Multiple Bioimaging Applications Based on the Excellent Properties of Nanodiamond: A Review
(This article belongs to the Section Materials Chemistry)
Abstract
1. Introduction
2. Fluorescence Imaging
2.1. Photoluminescence Spectroscopy

2.2. Imaging Microscope
2.2.1. Optical Microscope
2.2.2. Electron Microscope
2.2.3. Atomic Force Microscope
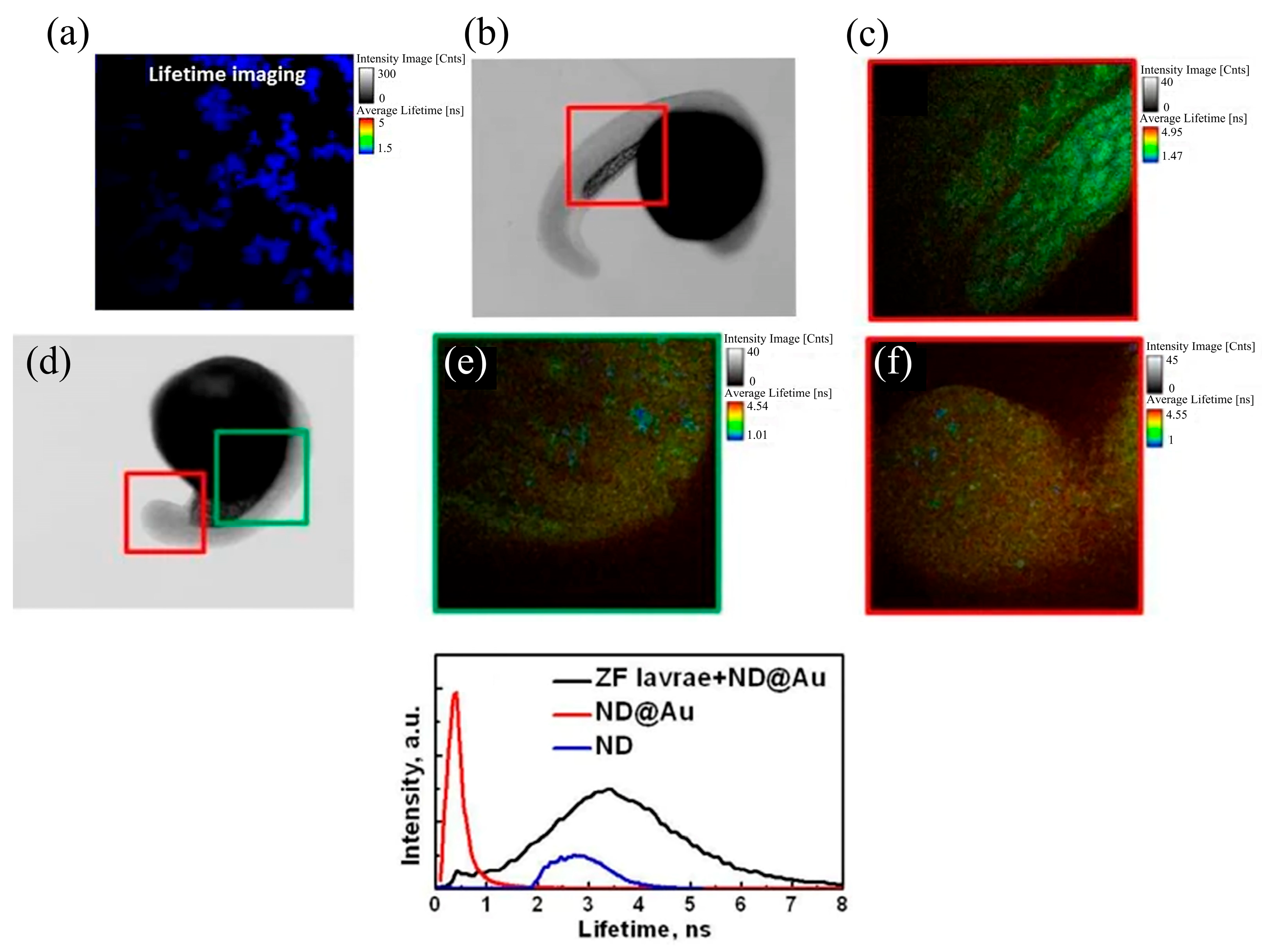
2.3. Fluorescence Lifetime Imaging
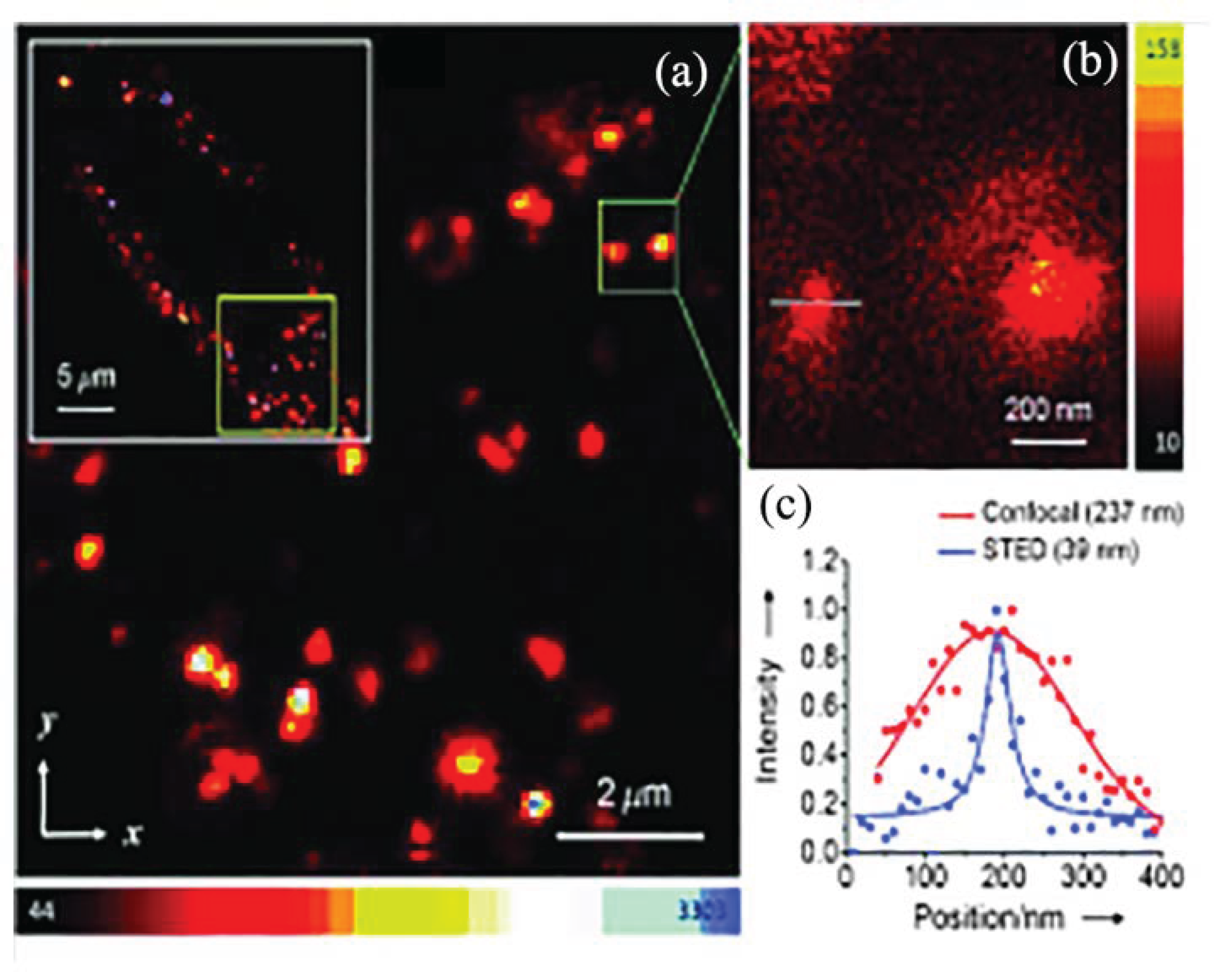
2.4. Super-Resolution Optical Imaging
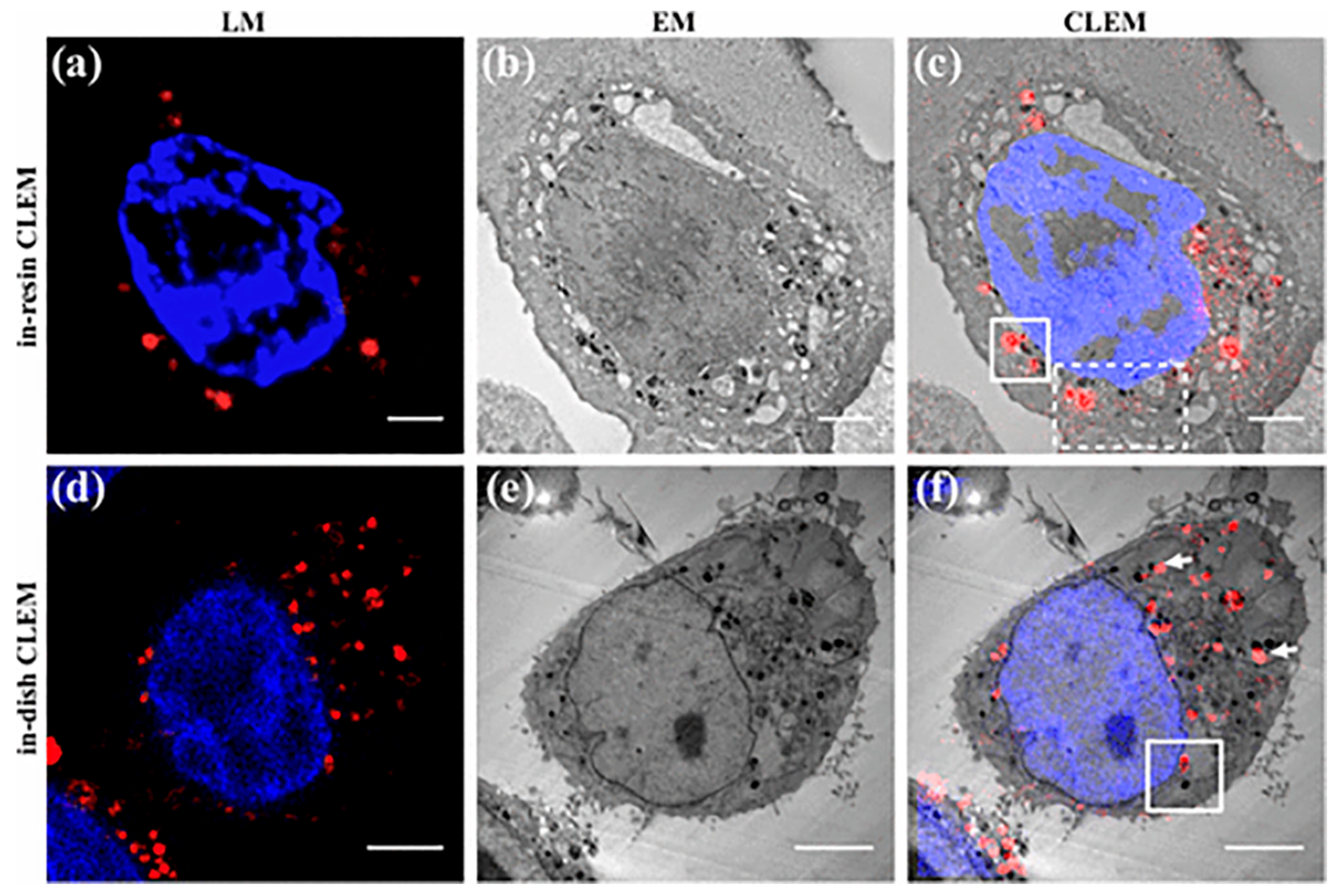
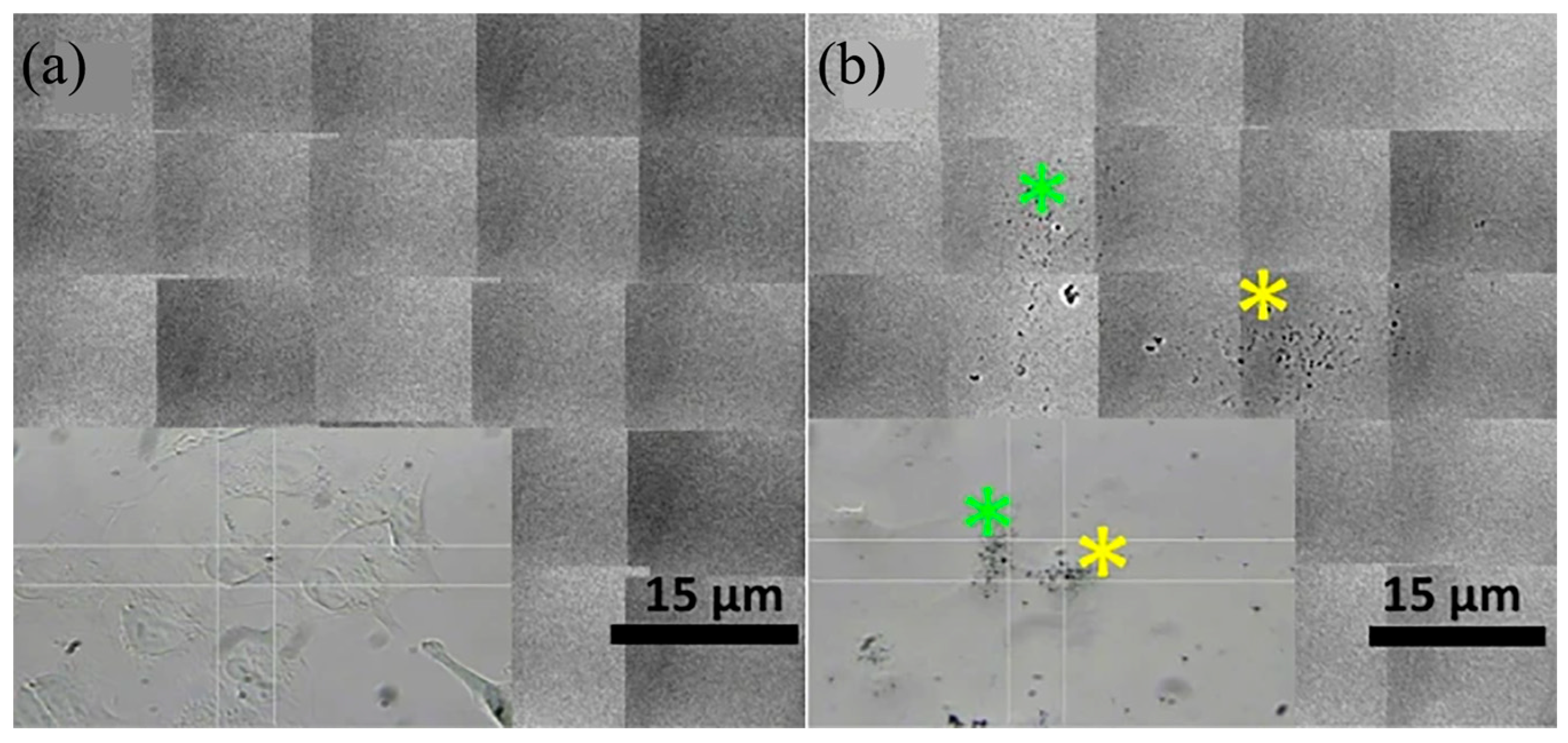
2.5. Correlative Light and Electron Microscope
3. Raman Imaging
4. X-ray Imaging
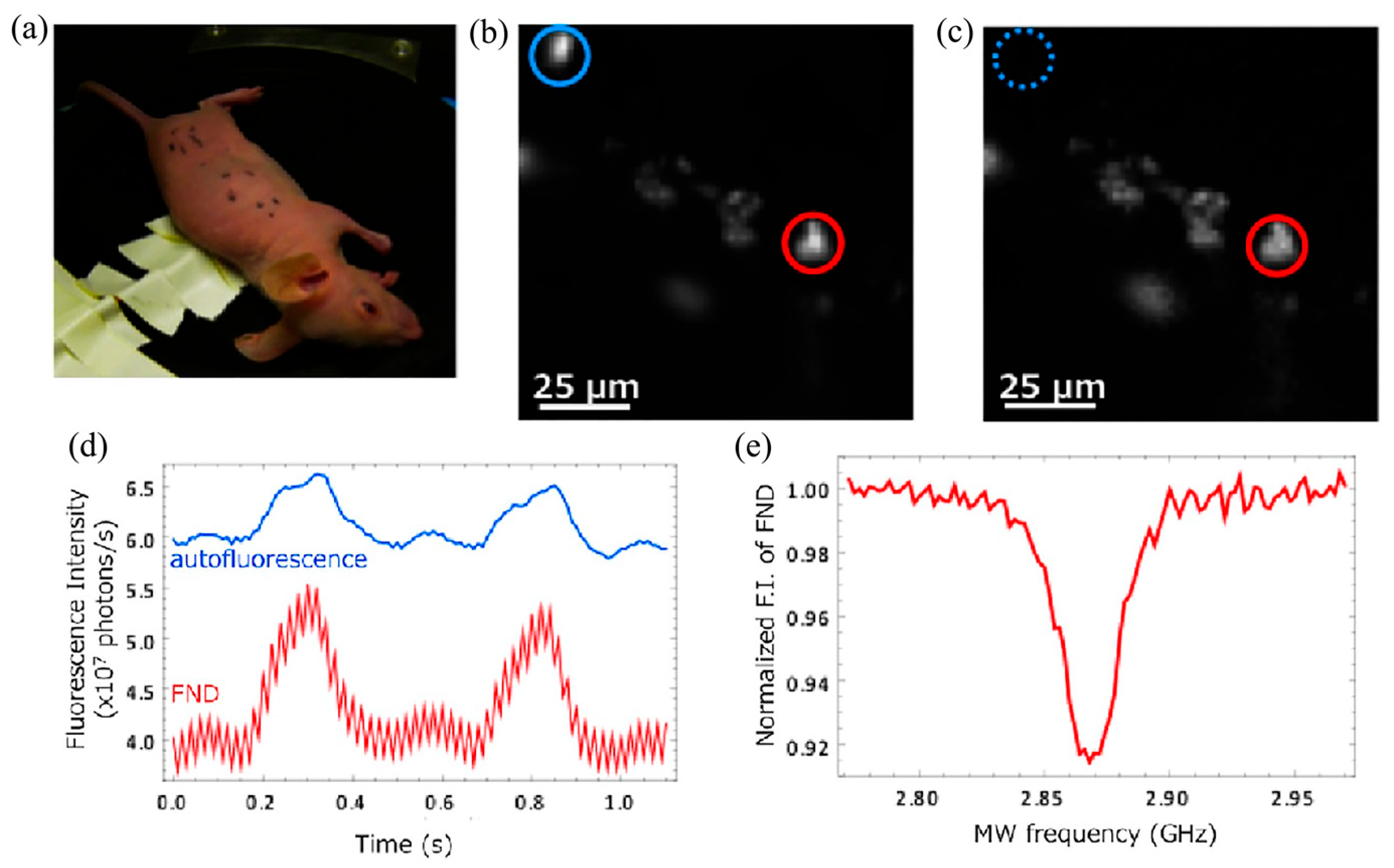
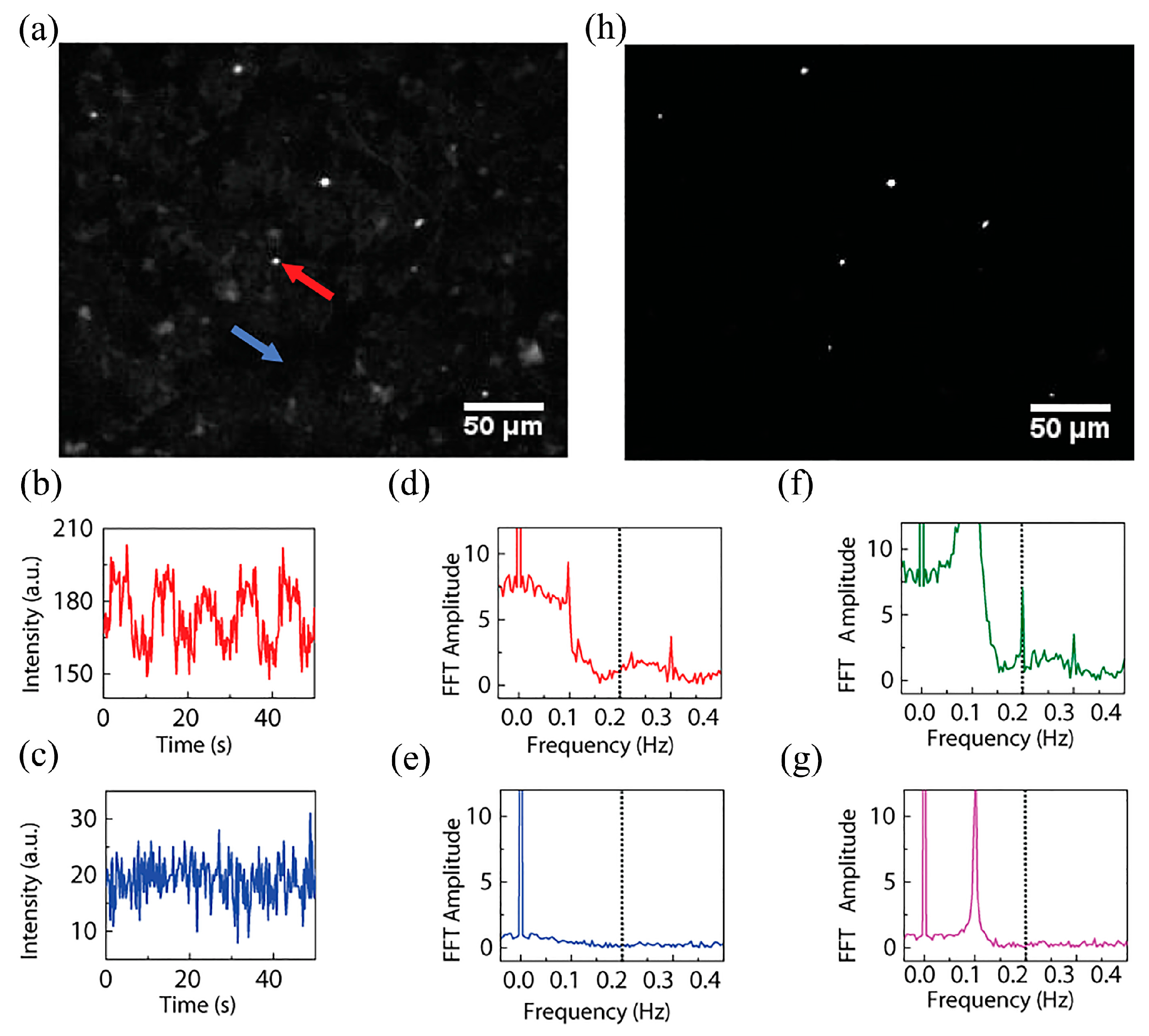
5. Magnetic Modulation Fluorescence Imaging
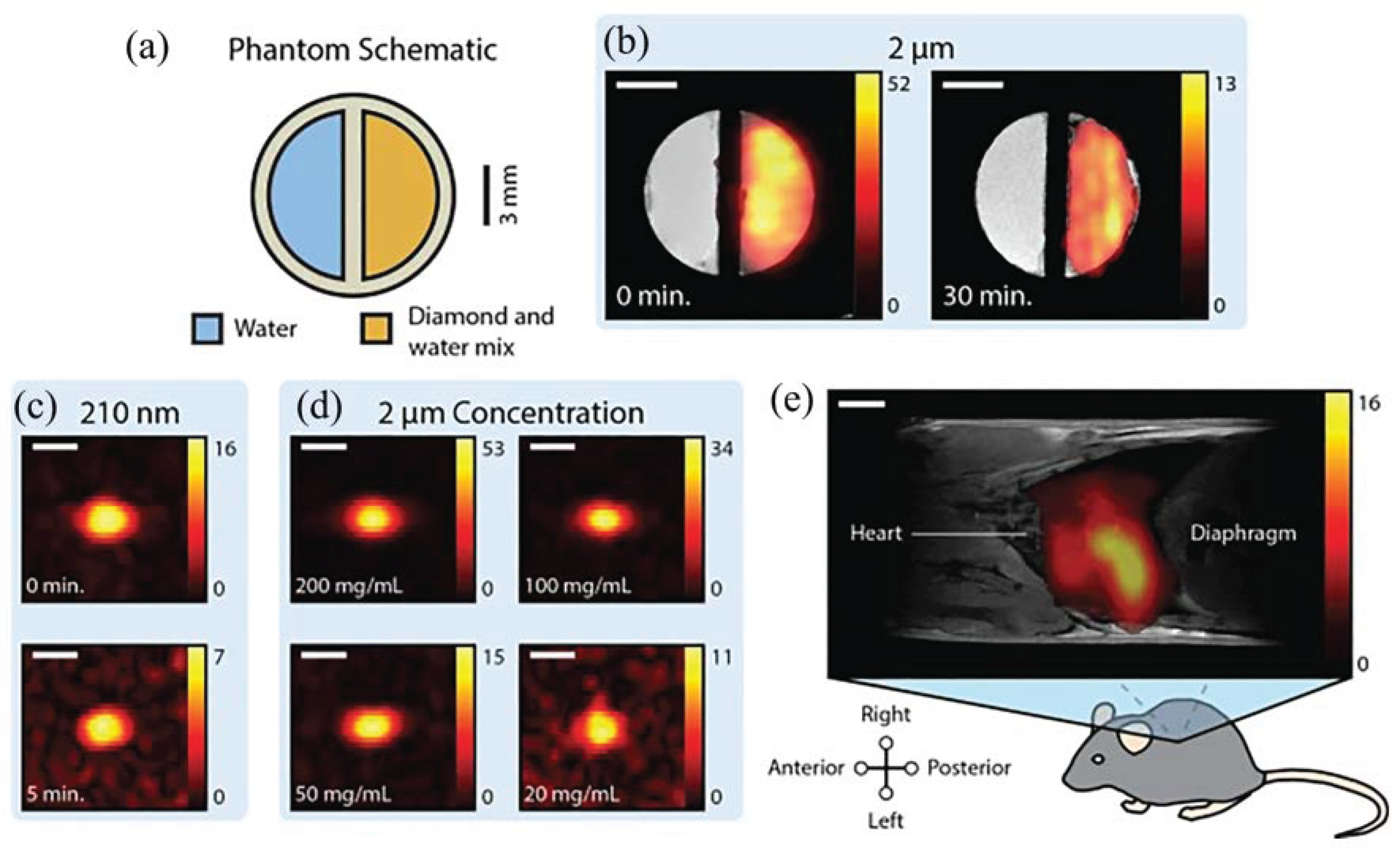
6. Magnetic Resonance Imaging
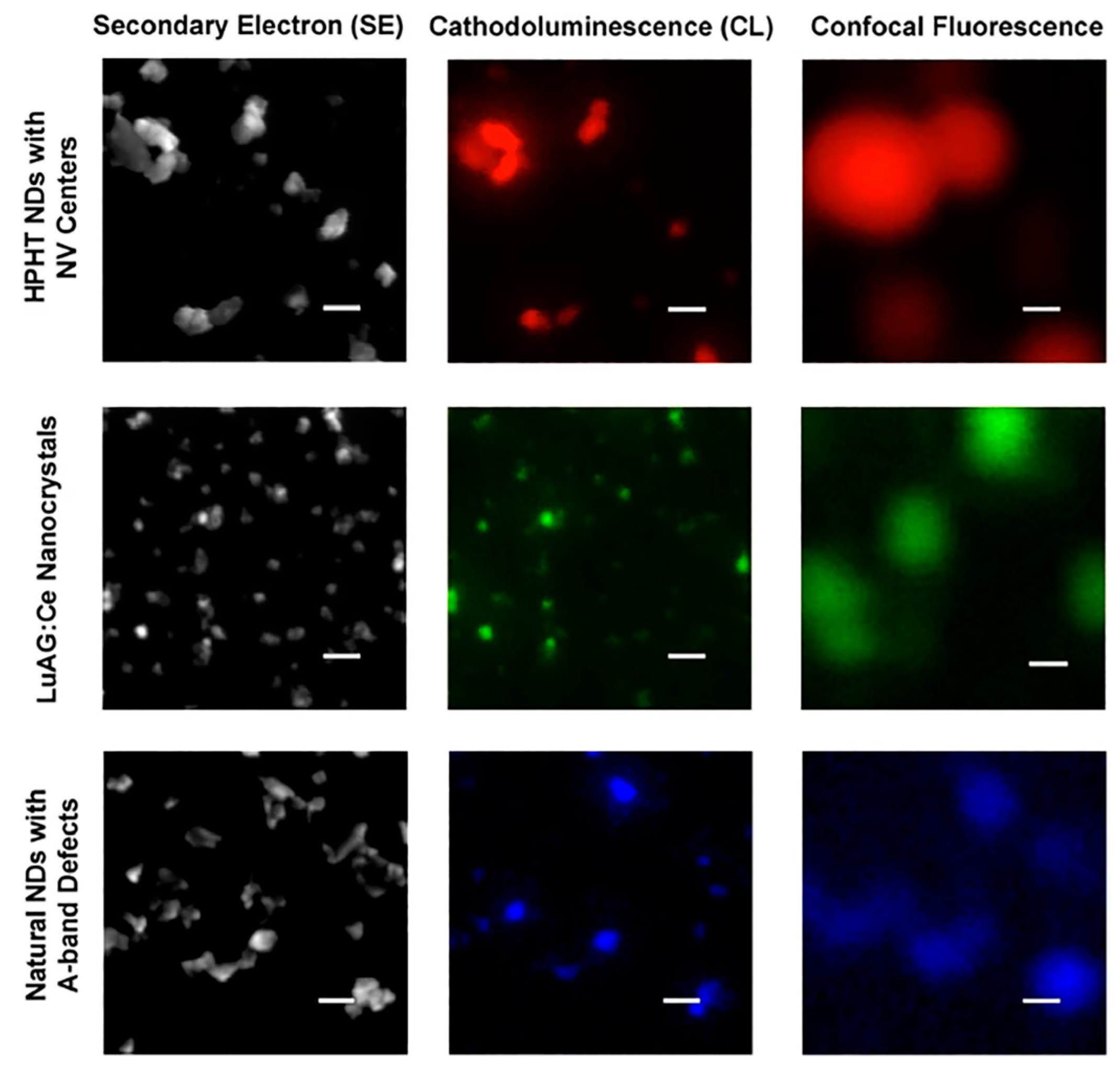
7. Cathodoluminescence Imaging
8. Optical Coherence Tomography Imaging
9. Conclusions and Perspective
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Turcheniuk, K.; Mochalin, V.N. Biomedical applications of nanodiamond. Nanotechnology 2017, 28, 252001. [Google Scholar] [CrossRef] [PubMed]
- Perevedentseva, E.; Lin, Y.C.; Jani, M.; Cheng, C.L. Biomedical applications of nanodiamonds in imaging and therapy. Nanomedicine 2013, 8, 2041–2060. [Google Scholar] [CrossRef]
- Sow, M.; Steuer, H.; Adekanye, S.; Ginés, L.; Mandal, S.; Gilboa, B.; Williams, O.A.; Smith, J.M.; Kapanidis, A.N. High-throughput nitrogen-vacancy center imaging for NDs photophysical characterization and pH nanosensing. Nanoscale 2020, 12, 21821–21831. [Google Scholar] [CrossRef] [PubMed]
- Zeleneev, A.I.; Bolshedvorskii, S.V.; Zhulikov, L.A.; Sochenko, V.V.; Rubinas, O.R.; Sorokin, V.N.; Smolyaninov, A.N.; Akimov, A.V. On studying the optical properties of NV/SiV color centers in ultrasmall nanodiamonds. In AIP Conference Proceedings; AIP Publishing LLC: Melville, NY, USA, 2022; Volume 2241, p. 020039. [Google Scholar]
- Lin, Y.C.; Tsai, L.W.; Perevedentseva, E.; Karmenyan, A.; Cheng, C.L. Near-infrared fluorescence from NDs for multimodal bioimaging. Sovrem. Tehnol. Med. 2018, 10, 49–56. [Google Scholar] [CrossRef]
- Qin, J.X.; Yang, X.G.; Lv, C.F.; Li, Y.Z.; Liu, K.K.; Zang, J.H.; Yang, X.; Dong, L.; Shan, C.X. Nanodiamonds: Synthesis, properties, and applications in nanomedicine. Mater. Des. 2021, 210, 110091. [Google Scholar] [CrossRef]
- Vervald, A.M.; Burikov, S.A.; Scherbakov, A.M.; Kudryavtsev, O.S.; Kalyagina, N.A.; Vlasov, I.I.; Ekimov, E.A.; Dolenko, T.A. Boron-doped nanodiamonds as anticancer agents: En route to hyperthermia/thermoablation therapy. ACS Biomater. Sci. Eng. 2020, 6, 4446–4453. [Google Scholar] [CrossRef]
- Kolarova, K.; Bydzovska, I.; Romanyuk, O.; Shagieva, E.; Ukraintsev, E.; Kromka, A.; Rezek, B.; Stehlik, S. Hydrogenation of HPHT nanodiamonds and their nanoscale interaction with chitosan. Diam. Relat. Mater. 2023, 134, 109754. [Google Scholar] [CrossRef]
- Zhang, K.; Guo, Q.; Zhao, Q.; Wang, F.; Wang, H.; Zhi, J.; Shan, C. Photosensitizer Functionalized NDs for Raman Imaging and Photodynamic Therapy of Cancer Cells. Langmuir 2021, 37, 4308–4315. [Google Scholar] [CrossRef]
- Wu, T.J.; Tzeng, Y.K.; Chang, W.W.; Cheng, C.A.; Kuo, Y.; Chien, C.H.; Chang, H.-C.; Yu, J. Tracking the engraftment and regenerative capabilities of transplanted lung stem cells using FNDs. Nat. Nanotechnol. 2013, 8, 682–689. [Google Scholar] [CrossRef]
- Han, S.; Raabe, M.; Hodgson, L.; Mantell, J.; Verkade, P.; Lasser, T.; Landfester, K.; Weil, T.; Lieberwirth, I. High-contrast imaging of NDs in cells by energy filtered and correlative light-electron microscopy: Toward a quantitative nanoparticle-cell analysis. Nano Lett. 2019, 19, 2178–2185. [Google Scholar] [CrossRef]
- Zhang, P.; Yang, J.; Li, W.; Wang, W.; Liu, C.; Griffith, M.; Liu, W. Cationic polymer brush grafted-nanodiamond via atom transfer radical polymerization for enhanced gene delivery and bioimaging. J. Mater. Chem. 2011, 21, 7755–7764. [Google Scholar] [CrossRef]
- Perevedentseva, E.; Ali, N.; Karmenyan, A.; Skovorodkin, I.; Prunskaite-Hyyryläinen, R.; Vainio, S.; Cheng, C.-L.; Kinnunen, M. Optical studies of NDs-tissue interaction: Skin penetration and localization. Materials 2019, 12, 3762. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Alam, M.N.A.; Balasubramanian, P.; Winterwerber, P.; Ermakova, A.; Müller, M.; Wagner, M.; Jelezko, F.; Raabe, M.; Weil, T. Fluorescent nanodiamond–nanogels for nanoscale sensing and photodynamic applications. Adv. NanoBiomed Res. 2021, 1, 2000101. [Google Scholar] [CrossRef]
- Perevedentseva, E.; Lin, Y.C.; Cheng, C.L. A review of recent advances in nanodiamond-mediated drug delivery in cancer. Expert Opin. Drug Deliv. 2021, 18, 369–382. [Google Scholar] [CrossRef] [PubMed]
- Avdeev, M.V.; Tomchuk, O.V.; Ivankov, O.I.; Alexenskii, A.E.; Dideikin, A.T.; Vul, A.Y. On the structure of concentrated detonation nanodiamond hydrosols with a positive ζ potential: Analysis of small-angle neutron scattering. Chem. Phys. Lett. 2016, 658, 58–62. [Google Scholar] [CrossRef]
- Knizhnik, A.A.; Polynskaya, Y.G.; Sinitsa, A.S.; Kuznetsov, N.M.; Belousov, S.I.; Chvalun, S.N.; Potapkin, B.V. Analysis of structural organization and interaction mechanisms of detonation nanodiamond particles in hydrosols. Phys. Chem. Chem. Phys. 2021, 23, 674–682. [Google Scholar] [CrossRef]
- Tomchuk, O.V.; Avdeev, M.V.; Aksenov, V.L.; Garamus, V.M.; Bulavin, L.A.; Ivashevskaya, S.N.; Rozhkova, N.N.; Schreiber, N.; Schreiber, J. Comparative structural characterization of the water dispersions of detonation nanodiamonds by small-angle neutron scattering. J. Surf. Investig. X-ray Synchrotron Neutron Tech. 2012, 6, 821–824. [Google Scholar] [CrossRef]
- Williams, O.A.; Hees, J.; Dieker, C.; Jager, W.; Kirste, L.; Nebel, C.E. Size-dependent reactivity of diamond nanoparticles. ACS Nano 2010, 4, 4824–4830. [Google Scholar] [CrossRef]
- Dideikin, A.T.; Aleksenskii, A.E.; Baidakova, M.V.; Brunkov, P.N.; Brzhezinskaya, M.; Davydov, V.Y.; Levitskii, V.S.; Kidalov, S.V.; Kukushkina, Y.A.; Vul, A.Y.; et al. Rehybridization of carbon on facets of detonation diamond nanocrystals and forming hydrosols of individual particles. Carbon 2017, 122, 737–745. [Google Scholar] [CrossRef]
- Aleksenskiy, A.E.; Eydelman, E.D.; Vul, A.Y. Deagglomeration of detonation nanodiamonds. Nanotechnol. Lett. 2011, 3, 68. [Google Scholar] [CrossRef]
- Petrakova, V.; Benson, V.; Buncek, M.; Fiserova, A.; Ledvina, M.; Stursa, J.; Cigler, P.; Nesladek, M. Imaging of transfection and intracellular release of intact, non-labeled DNA using FNDs. Nanoscale 2016, 8, 12002–12012. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Kaminski Schierle, G.S.; Lei, B.; Liu, Y.; Kaminski, C.F. Fluorescent nanoparticles for super-resolution imaging. Chem. Rev. 2022, 122, 12495–12543. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Liu, M.; Huang, Q.; Huang, L.; Huang, H.; Deng, F.; Wen, Y.; Tian, J.; Zhang, X.; Wei, Y. Facile preparation of fluorescent nanodiamond-based polymer composites through a metal-free photo-initiated RAFT process and their cellular imaging. Chem. Eng. J. 2018, 337, 82–90. [Google Scholar] [CrossRef]
- Osipov, V.Y.; Shakhov, F.M.; Bogdanov, K.V.; Takai, K.; Hayashi, T.; Treussart, F.; Baldycheva, A.; Hogan, B.T.; Jentgens, C. High-quality green-emitting NDs fabricated by HPHT sintering of polycrystalline shockwave diamonds. Nanoscale Res. Lett. 2020, 15, 209. [Google Scholar] [CrossRef]
- Su, L.X.; Lou, Q.; Zang, J.H.; Shan, C.X.; Gao, Y.F. Temperature-dependent fluorescence in NDs. Appl. Phys. Express 2017, 10, 025102. [Google Scholar] [CrossRef]
- Rondin, L.; Dantelle, G.; Slablab, A.; Grosshans, F.; Treussart, F.; Bergonzo, P.; Perruchas, S.; Gacoin, T.; Chaigneau, H.-C.; Chang, V.; et al. Surface-induced charge state conversion of nitrogen-vacancy defects in NDs. Phys. Rev. B 2010, 82, 115449. [Google Scholar] [CrossRef]
- Wang, B.K.; Barbiero, M.; Zhang, Q.M.; Gu, M. Super-resolution optical microscope: Principle, instrumentation, and application. Front. Inf. Technol. Electron. Eng. 2019, 20, 608–630. [Google Scholar] [CrossRef]
- Schmidheini, L.; Tiefenauer, R.F.; Gatterdam, V.; Frutiger, A.; Sannomiya, T.; Aramesh, M. Self-assembly of nanodiamonds and plasmonic nanoparticles for nanoscopy. Biosensors 2022, 12, 148. [Google Scholar] [CrossRef]
- Liu, W.; Naydenov, B.; Chakrabortty, S.; Wuensch, B.; Hubner, K.; Ritz, S.; Cölfen, H.; Barth, H.; Koynov, K.; Weil, T.; et al. Fluorescent nanodiamond–gold hybrid particles for multimodal optical and electron microscopy cellular imaging. Nano Lett. 2016, 16, 6236–6244. [Google Scholar] [CrossRef]
- Hubbuch, J.; Kula, M.R. Confocal laser scanning microscopy as an analytical tool in chromatographic research. Bioprocess Biosyst. Eng. 2008, 31, 241–259. [Google Scholar] [CrossRef]
- Kulvelis, Y.V.; Lebedev, V.T.; Yevlampieva, N.P.; Cherechukin, D.S.; Yudina, E.B. Enhancement of Singlet Oxygen Generation of Radachlorin® Conjugated with Polyvinylpyrrolidone and Nanodiamonds in Aqueous Media. In Green Photocatalytic Semiconductors: Recent Advances and Applications; Springer: Cham, Switzerland, 2022; pp. 281–306. [Google Scholar] [CrossRef]
- Lebedev, V.T.; Török, G.; Kulvelis, Y.V.; Soroka, M.A.; Ganzha, V.A.; Orlova, V.A.; Fomin, E.V.; Sharanova, L.V.; Shvidchenko, A.V. New Photocatalytic Materials Based on Complexes of Nanodiamonds with Diphthalocyanines of Rare Earth Elements. In Green Photocatalytic Semiconductors: Recent Advances and Applications; Springer: Cham, Switzerland, 2022; pp. 179–208. [Google Scholar] [CrossRef]
- Terada, D.; Genjo, T.; Segawa, T.F.; Igarashi, R.; Shirakawa, M. Nanodiamonds for bioapplications–specific targeting strategies. Biochim. Et Biophys. Acta (BBA)-Gen. Subj. 2020, 1864, 129354. [Google Scholar] [CrossRef] [PubMed]
- Khalid, A.; Mitropoulos, A.N.; Marelli, B.; Simpson, D.A.; Tran, P.A.; Omenetto, F.G.; Tomljenovic-Hanic, S. Fluorescent nanodiamond silk fibroin spheres: Advanced nanoscale bioimaging tool. ACS Biomater. Sci. Eng. 2015, 1, 1104–1113. [Google Scholar] [CrossRef]
- Morita, M.; Tachikawa, T.; Seino, S.; Tanaka, K.; Majima, T. Controlled synthesis of gold nanoparticles on fluorescent nanodiamond via electron-beam-induced reduction method for dual-modal optical and electron bioimaging. ACS Appl. Nano Mater. 2017, 1, 355–363. [Google Scholar] [CrossRef]
- Han, S.J.; Seo, M.; Cho, M.G.; Kwon, Y.W.; Lim, S.H.; Han, S.H. Sub-5 nm Nanodiamonds Fabricated by Plasma Immersion Ion Implantation as Fluorescent Probes. ACS Appl. Nano Mater. 2021, 4, 2238–2246. [Google Scholar] [CrossRef]
- Benson, V.; Amini, A. Why nanodiamond carriers manage to overcome drug resistance in cancer. Cancer Drug Resist. 2020, 3, 854. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zheng, Y.; Zhao, D.; Zhao, Z.; Liu, L.; Pliss, A.; Zhu, F.; Liu, J.; Qu, J.; Luan, P. Applications of fluorescence lifetime imaging in clinical medicine. J. Innov. Opt. Health Sci. 2018, 11, 1830001. [Google Scholar] [CrossRef]
- Alfonso-Garcia, A.; Bec, J.; Weyers, B.; Marsden, M.; Zhou, X.; Li, C.; Marcu, L. Mesoscopic fluorescence lifetime imaging: Fundamental principles, clinical applications and future directions. J. Biophotonics 2021, 14, e202000472. [Google Scholar] [CrossRef]
- Lin, Y.C.; Perevedentseva, E.; Lin, Z.R.; Chang, C.C.; Chen, H.H.; Yang, S.M.; Lin, M.-D.; Karmenyan, A.; Speranza, G.; Minati, L.; et al. Multimodal bioimaging using nanodiamond and gold hybrid nanoparticles. Sci. Rep. 2022, 12, 5331. [Google Scholar] [CrossRef]
- Liu, Y.Y.; Chang, B.M.; Chang, H.C. Nanodiamond-enabled biomedical imaging. Nanomedicine 2020, 15, 1599–1616. [Google Scholar] [CrossRef]
- Hui, Y.Y.; Su, L.J.; Chen, O.Y.; Chen, Y.T.; Liu, T.M.; Chang, H.C. Wide-field imaging and flow cytometric analysis of cancer cells in blood by fluorescent nanodiamond labeling and time gating. Sci. Rep. 2014, 4, 5574. [Google Scholar] [CrossRef]
- Su, L.J.; Wu, M.S.; Hui, Y.Y.; Chang, B.M.; Pan, L.; Hsu, P.C.; Chen, Y.-T.; Ho, H.-N.; Huang, Y.-H.; Ling, T.-Y.; et al. Fluorescent nanodiamonds enable quantitative tracking of human mesenchymal stem cells in miniature pigs. Sci. Rep. 2017, 7, 45607. [Google Scholar] [CrossRef] [PubMed]
- Torelli, M.D.; Nunn, N.A.; Shenderova, O.A. A perspective on fluorescent nanodiamond bioimaging. Small 2019, 15, 1902151. [Google Scholar] [CrossRef]
- Pfender, M.; Aslam, N.; Waldherr, G.; Neumann, P.; Wrachtrup, J. Single-spin stochastic optical reconstruction microscopy. Proc. Natl. Acad. Sci. USA 2014, 111, 14669–14674. [Google Scholar] [CrossRef]
- Castelletto, S.; Boretti, A. Color centers in wide-bandgap semiconductors for subdiffraction imaging: A review. Adv. Photonics 2021, 3, 054001. [Google Scholar] [CrossRef]
- Opstad, I.S.; Wolfson, D.L.; Øie, C.I.; Ahluwalia, B.S. Multi-color imaging of sub-mitochondrial structures in living cells using structured illumination microscopy. Nanophotonics 2018, 7, 935–947. [Google Scholar] [CrossRef]
- Melnikov, A.; Lun, D.P.K. 3D Imaging with Moving Fringe Structured Illumination Microscopy. IEEE Photonics J. 2022, 14, 1–10. [Google Scholar] [CrossRef]
- Sotoma, S.; Hsieh, F.J.; Chang, H.C. Biohybrid FNDs as dual-contrast markers for light and electron microscopies. J. Chin. Chem. Soc. 2018, 65, 1136–1146. [Google Scholar] [CrossRef]
- Xu, Y.; Gan, H.; Lu, Z.; Ye, R.; Chen, T.; Zhang, J.; Ying, T.; Xu, S. The fabrication of NDs doped tellurite germanate glass with robust structure and its luminescence. J. Am. Ceram. Soc. 2021, 104, 1698–1706. [Google Scholar] [CrossRef]
- Gao, G.; Guo, Q.; Zhi, J. NDs-based theranostic platform for drug delivery and bioimaging. Small 2019, 15, 1902238. [Google Scholar] [CrossRef]
- Liu, W.; Speranza, G. Functionalization of carbon nanomaterials for biomedical applications. C 2019, 5, 72. [Google Scholar] [CrossRef]
- Sundar, L.S.; Anjum, N.A.; Ferro, M.C.; Pereira, E.; Singh, M.K.; Sousa, A.C.M. Biocompatibility and biotoxicity of in-situ synthesized carboxylated NDs-cobalt oxide nanocomposite. J. Mater. Sci. Technol. 2017, 33, 879–888. [Google Scholar] [CrossRef]
- Shery, L.Y. Dynamic self-assembly of detonation NDs in water. Nanoscale 2020, 12, 5363–5367. [Google Scholar]
- Jung, H.S.; Neuman, K.C. Surface modification of fluorescent nanodiamonds for biological applications. Nanomaterials 2021, 11, 153. [Google Scholar] [CrossRef]
- Igarashi, R.; Yoshinari, Y.; Yokota, H.; Sugi, T.; Sugihara, F.; Ikeda, K.; Sumiya, H.; Tsuji, S.; Mori, I.; Tochio, H.; et al. Real-time background-free selective imaging of fluorescent nanodiamonds in vivo. Nano Lett. 2012, 12, 5726–5732. [Google Scholar] [CrossRef] [PubMed]
- Nagarajan, S.; Pioche-Durieu, C.; Tizei, L.H.; Fang, C.Y.; Bertrand, J.R.; Le Cam, E.; Chang, H.-C.; Treussart, F.; Kociak, M. Simultaneous cathodoluminescence and electron microscopy cytometry of cellular vesicles labeled with FNDs. Nanoscale 2016, 8, 11588–11594. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, S.K.; Bumb, A.; Wu, X.; Sochacki, K.A.; Kellman, P.; Brechbiel, M.W.; Neuman, K.C. Wide-field in vivo background free imaging by selective magnetic modulation of nanodiamond fluorescence. Biomed. Opt. Express 2014, 5, 1190–1202. [Google Scholar] [CrossRef]
- Mamin, H.J.; Kim, M.; Sherwood, M.H.; Rettner, C.T.; Ohno, K.; Awschalom, D.D.; Rugar, D. Nanoscale nuclear magnetic resonance with a nitrogen-vacancy spin sensor. Science 2013, 339, 557–560. [Google Scholar] [CrossRef]
- Panich, A.M.; Salti, M.; Prager, O.; Swissa, E.; Kulvelis, Y.V.; Yudina, E.B.; Alekesenskii, A.E.; Goren, S.D.; Vul, A.Y.; Shames, A.I. PVP-coated Gd-grafted nanodiamonds as a novel and potentially safer contrast agent for in vivo MRI. Magn. Reson. Med. 2021, 86, 935–942. [Google Scholar] [CrossRef]
- Lebedev, V.T.; Kulvelis, Y.V.; Peters, G.S.; Bolshakova, O.I.; Sarantseva, S.V.; Popova, M.V.; Vul, A.Y. Complexes of nanodiamonds with Gd-fullerenols for biomedicine. Fuller. Nanotub. Carbon Nanostructures 2022, 30, 36–45. [Google Scholar] [CrossRef]
- Lebedev, V.T.; Török, G.; Kulvelis, Y.V.; Bolshkova, O.I.; Yevlampieva, N.P.; Soroka, M.A.; Fomin, E.V.; Vul, A.Y.; Garg, S. Diamond-based nanostructures with metal-organic molecules. Soft Mater. 2022, 20 (Suppl. S1), S34–S43. [Google Scholar] [CrossRef]
- Yano, K.; Matsumoto, T.; Okamoto, Y.; Bito, K.; Kurokawa, N.; Hasebe, T.; Hotta, A. Gadolinium-complexed carboxylated nanodiamond particles for magnetic resonance imaging of the lymphatic system. ACS Appl. Nano Mater. 2021, 4, 1702–1711. [Google Scholar] [CrossRef]
- Panich, A.M.; Salti, M.; Aleksenskii, A.E.; Kulvelis, Y.V.; Chizhikova, A.; Vul, A.Y.; Shames, A.I. Suspensions of manganese-grafted nanodiamonds: Preparation, NMR, and MRI study. Diam. Relat. Mater. 2023, 131, 109591. [Google Scholar] [CrossRef]
- Panich, A.M.; Shames, A.I.; Aleksenskii, A.E.; Yudina, E.B.; Vul, A.Y. Manganese-grafted detonation nanodiamond, a novel potential MRI contrast agent. Diam. Relat. Mater. 2021, 119, 108590. [Google Scholar] [CrossRef]
- Glenn, D.R.; Zhang, H.; Kasthuri, N.; Schalek, R.; Lo, P.K.; Trifonov, A.S.; Park, H.; Lichtman, J.W.; Walsworth, R.L. Correlative light and electron microscopy using cathodoluminescence from nanoparticles with distinguishable colours. Sci. Rep. 2012, 2, 865. [Google Scholar] [CrossRef] [PubMed]
- Tizei, L.H.G.; Kociak, M. Spectrally and spatially resolved cathodoluminescence of NDs: Local variations of the NV0 emission properties. Nanotechnology 2012, 23, 175702. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Sang, D.; Zou, L.; Ge, S.; Yao, Y.; Fan, J.; Wang, Q. Multiple Bioimaging Applications Based on the Excellent Properties of Nanodiamond: A Review. Molecules 2023, 28, 4063. https://doi.org/10.3390/molecules28104063
Wang X, Sang D, Zou L, Ge S, Yao Y, Fan J, Wang Q. Multiple Bioimaging Applications Based on the Excellent Properties of Nanodiamond: A Review. Molecules. 2023; 28(10):4063. https://doi.org/10.3390/molecules28104063
Chicago/Turabian StyleWang, Xinyue, Dandan Sang, Liangrui Zou, Shunhao Ge, Yu Yao, Jianchao Fan, and Qinglin Wang. 2023. "Multiple Bioimaging Applications Based on the Excellent Properties of Nanodiamond: A Review" Molecules 28, no. 10: 4063. https://doi.org/10.3390/molecules28104063
APA StyleWang, X., Sang, D., Zou, L., Ge, S., Yao, Y., Fan, J., & Wang, Q. (2023). Multiple Bioimaging Applications Based on the Excellent Properties of Nanodiamond: A Review. Molecules, 28(10), 4063. https://doi.org/10.3390/molecules28104063






