Single Cell Determination of 7,8-dihydro-8-oxo-2′-deoxyguanosine by Fluorescence Techniques: Antibody vs. Avidin Labeling
Abstract
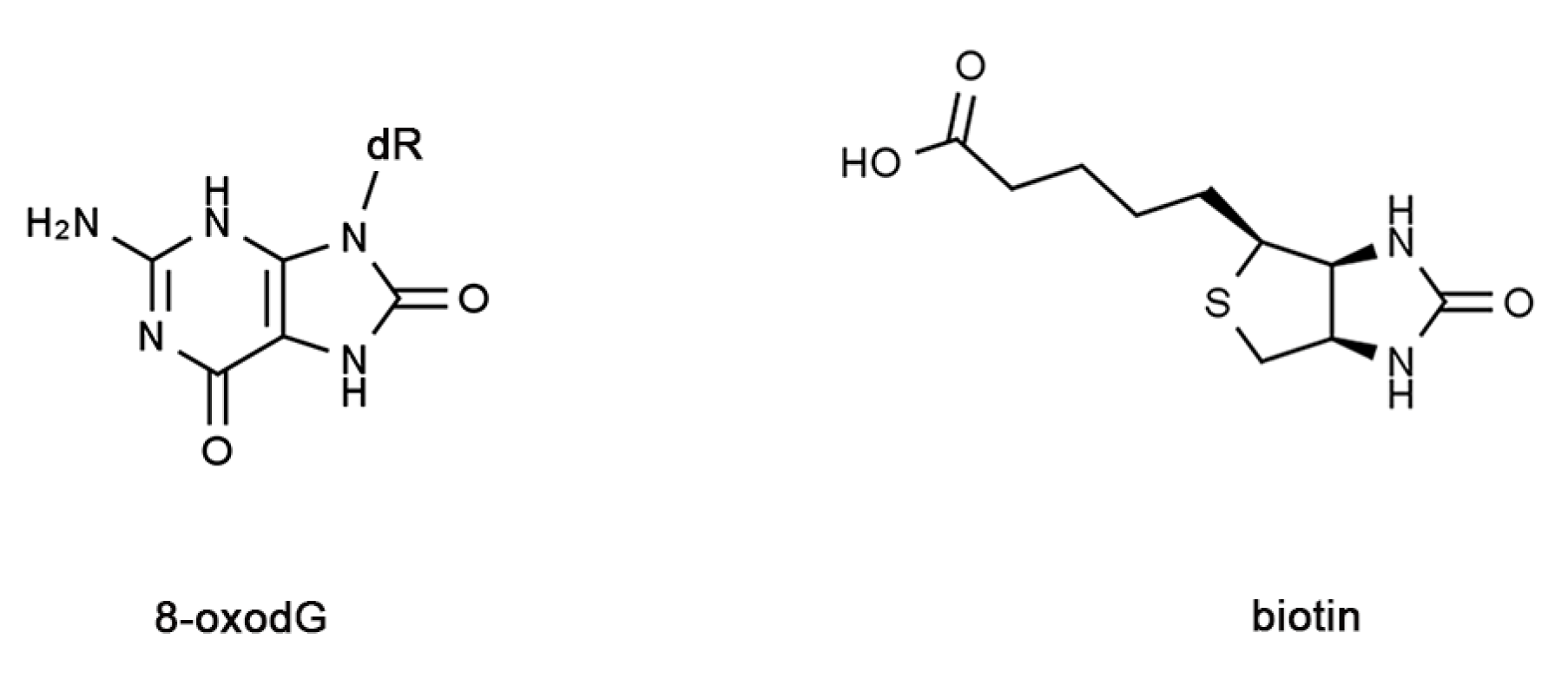
1. Introduction
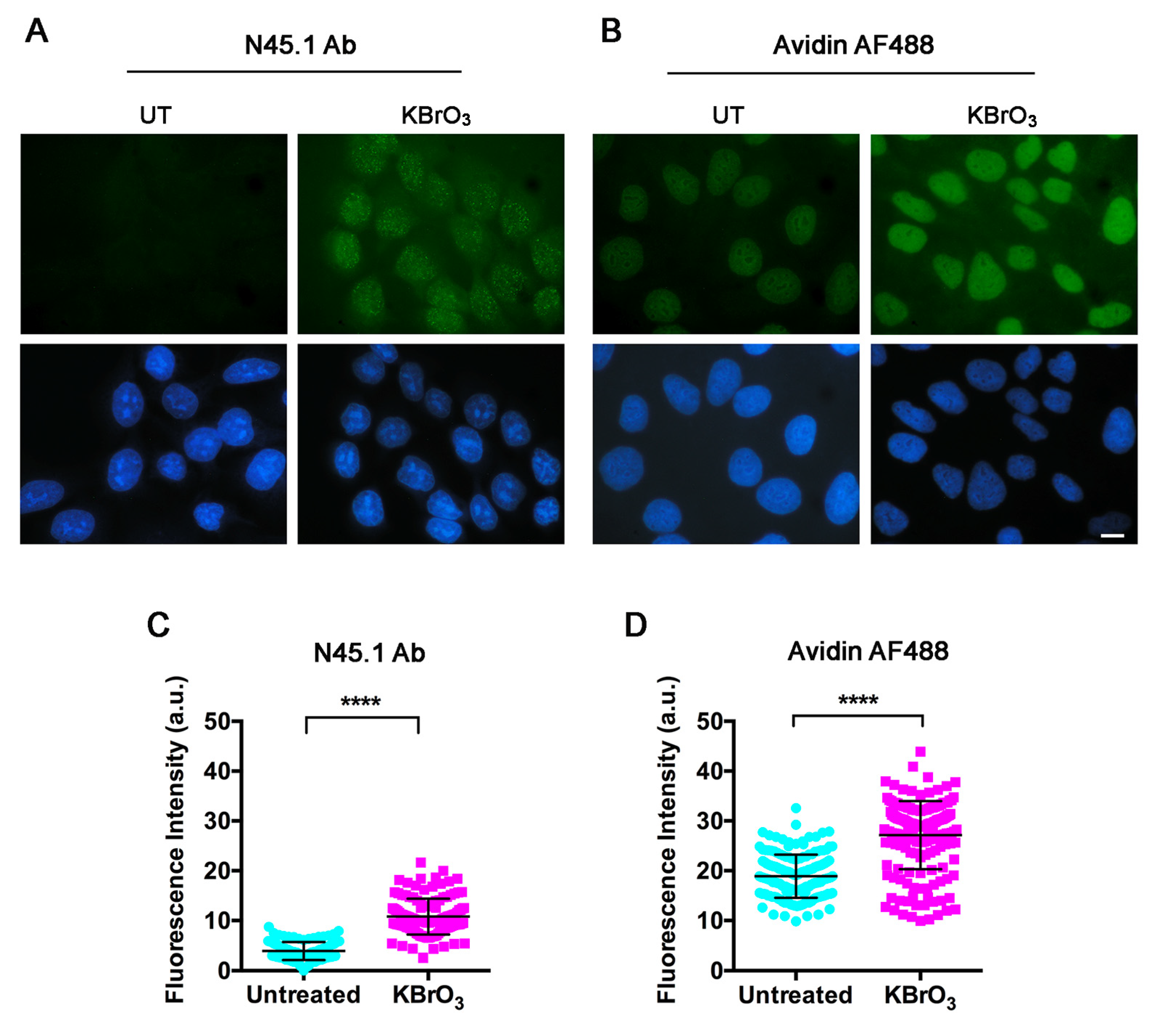
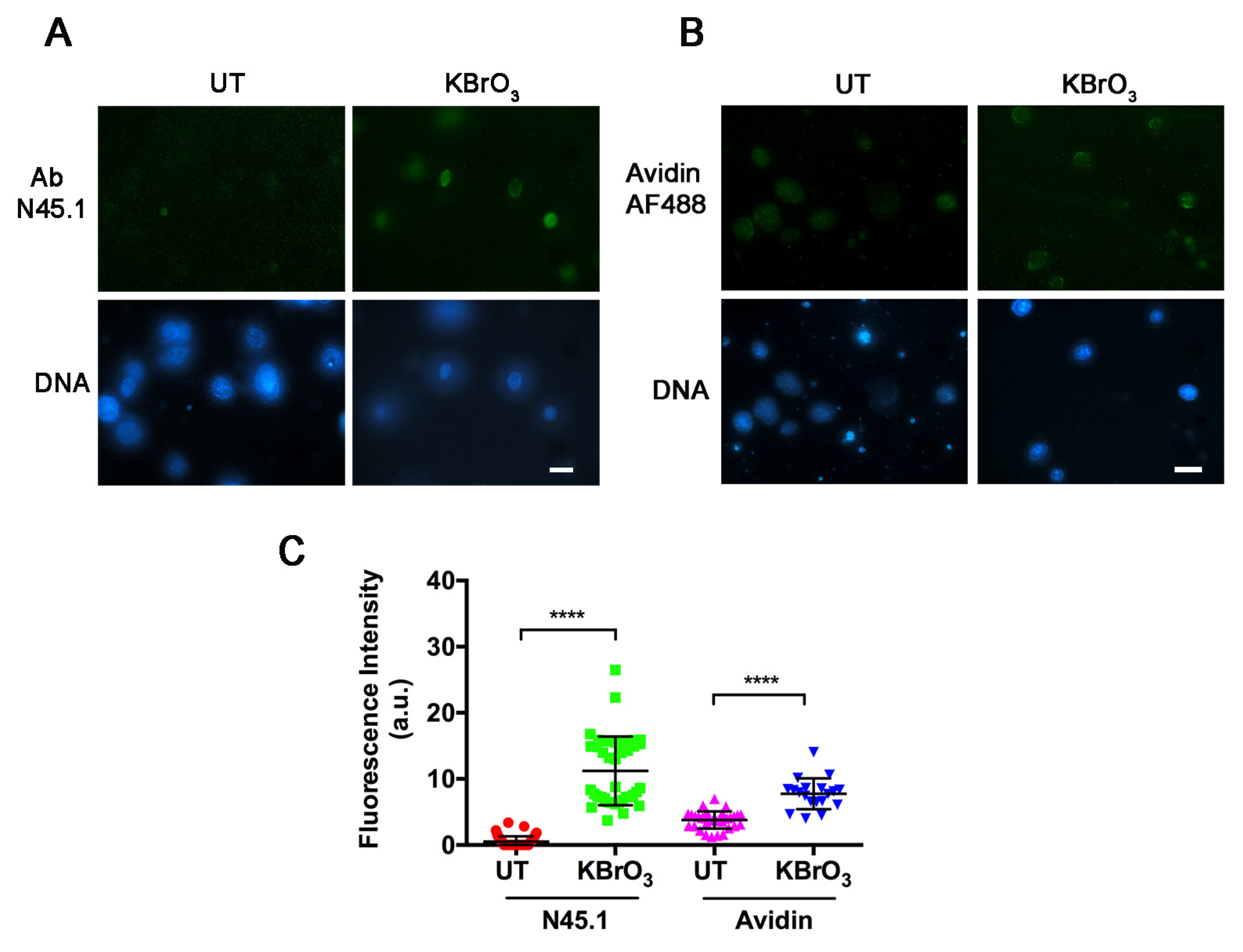
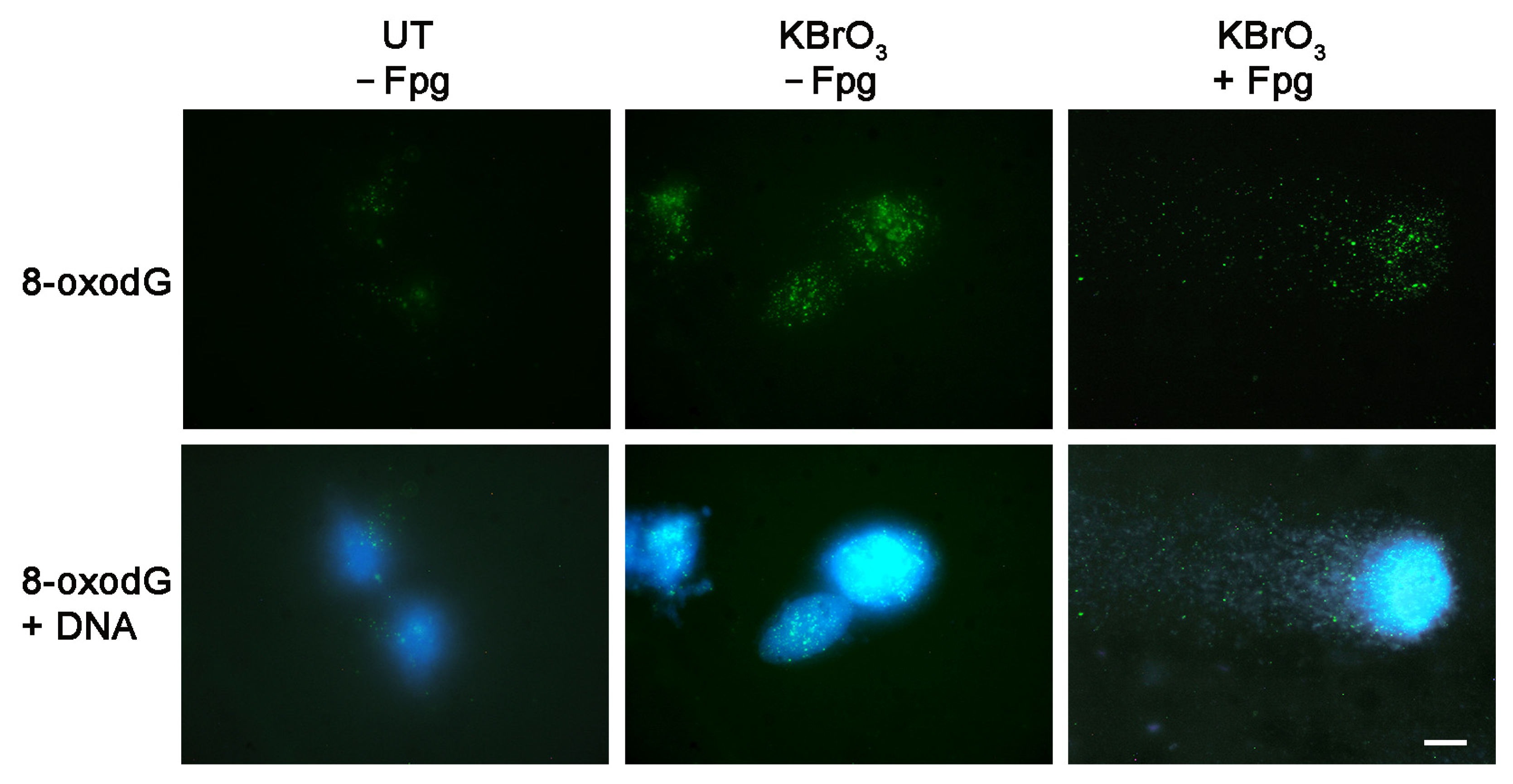
2. Results
3. Discussion
4. Materials and Methods
4.1. Cells and Reagents
4.2. Cell Treatments
4.3. Nucleoid Preparation
4.4. Immunofluorescence Labeling
4.5. Avidin-AF488 Labeling
4.6. Fluorescence Microscopy and Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cooke, M.S.; Evans, M.D.; Dizdaroglou, M.; Lunec, J. Oxidative DNA damage: Mechanisms, mutation, and disease. FASEB J. 2003, 17, 1195–1214. [Google Scholar] [CrossRef] [PubMed]
- Cadet, J.; Davies, K.J.A.; Medeiros, M.H.G.; Di Mascio, P.; Wagner, J.R. Formation and repair of oxidatively generated damage in cellular DNA. Free Radic. Biol. Med. 2017, 107, 13–34. [Google Scholar] [CrossRef] [PubMed]
- Culp, S.J.; Cho, B.P.; Kadlubar, F.F.; Evans, F.E. Structural and conformational analyses of 8-hydroxy-2′-deoxyguanosine. Chem. Res. Toxicol. 1989, 2, 416–422. [Google Scholar] [CrossRef] [PubMed]
- Neeley, W.L.; Essigmann, J.M. Mechanisms of formation, genotoxicity, and mutation of guanine oxidation products. Chem. Res. Toxicol. 2006, 19, 491–505. [Google Scholar] [CrossRef]
- Barnes, D.E.; Lindahl, T. Repair and genetic consequences of endogenous DNA base damage in mammalian cells. Annu. Rev. Genet. 2004, 38, 445–476. [Google Scholar] [CrossRef]
- Carter, R.J.; Parsons, J.L. Base excision repair, a pathway regulated by posttranslational modifications. Mol. Cell. Biol. 2016, 36, 1426–1437. [Google Scholar] [CrossRef]
- Fortini, P.; Pascucci, B.; Parlanti, E.; D’Errico, M.; Simonelli, V.; Dogliotti, E. 8-Oxoguanine DNA damage: At the crossroad of alternative repair pathways. Mutat. Res. 2003, 531, 127–139. [Google Scholar] [CrossRef]
- Li, C.; Xue, Y.; Ba, X.; Wang, R. The role of 8-oxoG repair systems in tumorigenesis and cancer therapy. Cells 2022, 11, 3798. [Google Scholar] [CrossRef]
- Scheijen, E.E.M.; Wilson, D.M., III. Genome integrity and neurological disease. Int. J. Mol. Sci. 2022, 23, 4142. [Google Scholar] [CrossRef]
- Welch, G.; Tsai, L.H. Mechanisms of DNA damage-mediated neurotoxicity in neurodegenerative disease. EMBO Rep. 2022, 23, e54217. [Google Scholar] [CrossRef]
- Behrouzi, A.; Kelley, M.R.; Fehrenbacher, J.C. Oxidative DNA damage: A role in altering neuronal function. J. Cell Signal. 2022, 3, 160–166. [Google Scholar]
- Souliotis, V.L.; Vlachogiannis, N.I.; Pappa, M.; Argyriou, A.; Ntouros, P.A.; Sfikakis, P.P. DNA damage response and oxidative stress in systemic autoimmunity. Int. J. Mol. Sci. 2019, 21, 55. [Google Scholar] [CrossRef]
- Di Minno, A.; Turnu, L.; Porro, B.; Squellerio, I.; Cavalca, V.; Tremoli, E.; Di Minno, M.N. 8-hydroxy-2-deoxyguanosine levels and cardiovascular disease: A systematic review and meta-analysis of the literature. Antioxid. Redox Signal. 2016, 24, 548–555. [Google Scholar] [CrossRef]
- Valavanidis, A.; Vlachogianni, T.; Fiotakis, C. 8-hydroxy-2′-deoxyguanosine (8-OHdG): A critical biomarker of oxidative stress and carcinogenesis. J. Environ. Sci. Health C 2009, 27, 120–139. [Google Scholar] [CrossRef]
- Korkmaz, K.S.; Butuner, B.D.; Roggenbuck, D. Detection of 8-OHdG as a diagnostic biomarker. J. Lab. Precis. Med. 2018, 3, 95. [Google Scholar] [CrossRef]
- Dizdaroglou, M.; Jaruga, P.; Rodriguez, H. Measurement of 8-hydroxy-2′-deoxyguanosine in DNA by high-performance liquid chromatography-mass spectrometry: Comparison with measurement by gas chromatography-mass spectrometry. Nucleic Acids Res. 2001, 29, e12. [Google Scholar] [CrossRef]
- Mangal, D.; Vudathala, D.; Park, J.H.; Lee, S.H.; Penning, T.M.; Blair, I.A. Analysis of 7,8-dihydro-8-oxo-2′-deoxyguanosine in cellular DNA during oxidative stress. Chem. Res. Toxicol. 2009, 22, 788–797. [Google Scholar] [CrossRef]
- Cadet, J.; Douki, T.; Ravanat, J.L.; Wagner, J.R. Measurement of oxidatively generated base damage to nucleic acids in cells: Facts and artifacts. Bioanal. Rev. 2012, 4, 55–74. [Google Scholar] [CrossRef]
- Chepelev, N.L.; Kennedy, D.A.; Gagnè, R.; White, T.; Long, A.S.; Yauk, C.L.; White, P.A. HPLC Measurement of the DNA oxidation biomarker, 8-oxo-7,8-dihydro-2′-deoxyguanosine, in cultured cells and animal tissues. J. Vis. Exp. 2015, 102, e52697. [Google Scholar]
- Chiorcea-Paquim, A.-M. 8-oxoguanine and 8-oxodeoxyguanosine biomarkers of oxidative DNA damage: A review on HPLC–ECD determination. Molecules 2022, 27, 1620. [Google Scholar] [CrossRef]
- Ravanat, J.L.; Douki, T.; Duez, P.; Gremaud, E.; Herbert, K.; Hofer, T.; Lasserre, L.; Pierre, C.S.; Favier, A.; Cadet, J. Cellular background level of 8-oxo-7,8-dihydro-2′-deoxyguanosine: An isotope based method to evaluate artefactual oxidation of DNA during its extraction and subsequent work-up. Carcinogenesis 2002, 23, 1911–1918. [Google Scholar] [CrossRef] [PubMed]
- Yarborough, A.; Zhang, Y.J.; Hsu, T.M.; Santella, R.M. Immunoperoxidase detection of 8-hydroxydeoxyguanosine in aflatoxin B1-treated rat liver and human oral mucosal cells. Cancer Res. 1996, 56, 683–688. [Google Scholar] [PubMed]
- Hattori, Y.; Nishigori, C.; Tanaka, T.; Uchida, K.; Nikaido, O.; Osawa, T.; Hiai, H.; Imamura, S.; Toyokuni, S. 8-hydroxy-2′-deoxyguanosine is increased in epidermal cells of hairless mice after chronic ultraviolet B exposure. J. Investig. Dermatol. 1996, 107, 733–737. [Google Scholar] [CrossRef] [PubMed]
- Toyokuni, S.; Tanaka, T.; Hattori, Y.; Nishiyama, Y.; Yoshida, A.; Uchida, K.; Hiai, H.; Ochi, H.; Osawa, T. Quantitative immunohistochemical determination of 8-hydroxy-2′-deoxyguanosine by a monoclonal antibody N45.1: Its application to ferric nitrilotriacetate-induced renal carcinogenesis model. Lab. Investig. 1997, 76, 365–374. [Google Scholar] [PubMed]
- Soultanakis, R.P.; Melamede, R.J.; Bespalov, I.A.; Wallace, S.S.; Beckman, K.B.; Ames, B.N.; Taatjes, D.J.; Janssen-Heininger, Y.M. Fluorescence detection of 8-oxoguanine in nuclear and mitochondrial DNA of cultured cells using a recombinant Fab and confocal scanning laser microscopy. Free Radic. Biol. Med. 2000, 28, 987–998. [Google Scholar] [CrossRef]
- Nakae, Y.; Stoward, P.J.; Bespalov, I.A.; Melamede, R.J.; Wallace, S.S. A new technique for the quantitative assessment of 8-oxoguanine in nuclear DNA as a marker of oxidative stress. Application to dystrophin-deficient DMD skeletal muscles. Histochem. Cell Biol. 2005, 124, 335–345. [Google Scholar] [CrossRef]
- Ohno, M.; Oka, S.; Nakabeppu, Y. Quantitative analysis of oxidized guanine, 8-oxoguanine, in mitochondrial DNA by immunofluorescence method. Methods Mol. Biol. 2009, 554, 199–212. [Google Scholar]
- Struthers, L.; Patel, R.; Clark, J.; Thomas, S. Direct detection of 8-oxodeoxyguanosine and 8-oxoguanine by avidin and its analogues. Anal. Biochem. 1998, 255, 20–31. [Google Scholar] [CrossRef]
- Bayer, E.A.; Wilchek, M. Biotin-binding proteins: Overview and prospects. Methods Enzymol. 1990, 184, 49–51. [Google Scholar]
- Chen, S.K.; Tsai, M.H.; Lin, C.H.; Hwang, J.J.; Chang, W.P. Determination of 8-oxoguanine in individual cell nucleus of gamma-irradiated mammalian cells. Radiat. Res. 2001, 155, 832–836. [Google Scholar] [CrossRef]
- Gad, H.; Koolmeister, T.; Jemth, A.S.; Eshtad, S.; Jacques, S.A.; Ström, C.E.; Svensson, L.M.; Schultz, N.; Lundbäck, T.; Einarsdottir, B.O.; et al. MTH1 inhibition eradicates cancer by preventing sanitation of the dNTP pool. Nature 2014, 508, 215–221. [Google Scholar] [CrossRef]
- Galanos, P.; Pappas, G.; Polyzos, A.; Kotsinas, A.; Svolaki, I.; Giakoumakis, N.N.; Glytsou, C.; Pateras, I.S.; Swain, U.; Souliotis, V.L.; et al. Mutational signatures reveal the role of RAD52 in p53-independent p21-driven genomic instability. Genome Biol. 2018, 19, 37. [Google Scholar] [CrossRef]
- Kawanishi, S.; Murata, M. Mechanism of DNA damage induced by bromate differs from general types of oxidative stress. Toxicology 2006, 221, 172–178. [Google Scholar] [CrossRef]
- Ballmaier, D.; Epe, B. DNA damage by bromate: Mechanisms and consequences. Toxicology 2006, 221, 166–171. [Google Scholar] [CrossRef]
- D’Errico, M.; Parlanti, E.; Teson, M.; Bernardes de Jesus, B.M.; Degan, P.; Calcagnile, A.; Jaruga, P.; Bjørås, M.; Crescenzi, M.; Pedrini, A.M.; et al. New functions of XPC in the protection of human skin cells from oxidative damage. EMBO J. 2006, 25, 4305–4315. [Google Scholar] [CrossRef]
- Collins, A.R. The comet assay for DNA damage and repair. Mol. Biotechnol. 2004, 26, 249–261. [Google Scholar] [CrossRef]
- Afanasieva, K.; Chopei, M.; Lozovik, A.; Semenova, A.; Lukash, L.; Sivolob, A. DNA loop organization in nucleoids from cells of different types. Biochem. Biophys. Res. Commun. 2017, 483, 142–146. [Google Scholar] [CrossRef]
- Conners, R.; Hooley, E.; Clarke, A.R.; Thomas, S.; Brady, R.L. Recognition of oxidatively modified bases within the biotin-binding site of avidin. J. Mol. Biol. 2006, 357, 263–274. [Google Scholar] [CrossRef]
- Campalans, A.; Kortulewski, T.; Amouroux, R.; Menoni, H.; Vermeulen, W.; Radicella, J.P. Distinct spatiotemporal patterns and PARP dependence of XRCC1 recruitment to single-strand break and base excision repair. Nucleic Acids Res. 2013, 41, 3115–3129. [Google Scholar] [CrossRef]
- Wienholz, F.; Vermeulen, W.; Marteijn, J.A. Amplification of unscheduled DNA synthesis signal enables fluorescence-based single cell quantification of transcription-coupled nucleotide excision repair. Nucleic Acids Res. 2017, 45, e68. [Google Scholar] [CrossRef]
- Kumar, N.; Theil, A.F.; Roginskaya, V.; Ali, Y.; Calderon, M.; Watkins, S.C.; Barnes, R.P.; Opresko, P.L.; Pines, A.; Lans, H.; et al. Global and transcription-coupled repair of 8-oxoG is initiated by nucleotide excision repair proteins. Nat. Commun. 2022, 13, 974. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Nakagawa, O.; Koga, Y.; Taniguchi, Y.; Sasaki, S. Synthesis of new derivatives of 8-oxoG-Clamp for better understanding the recognition mode and improvement of selective affinity. Bioorg. Med. Chem. 2010, 18, 3992–3998. [Google Scholar] [CrossRef] [PubMed]
- Furman, J.L.; Mok, P.W.; Badran, A.H.; Ghosh, I. Turn-on DNA damage sensors for the direct detection of 8-oxoguanine and photoproducts in native DNA. J. Am. Chem. Soc. 2011, 133, 12518–12527. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Wang, Y.; Meng, X.; Dhar, R.; Huang, H. Triple-stranded DNA containing 8-oxo-7,8-dihydro-2′-deoxyguanosine: Implication in the design of selective aptamer sensors for 8-oxo-7,8-dihydroguanine. Anal. Chem. 2013, 85, 201–207. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maraventano, G.; Ticli, G.; Cazzalini, O.; Stivala, L.A.; Ramos-Gonzalez, M.; Rodríguez, J.-L.; Prosperi, E. Single Cell Determination of 7,8-dihydro-8-oxo-2′-deoxyguanosine by Fluorescence Techniques: Antibody vs. Avidin Labeling. Molecules 2023, 28, 4326. https://doi.org/10.3390/molecules28114326
Maraventano G, Ticli G, Cazzalini O, Stivala LA, Ramos-Gonzalez M, Rodríguez J-L, Prosperi E. Single Cell Determination of 7,8-dihydro-8-oxo-2′-deoxyguanosine by Fluorescence Techniques: Antibody vs. Avidin Labeling. Molecules. 2023; 28(11):4326. https://doi.org/10.3390/molecules28114326
Chicago/Turabian StyleMaraventano, Giusy, Giulio Ticli, Ornella Cazzalini, Lucia A. Stivala, Mariella Ramos-Gonzalez, José-Luis Rodríguez, and Ennio Prosperi. 2023. "Single Cell Determination of 7,8-dihydro-8-oxo-2′-deoxyguanosine by Fluorescence Techniques: Antibody vs. Avidin Labeling" Molecules 28, no. 11: 4326. https://doi.org/10.3390/molecules28114326
APA StyleMaraventano, G., Ticli, G., Cazzalini, O., Stivala, L. A., Ramos-Gonzalez, M., Rodríguez, J.-L., & Prosperi, E. (2023). Single Cell Determination of 7,8-dihydro-8-oxo-2′-deoxyguanosine by Fluorescence Techniques: Antibody vs. Avidin Labeling. Molecules, 28(11), 4326. https://doi.org/10.3390/molecules28114326






