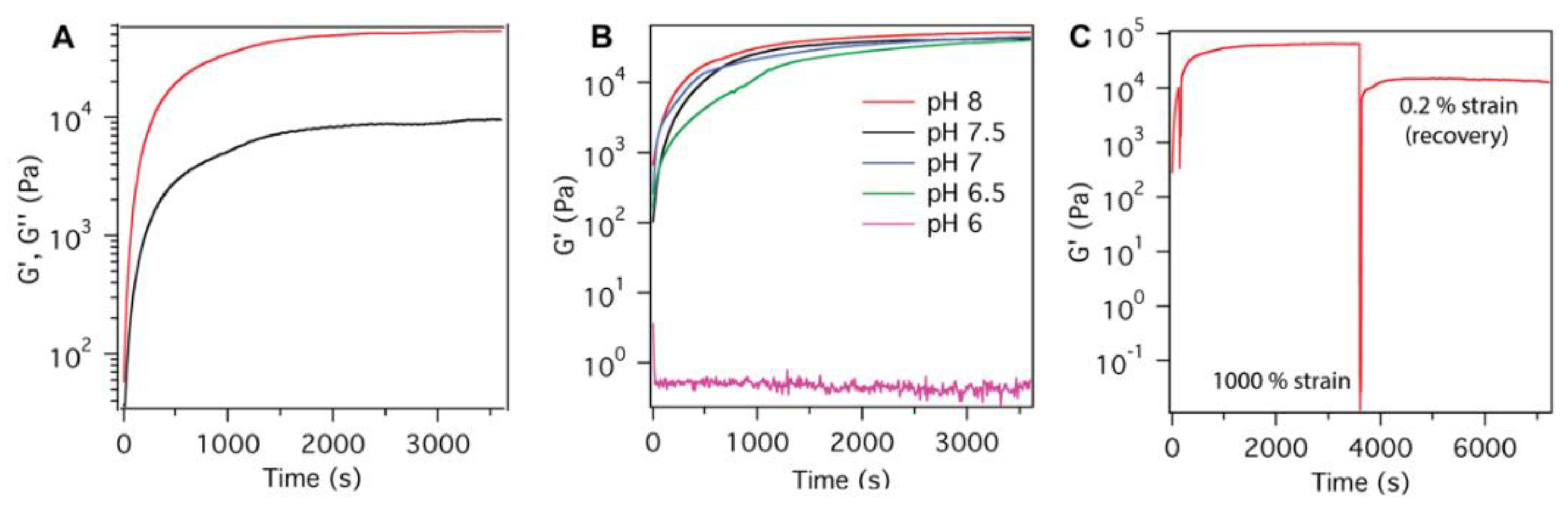

| Peptide Name | pH | Peptide Sequence | Shear Modulus G′ (Pa) |
|---|---|---|---|
| FHHF-11 | 8 | Ac-FHHFRFRFHHF-CONH2 | 51,466 ± 4800 |
| 7.5 | 42,000 ± 6850 | ||
| 7 | 40,100 ± 2250 | ||
| 6.5 | 35,350 ± 4950 | ||
| 6 | 0.7 ± 0.2 | ||
| FHHF-K | 8 | Ac-FHHFKFKFHHF-CONH2 | 94,000 ± 9000 |
| 7.5 | 89,200 ± 28,800 | ||
| 7 | 67,050 ± 10,550 | ||
| 6.5 | 25,350 ± 10,750 | ||
| 6 | 0.3 ± 0.1 | ||
| No-Ac FHHF-11 | 8 | NH2-FHHFRFRFHHF-CONH2 | 940 ± 340 |
| COOH FHHF-11 | 8 | Ac-FHHFRFRFHHF-COOH | 0 |
| HHF | 8 | Ac-HHFRFRFRFHH-CONH2 | 30 |
| HFH | 8 | Ac-HFHFRFRFHFH-CONH2 | 65 |
| FHFH | 8 | Ac-FHFHFRFRFHFHF-CONH2 | 60 |
| FHHF-GRGD | 8 | Ac-FHHFRFRFHHFGRGD-CONH2 | 2620 ±360 |
| W substitutions | 8 | Ac-WHHFRFRFHHF-CONH2 | 865 ± 126 |
| 8 | Ac-FHHFRFRFHHW-CONH2 | 125 ± 75 | |
| 8 | Ac-FHWFRFRFHHF-CONH2 | 19 ± 9 | |
| 8 | Ac-FHHFRWRFHHF-CONH2 | 5361 ± 3448 | |
| 8 | Ac-FHHFKWKFHHF-CONH2 | 4140 ± 2356 | |
| 8 | Ac-FHHFRFWFHHF-CONH2 | 435 ± 339 | |
| 8 | Ac-FHHFWFRFHHF-CONH2 | 205 ± 168 | |
| AzAla substitutions | 8 | Ac-FHHFR(AzAla)RFHHF-CONH2 | 10,520 ± 3790 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Edirisinghe, D.I.U.; D’Souza, A.; Ramezani, M.; Carroll, R.J.; Chicón, Q.; Muenzel, C.L.; Soule, J.; Monroe, M.B.B.; Patteson, A.E.; Makhlynets, O.V. Antibacterial and Cytocompatible pH-Responsive Peptide Hydrogel. Molecules 2023, 28, 4390. https://doi.org/10.3390/molecules28114390
Edirisinghe DIU, D’Souza A, Ramezani M, Carroll RJ, Chicón Q, Muenzel CL, Soule J, Monroe MBB, Patteson AE, Makhlynets OV. Antibacterial and Cytocompatible pH-Responsive Peptide Hydrogel. Molecules. 2023; 28(11):4390. https://doi.org/10.3390/molecules28114390
Chicago/Turabian StyleEdirisinghe, Dona Imanga Upamadi, Areetha D’Souza, Maryam Ramezani, Robert J. Carroll, Quenten Chicón, Cheyene L. Muenzel, Jonathan Soule, Mary Beth Browning Monroe, Alison E. Patteson, and Olga V. Makhlynets. 2023. "Antibacterial and Cytocompatible pH-Responsive Peptide Hydrogel" Molecules 28, no. 11: 4390. https://doi.org/10.3390/molecules28114390
APA StyleEdirisinghe, D. I. U., D’Souza, A., Ramezani, M., Carroll, R. J., Chicón, Q., Muenzel, C. L., Soule, J., Monroe, M. B. B., Patteson, A. E., & Makhlynets, O. V. (2023). Antibacterial and Cytocompatible pH-Responsive Peptide Hydrogel. Molecules, 28(11), 4390. https://doi.org/10.3390/molecules28114390






