MicroRNA Regulation in Infectious Diseases and Its Potential as a Biosensor in Future Aquaculture Industry: A Review
Abstract
1. Introduction
2. MicroRNA Biogenesis and General Mechanism
3. MicroRNA Regulation in Aquaculture Responses to Infectious Diseases
3.1. Innate Immunity
3.1.1. Autophagy
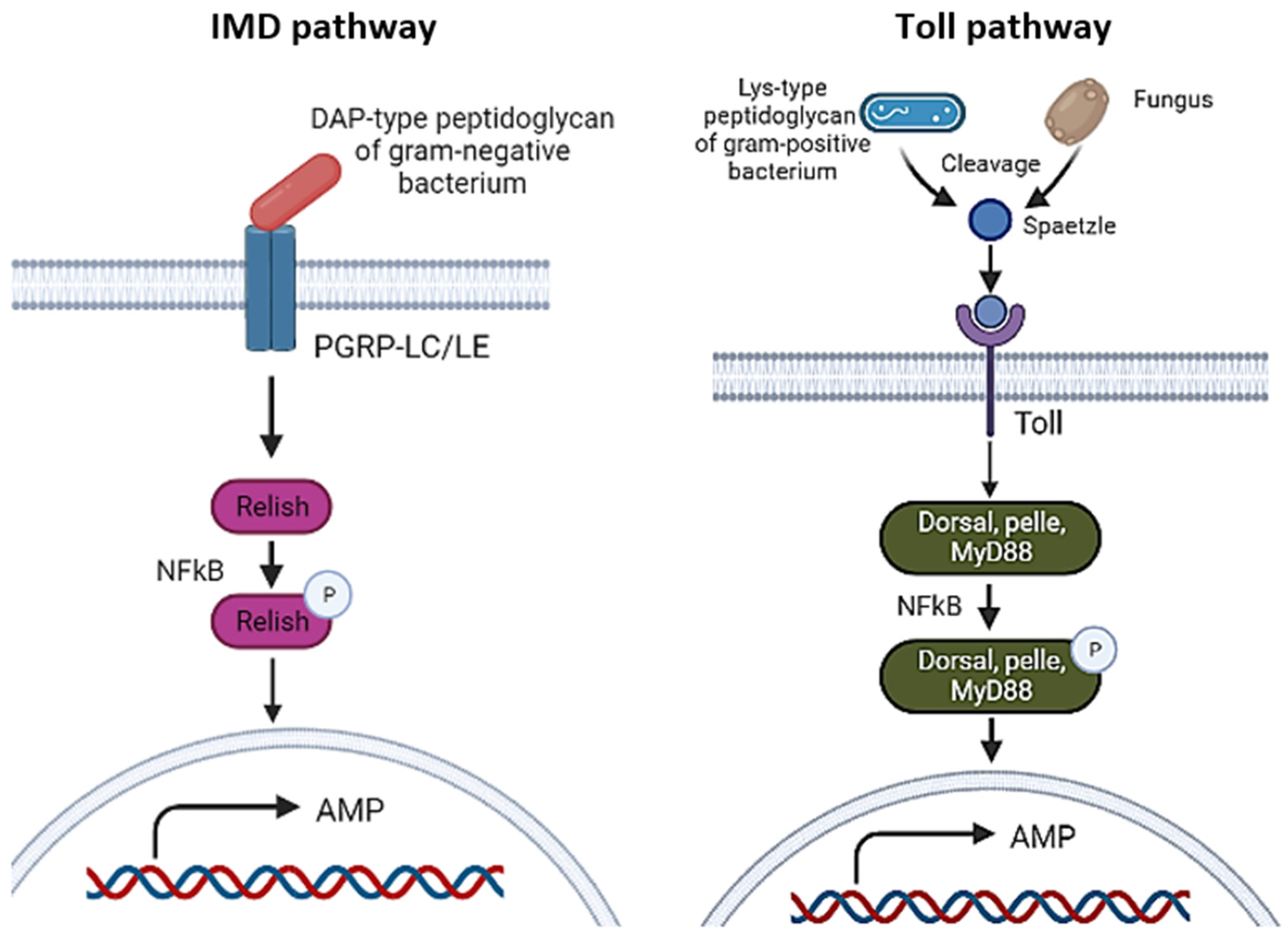
3.1.2. Toll and IMD Pathways
3.1.3. Vascular Endothelial Growth Factor (VEGF) Pathway
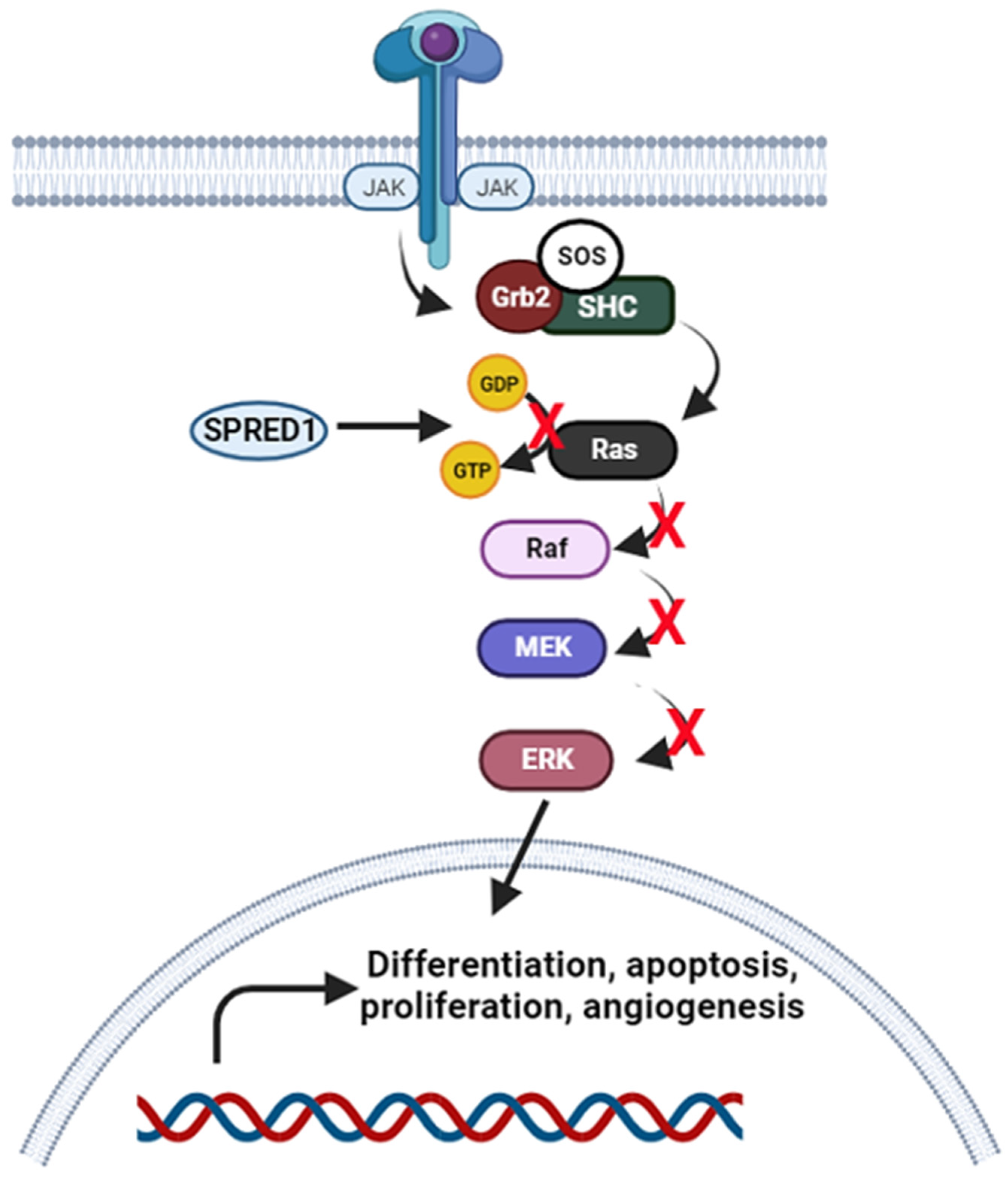
3.1.4. Janus Kinase-Signal Transducer and Activator of Transcription (JAK/STAT) Signalling Pathway
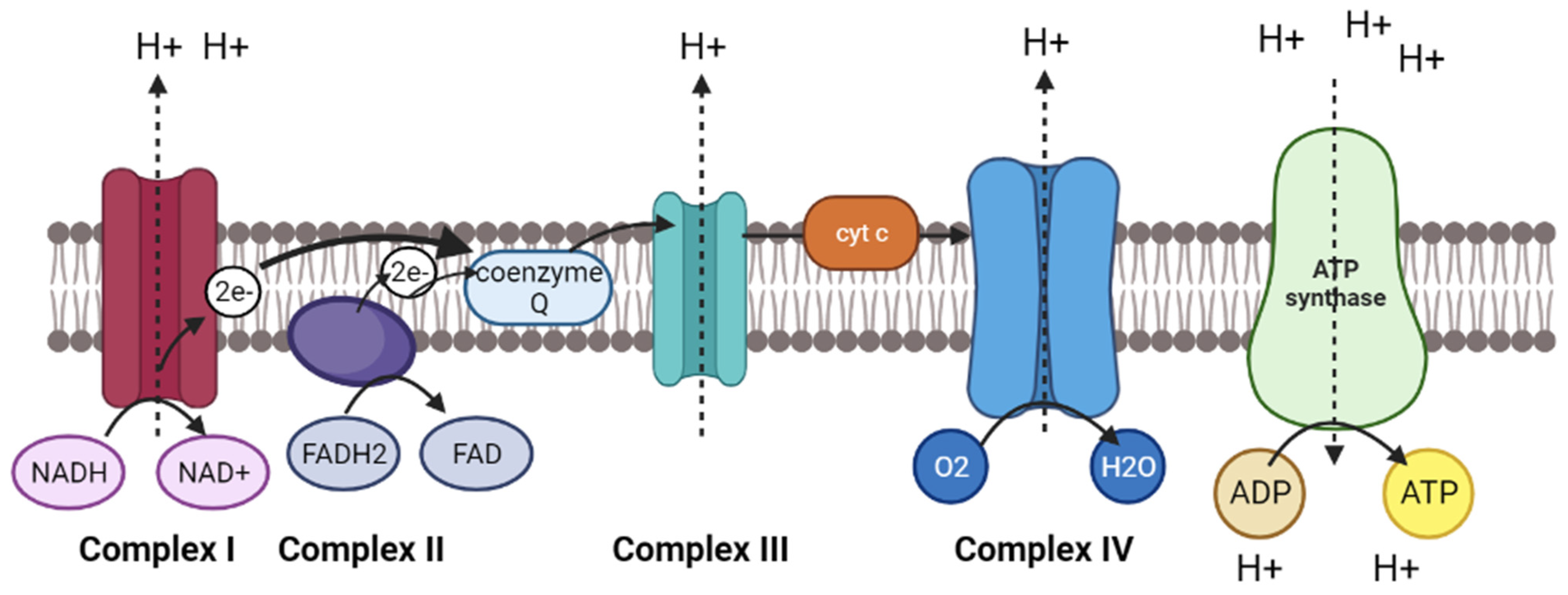
3.2. Oxidative Phosphorylation Pathway
3.3. Apoptosis
4. MicroRNAs Enhance Replication of Pathogens in Host
5. Current Applications of MicroRNAs
5.1. MicroRNAs Used as Biomarkers
5.2. MicroRNAs Used as Therapeutic Agents
6. Current Trends of miRNA and Its Future Perspectives Used in Aquaculture
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Global Seafood Alliance. What Is Aquaculture and Why Do We Need It? 2019. Available online: https://www.globalseafood.org/blog/what-is-aquaculture-why-do-we-need-it/ (accessed on 19 April 2023).
- FAO. The State of World Fisheries and Aquaculture 2022. Towards Blue Transformation; FAO: Rome, Italy, 2022. [Google Scholar]
- Kibenge, F.S. Emerging viruses in aquaculture. Curr. Opin. Virol. 2019, 34, 97–103. [Google Scholar] [CrossRef]
- Watanabe, S.; Sakami, T. Problems and challenges of aquaculture in Japan. In Proceedings of the International Workshop on the Promotion of Sustainable Aquaculture, Aquatic Animal Health, and Resource Enhancement in Southeast Asia, Aquaculture Department, Southeast Asian Fisheries Development Center. Iloilo City, Philippines, 25–27 June 2019; 2021; pp. 64–69. [Google Scholar]
- Sundaray, J.K.; Dixit, S.; Rather, A.; Rasal, K.D.; Sahoo, L. Aquaculture omics: An update on the current status of research and data analysis. Mar. Genom. 2022, 64, 100967. [Google Scholar] [CrossRef]
- Wang, K.C.; Chang, H.Y. Molecular Mechanisms of Long Noncoding RNAs. Mol. Cell 2011, 43, 904–914. [Google Scholar] [CrossRef]
- Deng, Q.; Zhao, N.; Zhu, C.; Zhang, B. Long non-coding RNAs in the physiology of aquaculture animals: A perspective update. Rev. Fish Biol. Fish. 2022, 32, 1103–1122. [Google Scholar] [CrossRef]
- Zhou, Z.; Wang, Z.; Long, L.; Lv, Y.; Gao, Z.; Wang, Y.; Li, P. The potential regulatory role of the lncRNA-miRNA-mRNA axis in teleost fish. Front. Immunol. 2023, 14, 723. [Google Scholar] [CrossRef]
- Xu, X.; Liang, Y.; Gareev, I.; Liang, Y.; Liu, R.; Wang, N.; Yang, G. LncRNA as potential biomarker and therapeutic target in glioma. Mol. Biol. Rep. 2023, 50, 841–851. [Google Scholar] [CrossRef]
- Gao, N.; Li, Y.; Li, J.; Gao, Z.; Yang, Z.; Li, Y.; Liu, H.; Fan, T. Long Non-Coding RNAs: The Regulatory Mechanisms, Research Strategies, and Future Directions in Cancers. Front. Oncol. 2020, 10, 598817. [Google Scholar] [CrossRef]
- Ou, J.; Chen, H.; Luan, X.; Ju, R.; Sun, Y.; Zhang, B.; Bian, Y.; Meng, Y.; Ji, H.; Wang, Z.; et al. Leveraging lncRNA-miRNA-mRNA network to reveal anti-Spiroplasma eriocheiris infection mechanisms in Macrobrachium nipponense. Aquaculture 2022, 557, 738286. [Google Scholar] [CrossRef]
- Zheng, W.; Cui, J.; Lv, X.; Xin, S.; Xu, T. LncRNA MIR144HG-derived miR-144 suppresses antibacterial signaling and facilitates bacteria escape in fish. Water Biol. Secur. 2023, 2, 100093. [Google Scholar] [CrossRef]
- Chen, Y.; Wan, S.; Li, Q.; Dong, X.; Diao, J.; Liao, Q.; Wang, G.-Y.; Gao, Z.-X. Genome-Wide Integrated Analysis Revealed Functions of lncRNA–miRNA–mRNA Interaction in Growth of Intermuscular Bones in Megalobrama amblycephala. Front. Cell Dev. Biol. 2021, 8, 603815. [Google Scholar] [CrossRef]
- Cui, J.; Zheng, W.; Xu, T.; Sun, Y. Long Noncoding RNA MIR122HG Inhibits MAVS-Mediated Antiviral Immune Response by Deriving miR-122 in Miiuy Croaker (Miichthys miiuy). Viruses 2022, 14, 930. [Google Scholar] [CrossRef]
- Li, T.; Mo, X.; Fu, L.; Xiao, B.; Guo, J. Molecular mechanisms of long noncoding RNAs on gastric cancer. Oncotarget 2016, 7, 8601–8612. [Google Scholar] [CrossRef]
- Rasal, K.D.; Nandanpawar, P.C.; Swain, P.; Badhe, M.R.; Sundaray, J.K.; Jayasankar, P. MicroRNA in aquaculture fishes: A way forward with high-throughput sequencing and a computational approach. Rev. Fish Biol. Fish. 2016, 26, 199–212. [Google Scholar] [CrossRef]
- Cao, Q.; Zhang, H.; Li, T.; He, L.; Zong, J.; Shan, H.; Huang, L.; Zhang, Y.; Liu, H.; Jiang, J. Profiling miRNAs of Teleost Fish in Responses to Environmental Stress: A Review. Biology 2023, 12, 388. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, Z.; Xu, S.; Zhang, X.; Liu, X. Transcriptome-wide identification and characterization of the Macrobrachium rosenbergii microRNAs potentially related to immunity against non-O1 Vibrio cholerae infection. Fish Shellfish Immunol. 2023, 135, 108692. [Google Scholar] [CrossRef]
- Fu, C.; Fu, X.; Li, F.; Li, Z.; Wang, A.; Jiang, S.; Liu, C.; Wang, H. Integrated microRNA-mRNA analysis reveals a possible molecular mechanism of enteritis susceptibility in Litopenaeus vannamei. Fish Shellfish Immunol. 2023, 136, 108699. [Google Scholar] [CrossRef]
- Zhang, J.; Sun, S.; Mao, Y.; Qiao, G.; Li, Q. Identification and analysis of differentially expressed microRNAs in gibel carp Carassius auratus gibelio responding to Polyinosinic-polycytidylic acid (poly I:C) stimulation. Fish Shellfish Immunol. Rep. 2023, 4, 100083. [Google Scholar] [CrossRef]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef]
- Vasudevan, S. Posttranscriptional Upregulation by MicroRNAs. Wiley Interdiscip. Rev. RNA 2012, 3, 311–330. [Google Scholar] [CrossRef]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Wightman, B.; Ha, I.; Ruvkun, G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 1993, 75, 855–862. [Google Scholar] [CrossRef]
- Pasquinelli, A.E.; Reinhart, B.J.; Slack, F.; Martindale, M.Q.; Kuroda, M.I.; Maller, B.; Hayward, D.C.; Ball, E.E.; Degnan, B.; Müller, P.; et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature 2000, 408, 86–89. [Google Scholar] [CrossRef]
- Li, S.-C.; Chan, W.-C.; Hu, L.-Y.; Lai, C.-H.; Hsu, C.-N.; Lin, W.-C. Identification of homologous microRNAs in 56 animal genomes. Genomics 2010, 96, 1–9. [Google Scholar] [CrossRef]
- Friedländer, M.R.; Lizano, E.; Houben, A.J.S.; Bezdan, D.; Báñez-Coronel, M.; Kudla, G.; Mateu-Huertas, E.; Kagerbauer, B.; González, J.; Chen, K.C.; et al. Evidence for the biogenesis of more than 1,000 novel human microRNAs. Genome Biol. 2014, 15, R57. [Google Scholar] [CrossRef]
- Yao, S. MicroRNA biogenesis and their functions in regulating stem cell potency and differentiation. Biol. Proced. Online 2016, 18, 1–10. [Google Scholar] [CrossRef]
- MacFarlane, L.A.; Murphy, P.R. MicroRNA: Biogenesis, function and role in cancer. Curr. Genom. 2010, 11, 537–561. [Google Scholar] [CrossRef]
- Niu, J.; Sun, M.; Li, Z.; Wang, Z.; Kong, M.; Wang, Y.; Song, J.; Zhang, Q.; He, Y.; Qi, J. Whole transcriptome analysis provides new insight on immune response mechanism of golden pompano (Trachinotus ovatus) to Amyloodinium ocellatum infestation. Aquaculture 2022, 560, 738396. [Google Scholar] [CrossRef]
- Du, H.; Cao, Z.; Liu, Z.; Wang, G.; Wu, Y.; Du, X.; Wei, C.; Sun, Y.; Zhou, Y. Cromileptes altivelis microRNA Transcriptome Analysis upon Nervous Necrosis Virus (NNV) Infection and the Effect of cal-miR-155 on Cells Apoptosis and Virus Replication. Viruses 2022, 14, 2184. [Google Scholar] [CrossRef]
- Zhou, Y.; Wang, Y.-Y.; Fu, H.-C.; Huang, H.-Z. MicroRNA expression and analysis of immune-related putative target genes in ISKNV-infected spleen of mandarin fish (Siniperca chuatsi). Aquaculture 2022, 547, 737450. [Google Scholar] [CrossRef]
- Ou, J.; Liu, Q.; Bian, Y.; Luan, X.; Meng, Y.; Dong, H.; Cao, M.; Zhang, B.; Wang, Z.; Zhao, W. Integrated analysis of mRNA and microRNA transcriptome related to immunity and autophagy in shrimp hemocytes infected with Spiroplasma eriocheiris. Fish Shellfish Immunol. 2022, 130, 436–452. [Google Scholar] [CrossRef]
- Xia, Y.Q.; Cheng, J.X.; Liu, Y.F.; Li, C.H.; Liu, Y.; Liu, P.F. Genome-wide integrated analysis reveals functions of lncRNA-miRNA-mRNA interactions in Atlantic salmon challenged by Aeromonas salmonicida. Genomics 2022, 114, 328–339. [Google Scholar] [CrossRef]
- Hultmark, D. Drosophila immunity: Paths and patterns. Curr. Opin. Immunol. 2003, 15, 12–19. [Google Scholar] [CrossRef]
- Firdaus-Nawi, M.; Zamri-Saad, M. Major Components of Fish Immunity: A Review. Pertanika J. Trop. Agric. Sci. 2016, 39, 393–420. [Google Scholar]
- Musthaq, S.K.S.; Kwang, J. Evolution of specific immunity in shrimp—A vaccination perspective against white spot syndrome virus. Dev. Comp. Immunol. 2014, 46, 279–290. [Google Scholar] [CrossRef]
- Kulkarni, A.; Krishnan, S.; Anand, D.; Uthaman, S.K.; Otta, S.K.; Karunasagar, I.; Valappil, R.K. Immune responses and immunoprotection in crustaceans with special reference to shrimp. Rev. Aquac. 2021, 13, 431–459. [Google Scholar] [CrossRef]
- Liu, L.; Zhu, B.; Wu, S.; Lin, L.; Liu, G.; Zhou, Y.; Wang, W.; Asim, M.; Yuan, J.; Li, L.; et al. Spring viraemia of carp virus induces autophagy for necessary viral replication. Cell. Microbiol. 2015, 17, 595–605. [Google Scholar] [CrossRef]
- Ezzati-Tabrizi, R.; Farrokhi, N.; Talaei-Hassanloui, R.; Mehdi Alavi, S.; Hosseininaveh, V. Insect inducible antimicrobial peptides and their applications. Curr. Protein Pept. Sci. 2013, 14, 698–710. [Google Scholar] [CrossRef]
- Tanji, T.; Hu, X.; Weber, A.N.R.; Ip, Y.T. Toll and IMD Pathways Synergistically Activate an Innate Immune Response in Drosophila melanogaster. Mol. Cell. Biol. 2007, 27, 4578–4588. [Google Scholar] [CrossRef]
- Ashok, Y. Drosophila Toll Pathway: The New Model. Sci. Signal. 2009, 2, jc1. [Google Scholar] [CrossRef]
- Hoffmann, A.; Natoli, G.; Ghosh, G. Transcriptional regulation via the NF-κB signaling module. Oncogene 2006, 25, 6706–6716. [Google Scholar] [CrossRef]
- Pazarentzos, E.; Mahul-Mellier, A.L.; Datler, C.; Chaisaklert, W.; Hwang, M.S.; Kroon, J.; Qize, D.; Osborne, F.; Al-Rubaish, A.; Al-Ali, A.; et al. IκΒα inhibits apoptosis at the outer mitochondrial membrane independently of NF-κB retention. EMBO J. 2014, 33, 2814–2828. [Google Scholar] [CrossRef]
- Luo, J.L.; Kamata, H.; Karin, M. The anti-death machinery in IKK/NF-κB signaling. J. Clin. Immunol. 2005, 25, 541–550. [Google Scholar] [CrossRef]
- Shibuya, M. Vascular endothelial growth factor (VEGF) and its receptor (VEGFR) signaling in angiogenesis: A crucial target for anti- and pro-angiogenic therapies. Genes Cancer 2011, 2, 1097–1105. [Google Scholar] [CrossRef]
- Li, S.; Zhang, X.; Sun, Z.; Li, F.; Xiang, J. Transcriptome Analysis on Chinese Shrimp Fenneropenaeus chinensis during WSSV Acute Infection. PLoS ONE 2013, 8, e58627. [Google Scholar] [CrossRef]
- Li, S.; Wang, Z.; Li, F.; Xiang, J. One type of VEGFR is involved in WSSV infection to the Pacific whiteleg shrimp Litopenaeus vannamei. Dev. Comp. Immunol. 2015, 50, 1–8. [Google Scholar] [CrossRef]
- Wang, Z.; Li, S.; Li, F.; Yang, H.; Yang, F.; Xiang, J. Characterization of two types of vascular endothelial growth factor from Litopenaeus vannamei and their involvements during WSSV infection. Fish Shellfish Immunol. 2015, 47, 824–832. [Google Scholar] [CrossRef]
- Wang, Z.; Li, S.; Li, F.; Xie, S.; Xiang, J. Identification and function analysis of a novel vascular endothelial growth factor, LvVEGF3, in the Pacific whiteleg shrimp Litopenaeus vannamei. Dev. Comp. Immunol. 2016, 63, 111–120. [Google Scholar] [CrossRef]
- Li, S.; Wang, Z.; Li, F.; Yu, K.; Xiang, J. A Novel Vascular Endothelial Growth Factor Receptor Participates in White Spot Syndrome Virus Infection in Litopenaeus vannamei. Front. Immunol. 2017, 8, 1457. [Google Scholar] [CrossRef]
- Wang, Z.; Li, S.; Yu, Y.; Yu, K.; Zhang, X.; Xiang, J.; Li, F. Identification and characterization of two novel vascular endothelial growth factor genes in Litopenaeus vannamei. Fish Shellfish Immunol. 2019, 84, 259–268. [Google Scholar] [CrossRef]
- Yu, D.; Zhai, Y.; He, P.; Jia, R. Comprehensive Transcriptomic and Metabolomic Analysis of the Litopenaeus vannamei Hepatopancreas After WSSV Challenge. Front. Immunol. 2022, 13, 826794. [Google Scholar] [CrossRef]
- Geindreau, M.; Ghiringhelli, F.; Bruchard, M. Vascular Endothelial Growth Factor, a Key Modulator of the Anti-Tumor Immune Response. Int. J. Mol. Sci. 2021, 22, 4871. [Google Scholar] [CrossRef]
- Troemel, E.R.; Chu, S.W.; Reinke, V.; Lee, S.S.; Ausubel, F.M.; Kim, D.H. p38 MAPK regulates expression of immune response genes and contributes to longevity in C. elegans. PLoS Genet. 2006, 2, e183. [Google Scholar] [CrossRef]
- Inada, M.; Yui, T.; Kono, T.; Yoshida, T.; Sakai, M.; Itami, T. Novel cytokine genes from Kuruma shrimp Marsupenaeus japonicus: MIF and VEGF are important in the innate immunity. Fish Shellfish Immunol. 2013, 34, 1656–1657. [Google Scholar] [CrossRef]
- Ou, J.; Chen, H.; Liu, Q.; Bian, Y.; Luan, X.; Jiang, Q.; Ji, H.; Wang, Z.; Lv, L.; Dong, X.; et al. Integrated transcriptome analysis of immune-related mRNAs and microRNAs in Macrobrachium rosenbergii infected with Spiroplasma eriocheiris. Fish Shellfish Immunol. 2021, 119, 651–669. [Google Scholar] [CrossRef]
- Chang, F.; Steelman, L.S.; Lee, J.T.; Shelton, J.G.; Navolanic, P.M.; Blalock, W.L.; Franklin, R.A.; McCubrey, J. Signal transduction mediated by the Ras/Raf/MEK/ERK pathway from cytokine receptors to transcription factors: Potential targeting for therapeutic intervention. Leukemia 2003, 17, 1263–1293. [Google Scholar] [CrossRef]
- Yoshimura, A. Regulation of Cytokine Signaling by the SOCS and Spred Family Proteins. Keio J. Med. 2009, 58, 73–83. [Google Scholar] [CrossRef]
- Angulo, P.; Kaushik, G.; Subramaniam, D.; Dandawate, P.; Neville, K.; Chastain, K.; Anant, S. Natural compounds targeting major cell signaling pathways: A novel paradigm for osteosarcoma therapy. J. Hematol. Oncol. 2017, 10, 1–13. [Google Scholar] [CrossRef]
- Wang, S.; Aurora, A.B.; Johnson, B.A.; Qi, X.; McAnally, J.; Hill, J.A.; Richardson, J.A.; Bassel-Duby, R.; Olson, E.N. The Endothelial-Specific MicroRNA miR-126 Governs Vascular Integrity and Angiogenesis. Dev. Cell 2008, 15, 261–271. [Google Scholar] [CrossRef]
- Heales, S.J.; Gegg, M.E.; Clark, J.B. Oxidative phosphorylation: Structure, function, and intermediary metabolism. Int. Rev. Neurobiol. 2002, 53, 25–56. [Google Scholar]
- Tan, T.T.; Chen, M.; Harikrishna, J.A.; Khairuddin, N.; Shamsudin, M.I.M.; Zhang, G.; Bhassu, S. Deep parallel sequencing reveals conserved and novel miRNAs in gill and hepatopancreas of giant freshwater prawn. Fish Shellfish Immunol. 2013, 35, 1061–1069. [Google Scholar] [CrossRef]
- Sun, X.; Liu, Q.-H.; Yang, B.; Huang, J. Differential expression of microRNAs of Litopenaeus vannamei in response to different virulence WSSV infection. Fish Shellfish Immunol. 2016, 58, 18–23. [Google Scholar] [CrossRef] [PubMed]
- Shekhar, M.; Karthic, K.; Kumar, K.V.; Kumar, J.A.; Swathi, A.; Hauton, C.; Peruzza, L.; Vijayan, K. Comparative analysis of shrimp (Penaeus vannamei) miRNAs expression profiles during WSSV infection under experimental conditions and in pond culture. Fish Shellfish Immunol. 2019, 93, 288–295. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Wang, Y.; Fu, X.; Tao, W.; Li, C. circRNA1149 from Apostichopus japonicus suppresses coelomocyte apoptosis act as miR-92a sponge to regulate Bax expression in response to Vibrio splendidus infection. Aquaculture 2023, 562, 738812. [Google Scholar] [CrossRef]
- Liu, J.; Zhao, X.; Duan, X.; Zhang, W.; Li, C. CircRNA75 and CircRNA72 function as the sponge of MicroRNA-200 to suppress coelomocyte apoptosis via targeting Tollip in Apostichopus japonicus. Front. Immunol. 2021, 12, 770055. [Google Scholar] [CrossRef] [PubMed]
- Qiao, X.; Lu, Y.; Xu, J.; Deng, N.; Lai, W.; Wu, Z.; Lin, H.; Zhang, Y.; Lu, D. Integrative analyses of mRNA and microRNA expression profiles reveal the innate immune mechanism for the resistance to Vibrio parahaemolyticus infection in Epinephelus coioides. Front. Immunol. 2022, 13, 982973. [Google Scholar] [CrossRef]
- Kaewkascholkul, N.; Somboonviwat, K.; Asakawa, S.; Hirono, I.; Tassanakajon, A.; Somboonwiwat, K. Shrimp miRNAs regulate innate immune response against white spot syndrome virus infection. Dev. Comp. Immunol. 2016, 60, 191–201. [Google Scholar] [CrossRef]
- Soo, T.C.C.; See, S.A.; Bhassu, S. Potential muscle activity disturbance in Penaeus monodon during Acute Hepatopancreatic Necrosis Disease (AHPND) infection: Inference through gene expression, calcium concentration, and MicroRNA. J. Invertebr. Pathol. 2020, 177, 107497. [Google Scholar] [CrossRef]
- Huang, Y.; Wang, W.; Ren, Q. Two host microRNAs influence WSSV replication via STAT gene regulation. Sci. Rep. 2016, 6, 23643. [Google Scholar] [CrossRef]
- Mangukia, N.; Rao, P.; Patel, K.; Pandya, H.; Rawal, R.M. Identifying potential human and medicinal plant microRNAs against SARS-CoV-2 3′ UTR region: A computational genomics assessment. Comput. Biol. Med. 2021, 136, 104662. [Google Scholar] [CrossRef]
- Bamunuarachchi, G.; Yang, X.; Huang, C.; Liang, Y.; Guo, Y.; Liu, L. MicroRNA -206 inhibits influenza A virus replication by targeting tankyrase 2. Cell. Microbiol. 2021, 23, e13281. [Google Scholar] [CrossRef]
- Huang, T.; Zhang, X. Functional Analysis of a Crustacean MicroRNA in Host-Virus Interactions. J. Virol. 2012, 86, 12997–13004. [Google Scholar] [CrossRef]
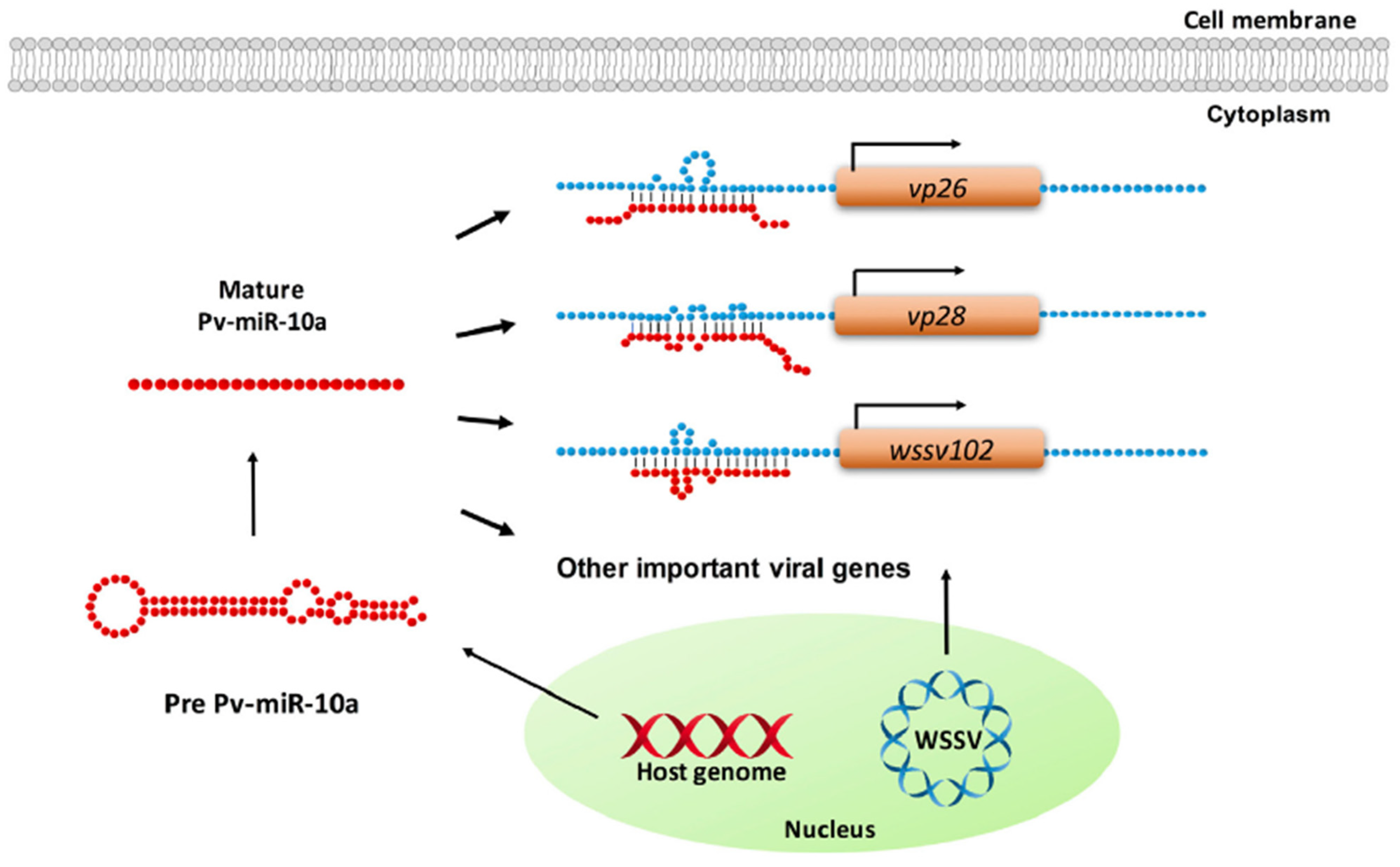
- Huang, J.-Y.; Kang, S.-T.; Chen, I.-T.; Chang, L.-K.; Lin, S.-S.; Kou, G.-H.; Chu, C.-Y.; Lo, C.-F. Shrimp miR-10a Is Co-opted by White Spot Syndrome Virus to Increase Viral Gene Expression and Viral Replication. Front. Immunol. 2017, 8, 1084. [Google Scholar] [CrossRef]
- Jaree, P.; Wongdontri, C.; Somboonwiwat, K. White Spot Syndrome Virus-Induced Shrimp miR-315 Attenuates Prophenoloxidase Activation via PPAE3 Gene Suppression. Front. Immunol. 2018, 9, 2184. [Google Scholar] [CrossRef]
- Sugumaran, M. Comparative Biochemistry of Eumelanogenesis and the Protective Roles of Phenoloxidase and Melanin in Insects. Pigment. Cell Res. 2002, 15, 2–9. [Google Scholar] [CrossRef]
- Lee, Y.J.; Park, E.G.; Kim, W.R.; Bae, W.H.; Lee, D.H.; Lee, Y.; Kim, D.-H.; Choi, Y.H.; Cha, H.-J.; Kim, S.; et al. Downregulated miR-15b-5p induces suppressor of cytokine signaling 6 (SOCS6) expression during viral hemorrhagic septicemia virus infection in olive flounder (Paralichthys olivaceus). Aquaculture 2023, 562, 738811. [Google Scholar] [CrossRef]
- Wang, Z.; Zhu, F. MicroRNA-100 is involved in shrimp immune response to white spot syndrome virus (WSSV) and Vibrio alginolyticus infection. Sci. Rep. 2017, 7, srep42334. [Google Scholar] [CrossRef]
- Ren, Q.; Huang, Y.; He, Y.; Wang, W.; Zhang, X. A white spot syndrome virus microRNA promotes the virus infection by targeting the host STAT. Sci. Rep. 2015, 5, 18384. [Google Scholar] [CrossRef]
- Nantapojd, T.; Panyim, S.; Ongvarrasopone, C. White spot syndrome virus-encoded microRNA promotes viral replication by maintaining viral early gene expression. Aquaculture 2022, 546, 737284. [Google Scholar] [CrossRef]
- Singh, M.; Chazal, M.; Quarato, P.; Bourdon, L.; Malabat, C.; Vallet, T.; Vignuzzi, M.; van der Werf, S.; Behillil, S.; Donati, F.; et al. A virus-encoded microRNA contributes to evade innate immune response during SARS-CoV-2 infection. bioRxiv 2021. [Google Scholar] [CrossRef]
- Chavalit, T.; Nimsamer, P.; Sirivassanametha, K.; Anuntakarun, S.; Saengchoowong, S.; Tangkijvanich, P.; Payungporn, S. Hepatitis B virus-encoded microRNA (HBV-miR-3) regulates host gene PPM1A related to hepatocellular carcinoma. Microrna 2020, 9, 232–239. [Google Scholar] [CrossRef]
- Hennig, T.; Prusty, A.B.; Kaufer, B.B.; Whisnant, A.W.; Lodha, M.; Enders, A.; Thomas, J.; Kasimir, F.; Grothey, A.; Klein, T.; et al. Selective inhibition of miRNA processing by a herpesvirus-encoded miRNA. Nature 2022, 605, 539–544. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Yang, K.; Zhang, X. Viral MicroRNAs Targeting Virus Genes Promote Virus Infection in Shrimp In Vivo. J. Virol. 2014, 88, 1104–1112. [Google Scholar] [CrossRef] [PubMed]
- He, J.-H.; Xia, Q.; Weng, S.; He, J.; Xu, X. Identification of infectious spleen and kidney necrosis virus (ISKNV)-encoded microRNAs. Virus Genes 2020, 56, 724–733. [Google Scholar] [CrossRef]
- Chen, Q.; Luo, Y.; Fu, Y.; Feng, Z.; Lu, L.; Jiang, Y.; Xu, D. microRNA (miR-KT-635) encoded by Cyprinid herpesvirus 2 regulates the viral replication with targeting to the ORF23. J. Fish Dis. 2022, 45, 631–639. [Google Scholar] [CrossRef] [PubMed]
- El-Sebaey, A.M.; Abramov, P.N.; Borunova, S.M. Cfa-miRNAs-122 and-21 as modern biomarkers of primary hepatitis in dogs. RUDN J. Agron. Anim. Ind. 2020, 15, 294–307. [Google Scholar] [CrossRef]
- Nguyen, T.B.; Do, D.N.; Nguyen, T.T.; Nguyen, T.L.; Nguyen-Thanh, T.; Nguyen, H.T. Immune–related biomarkers shared by inflammatory bowel disease and liver cancer. PLoS ONE 2022, 17, e0267358. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.T.P.; Suman, K.H.; Nguyen, T.B.; Nguyen, H.T.; Do, D.N. The Role of miR-29s in Human Cancers—An Update. Biomedicines 2022, 10, 2121. [Google Scholar] [CrossRef]
- Zhang, B.; Zhao, N.; Jia, L.; Peng, K.; Che, J.; Li, K.; He, X.; Sun, J.; Bao, B. Seminal Plasma Exosomes: Promising Biomarkers for Identification of Male and Pseudo-Males in Cynoglossus semilaevis. Mar. Biotechnol. 2019, 21, 310–319. [Google Scholar] [CrossRef]
- Nie, M.; Tan, X.; Lu, Y.; Wu, Z.; Li, J.; Xu, D.; Zhang, P.; You, F. Network of microRNA-transcriptional factor-mRNA in cold response of turbot Scophthalmus maximus. Fish Physiol. Biochem. 2019, 45, 583–597. [Google Scholar] [CrossRef]
- Ikert, H.; Lynch, M.D.J.; Doxey, A.C.; Giesy, J.P.; Servos, M.R.; Katzenback, B.A.; Craig, P.M. High Throughput Sequencing of MicroRNA in Rainbow Trout Plasma, Mucus, and Surrounding Water Following Acute Stress. Front. Physiol. 2021, 11, 1821. [Google Scholar] [CrossRef]
- Cui, C.; Cui, Q. The relationship of human tissue microRNAs with those from body fluids. Sci. Rep. 2020, 10, 5644. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Ba, Y.; Ma, L.; Cai, X.; Yin, Y.; Wang, K.; Guo, J.; Zhang, Y.; Chen, J.; Guo, X.; et al. Characterization of microRNAs in serum: A novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008, 18, 997–1006. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.Y.; Shen, Y.B.; Fu, J.J.; Yu, H.Y.; Huang, W.J.; Lu, L.Q.; Li, J.L. MicroRNA-induced negative regulation of TLR-5 in grass carp, Ctenopharyngodon idella. Sci. Rep. 2016, 6, 18595. [Google Scholar] [CrossRef]
- Zhao, N.; Jia, L.; Li, G.; He, X.; Zhu, C.; Zhang, B. Comparative Mucous miRomics in Cynoglossus semilaevis Related to Vibrio harveyi Caused Infection. Mar. Biotechnol. 2021, 23, 766–776. [Google Scholar] [CrossRef]
- Zhao, N.; Zhang, B.; Xu, Z.; Jia, L.; Li, M.; He, X.; Bao, B. Detecting Cynoglossus semilaevis infected with Vibrio harveyi using micro RNAs from mucous exosomes. Mol. Immunol. 2020, 128, 268–276. [Google Scholar] [CrossRef]
- Bhat, R.A.; Priyam, M.; Foysal, M.J.; Gupta, S.K.; Sundaray, J.K. Role of sex-biased miRNAs in teleosts—A review. Rev. Aquac. 2021, 13, 269–281. [Google Scholar] [CrossRef]
- Ahluwalia, J.K.; Khan, S.Z.; Soni, K.; Rawat, P.; Gupta, A.; Hariharan, M.; Scaria, V.; Lalwani, M.; Pillai, B.; Mitra, D.; et al. Human cellular microRNA hsa-miR-29a interferes with viral nef protein expression and HIV-1 replication. Retrovirology 2008, 5, 1–10. [Google Scholar] [CrossRef]
- Floch, P.; Capdevielle, C.; Staedel, C.; Izotte, J.; Sifré, E.; Laur, A.M.; Giese, A.; Korolik, V.; Dubus, P.; Mégraud, F.; et al. Deregulation of MicroRNAs in Gastric Lymphomagenesis Induced in the d3Tx Mouse Model of Helicobacter pylori Infection. Front. Cell. Infect. Microbiol. 2017, 7, 185. [Google Scholar] [CrossRef]
- Fatmi, A.; Rebiahi, S.A.; Chabni, N.; Zerrouki, H.; Azzaoui, H.; Elhabiri, Y.; Benmansour, S.; Ibáñez-Cabellos, J.S.; Smahi, M.C.-E.; Aribi, M.; et al. miRNA-23b as a biomarker of culture-positive neonatal sepsis. Mol. Med. 2020, 26, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Han, S.A.; Jhun, B.W.; Kim, S.-Y.; Moon, S.M.; Yang, B.; Kwon, O.J.; Daley, C.L.; Shin, S.J.; Koh, W.-J. miRNA Expression Profiles and Potential as Biomarkers in Nontuberculous Mycobacterial Pulmonary Disease. Sci. Rep. 2020, 10, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Yousefpouran, S.; Mostafaei, S.; Manesh, P.V.; Iranifar, E.; Bokharaei-Salim, F.; Nahand, J.S.; Mirzaei, H.; Taran, M.; Babaei, F.; Sayad, B.; et al. The assessment of selected MiRNAs profile in HIV, HBV, HCV, HIV/HCV, HIV/HBV Co-infection and elite controllers for determination of biomarker. Microb. Pathog. 2020, 147, 104355. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Ning, X.-H.; Guan, X.-L.; Li, X.-P.; Sun, L. Characterization of Streptococcus iniae-induced microRNA profiles in Paralichthys olivaceus and identification of pol-3p-10740_175 as a regulator of antibacterial immune response. Fish Shellfish Immunol. 2020, 98, 860–867. [Google Scholar] [CrossRef]
- Li, W.-R.; Hu, Y.-H.; Jiang, S.; Sun, L. Global profiling and characterization of Japanese flounder (Paralichthys olivaceus) kidney microRNAs regulated by Edwardsiella tarda infection in a time-dependent fashion. Fish Shellfish Immunol. 2019, 93, 766–780. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.-C.; Zhang, J.; Sun, L. In-depth profiling and analysis of host and viral microRNAs in Japanese flounder (Paralichthys olivaceus) infected with megalocytivirus reveal involvement of microRNAs in host-virus interaction in teleost fish. BMC Genom. 2014, 15, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Su, L.; Guo, B.; Zhou, S.; Li, C.; Xiu, Y. Bacterial induced miR-144-5p modulates intestinal inflammatory response of Japanese flounder by targeting Hsp90α-dependent NLR signaling pathway. Aquaculture 2023, 564, 739053. [Google Scholar] [CrossRef]
- Samir, M.; Pessler, F. Small Non-coding RNAs Associated with Viral Infectious Diseases of Veterinary Importance: Potential Clinical Applications. Front. Veter. Sci. 2016, 3, 22. [Google Scholar] [CrossRef] [PubMed]
- Katoh, T.; Sakaguchi, Y.; Miyauchi, K.; Suzuki, T.; Kashiwabara, S.-I.; Baba, T.; Suzuki, T. Selective stabilization of mammalian microRNAs by 3′ adenylation mediated by the cytoplasmic poly(A) polymerase GLD-2. Genes Dev. 2009, 23, 433–438. [Google Scholar] [CrossRef] [PubMed]
- Lam, J.K.W.; Chow, M.Y.T.; Zhang, Y.; Leung, S.W.S. siRNA Versus miRNA as Therapeutics for Gene Silencing. Mol. Ther. Nucleic Acids 2015, 4, e252. [Google Scholar] [CrossRef]
- Stenvang, J.; Petri, A.; Lindow, M.; Obad, S.; Kauppinen, S. Inhibition of microRNA function by antimiR oligonucleotides. Silence 2012, 3, 1–17. [Google Scholar] [CrossRef]
- Liu, L.; Bi, N.; Wu, L.; Ding, X.; Men, Y.; Zhou, W.; Li, L.; Zhang, W.; Shi, S.; Song, Y.; et al. MicroRNA-29c functions as a tumor suppressor by targeting VEGFA in lung adenocarcinoma. Mol. Cancer 2017, 16, 1–12. [Google Scholar] [CrossRef]
- Garofalo, M.; Quintavalle, C.; Romano, G.; Mcroce, C.; Condorelli, G. miR221/222 in cancer: Their role in tumor progression and response to therapy. Curr. Mol. Med. 2012, 12, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Arisan, E.D.; Freitas, I.L.; Pazoki, K.; Rahim, S.; Uysal Onganer, P. The Role of Differentially Expressed miRNAs and Potential miRNA-mRNA Regulatory Network in Prostate Cancer Progression and Metastasis. Biomed. J. Tech. Sci. Res. 2020, 25, 19382–19387. [Google Scholar]
- Kong, A.S.-Y.; Lai, K.-S.; Lim, S.-H.E.; Sivalingam, S.; Loh, J.-Y.; Maran, S. miRNA in Ischemic Heart Disease and Its Potential as Biomarkers: A Comprehensive Review. Int. J. Mol. Sci. 2022, 23, 9001. [Google Scholar] [CrossRef] [PubMed]
- Bernardo, B.C.; Nguyen, S.S.; Winbanks, C.E.; Gao, X.-M.; Boey, E.J.H.; Tham, Y.K.; Kiriazis, H.; Ooi, J.Y.Y.; Porrello, E.R.; Igoor, S.; et al. Therapeutic silencing of miR-652 restores heart function and attenuates adverse remodeling in a setting of established pathological hypertrophy. FASEB J. 2014, 28, 5097–5110. [Google Scholar] [CrossRef]
- Bansal, P.; Christopher, A.F.; Kaur, R.P.; Kaur, G.; Kaur, A.; Gupta, V. MicroRNA therapeutics: Discovering novel targets and developing specific therapy. Perspect. Clin. Res. 2016, 7, 68–74. [Google Scholar] [CrossRef]
- Dang, L.T.; Kondo, H.; Aoki, T.; Hirono, I. Engineered virus-encoded pre-microRNA (pre-miRNA) induces sequence-specific antiviral response in addition to nonspecific immunity in a fish cell line: Convergence of RNAi-related pathways and IFN-related pathways in antiviral response. Antivir. Res. 2008, 80, 316–323. [Google Scholar] [CrossRef]
- Kim, Y.-S.; Ke, F.; Lei, X.-Y.; Zhu, R.; Zhang, Q.-Y. Viral Envelope Protein 53R Gene Highly Specific Silencing and Iridovirus Resistance in Fish Cells by AmiRNA. PLoS ONE 2010, 5, e10308. [Google Scholar] [CrossRef]
- Cui, Y.; Wan, H.; Zhang, X. miRNA in food simultaneously controls animal viral disease and human tumorigenesis. Mol. Ther. Nucleic Acids 2021, 23, 995–1006. [Google Scholar] [CrossRef]
- Tribolet, L.; Kerr, E.; Cowled, C.; Bean, A.G.D.; Stewart, C.R.; Dearnley, M.; Farr, R.J. MicroRNA Biomarkers for Infectious Diseases: From Basic Research to Biosensing. Front. Microbiol. 2020, 11, 1197. [Google Scholar] [CrossRef]
- Zheng, W.; Yao, L.; Teng, J.; Yan, C.; Qin, P.; Liu, G.; Chen, W. Lateral flow test for visual detection of multiple MicroRNAs. Sens. Actuators B Chem. 2018, 264, 320–326. [Google Scholar] [CrossRef]
- Shamsi, M.H.; Choi, K.; Ng, A.H.; Chamberlain, M.D.; Wheeler, A.R. Electrochemiluminescence on digital microfluidics for microRNA analysis. Biosens. Bioelectron. 2016, 77, 845–852. [Google Scholar] [CrossRef] [PubMed]
- Miti, A.; Thamm, S.; Müller, P.; Csáki, A.; Fritzsche, W.; Zuccheri, G. A miRNA biosensor based on localized surface plasmon resonance enhanced by surface-bound hybridization chain reaction. Biosens. Bioelectron. 2020, 167, 112465. [Google Scholar] [CrossRef] [PubMed]
- Nehra, A.; Kumar, A.; Ahlawat, S.; Kumar, V.; Singh, K.P. Substrate-Free Untagged Detection of miR393a Using an Ultrasensitive Electrochemical Biosensor. ACS Omega 2022, 7, 5176–5189. [Google Scholar] [CrossRef]
- Guan, Y.; Wang, F.P.; Chen, Z.X.; Yang, Y.H.; Yang, T.; Hu, R. Ratiometrically homogeneous electrochemical biosensor based on the signal amplified strategy of dual DNA nanomachines for microRNA analysis. Talanta 2023, 254, 124191. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, N.; Yang, M.; Hou, C.; Huo, D. An ultrasensitive electrochemical biosensor for simultaneously detect microRNA-21 and microRNA-155 based on specific interaction of antimonide quantum dot with RNA. Microchem. J. 2023, 185, 108173. [Google Scholar] [CrossRef]
- He, Y.; Liao, X.; Wu, H.; Huang, J.; Zhang, Y.; Peng, Y.; Wang, Z.; Cao, X.; Wu, C.; Luo, X. A controllable SERS biosensor for ultrasensitive detection of miRNAs based on porous MOFs and subject-object recognition ability. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2023, 289, 122134. [Google Scholar] [CrossRef]
- Chan, Y.Y.; Bhassu, S.; Periasamy, V. Investigating the electronic properties of shrimp synthetic viral DNAs integrated within Schottky junctions. Appl. Phys. Express 2020, 13, 041005. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
SiouNing, A.S.; Seong, T.S.; Kondo, H.; Bhassu, S. MicroRNA Regulation in Infectious Diseases and Its Potential as a Biosensor in Future Aquaculture Industry: A Review. Molecules 2023, 28, 4357. https://doi.org/10.3390/molecules28114357
SiouNing AS, Seong TS, Kondo H, Bhassu S. MicroRNA Regulation in Infectious Diseases and Its Potential as a Biosensor in Future Aquaculture Industry: A Review. Molecules. 2023; 28(11):4357. https://doi.org/10.3390/molecules28114357
Chicago/Turabian StyleSiouNing, Aileen See, Tang Swee Seong, Hidehiro Kondo, and Subha Bhassu. 2023. "MicroRNA Regulation in Infectious Diseases and Its Potential as a Biosensor in Future Aquaculture Industry: A Review" Molecules 28, no. 11: 4357. https://doi.org/10.3390/molecules28114357
APA StyleSiouNing, A. S., Seong, T. S., Kondo, H., & Bhassu, S. (2023). MicroRNA Regulation in Infectious Diseases and Its Potential as a Biosensor in Future Aquaculture Industry: A Review. Molecules, 28(11), 4357. https://doi.org/10.3390/molecules28114357





