Abstract
Delineation of clinical complications secondary to fungal infections, such as cryptococcal meningitis, and the concurrent emergence of multidrug resistance in large population subsets necessitates the need for the development of new classes of antifungals. Herein, we report a series of ring-modified histidine-containing short cationic peptides exhibiting anticryptococcal activity via membrane lysis. The N-1 position of histidine was benzylated, followed by iodination at the C-5 position via electrophilic iodination, and the dipeptides were obtained after coupling with tryptophan. In vitro analysis revealed that peptides Trp-His[1-(3,5-di-tert-butylbenzyl)-5-iodo]-OMe (10d, IC50 = 2.20 μg/mL; MIC = 4.01 μg/mL) and Trp-His[1-(2-iodophenyl)-5-iodo)]-OMe (10o, IC50 = 2.52 μg/mL; MIC = 4.59 μg/mL) exhibit promising antifungal activities against C. neoformans. When administered in combination with standard drug amphotericin B (Amp B), a significant synergism was observed, with 4- to 16-fold increase in the potencies of both peptides and Amp B. Electron microscopy analysis with SEM and TEM showed that the dipeptides primarily act via membrane disruption, leading to pore formation and causing cell lysis. After entering the cells, the peptides interact with the intracellular components as demonstrated by confocal laser scanning microscopy (CLSM).
1. Introduction
Multidrug resistance and a compromised immune system due to underlying diseases such as HIV, cancer and COVID-19 infection have rendered easily treatable fungal infections as life-threatening. Cryptococcal meningitis is one such condition, and caused 580,000 deaths in 2020, accounting for 19% of the AIDS-related deaths globally [1]. Various classes of drugs approved for the treatment of invasive fungal infections act on the fungal membrane (polyenes, azoles and echinocandins) and DNA (pyrimidines). For the treatment of cryptococcal meningitis, the first line of treatment includes induction, consolidation and maintenance therapy which primarily relies on the administration of amphotericin B, fluconazole and flucytosine in different combinations for varied time durations [2]. Moreover, various formulations of the most potent and commonly used drug amphotericin B have been developed over a period of time that have shown promising results as compared to the conventional therapy [3]. However, due to acute and chronic toxicities, such as nephrotoxicity associated with amphotericin B and flucytosine, neurotoxicity with fluconazole and various gastrointestinal complications, along with development of resistance in microbes due to extensive and reckless use of antifungal drugs, there is a need for the development of newer classes of antifungals [4]. Hence, various new chemical structural classes are being explored and different anticryptococcal agents are being developed. Some of them act via interactions with various components of the fungal cells such as inhibition of glycosyl phosphatidylinositol (fosmanogepix), fungal-selective calcineurin inhibitor (APX879) and fungal-selective Hsp90 inhibitor (resorcylate aminopyrazoles). However, anticryptococcal agents acting via membrane disruption and lysis are quite popular, for example peptides [5,6], benzothioureas, ibomycin and hydrazycins [7].
Antifungal peptides (AFPs) primarily act via membrane disruption; however, their mechanism may involve complex secondary actions, such as reactive oxygen species (ROS) generation, mitochondrial dysfunction, induction of apoptosis, cell cycle interruption and interaction with genetic material [8]. The membrane interacting properties of AFPs basically depend on a few structural characteristics of the peptide such as positive charge and hydrophobicity [9,10,11,12,13]. The choice of amino acids in a peptide sequence specifically determines its toxicity and pharmacokinetic profiles along with biological activities. Peptides with more hydrophobic residues may lead to hemolysis whereas hydrophilic amino acid containing peptides may not be able to pass through the cell membrane [14]. The advantages associated with peptides, such as less systemic toxicity and lower potential for the development of resistance, render them a promising alternative to small antibiotics [15]. However, certain limitations such as enzymatic degradation, short half-life and poor oral bioavailability can be easily circumvented using modified/unnatural amino acids [16,17,18] or backbone modification [19,20].
Various reports suggest that introduction of a halogen group imparts overall hydrophobicity to a molecule, apart from inducing stability and modulating the biological activity. Jia et al. demonstrated that introduction of a halogen on naturally occurring Jelleine-I enhanced the antimicrobial and anti-biofilm activity of the peptide, with iodinated Jelleine-I showing the strongest in vitro activity while its chloro and bromo counterparts showed the best in vivo efficacies [21]. Similarly, another study demonstrated that addition of a fluorine atom on the C-terminal amidated tritrpticin analogue, tritrp1, showed similar activities against E. coli when compared to the parent peptide [22]. Cruz et al. demonstrated that the replacement of chlorine with bromine produced a variant of lantibiotic NAI-107 with improved bactericidal activity against gram-positive pathogens [23]. On a similar note, Molchanova et al. demonstrated that incorporation of halogen atoms into otherwise inactive peptoids led to improved antimicrobial activities. However, the activities were strongly dependent on the choice of halogen atom, as chlorine or bromine led to higher activity than the parent peptoid while introduction of fluorine did not alter the activity [24]. In line with this, Gottler et al. reported that a hexafluoroleucine-containing variant of protegrin showed diminished activity against various bacterial strains [25]. Hence, the introduction of a halogen atom to peptides modulates the activities but the altered activities are largely dependent on the choice of halogen atom, type of peptide and the hydrophobic-hydrophilic balance obtained after the modifications.
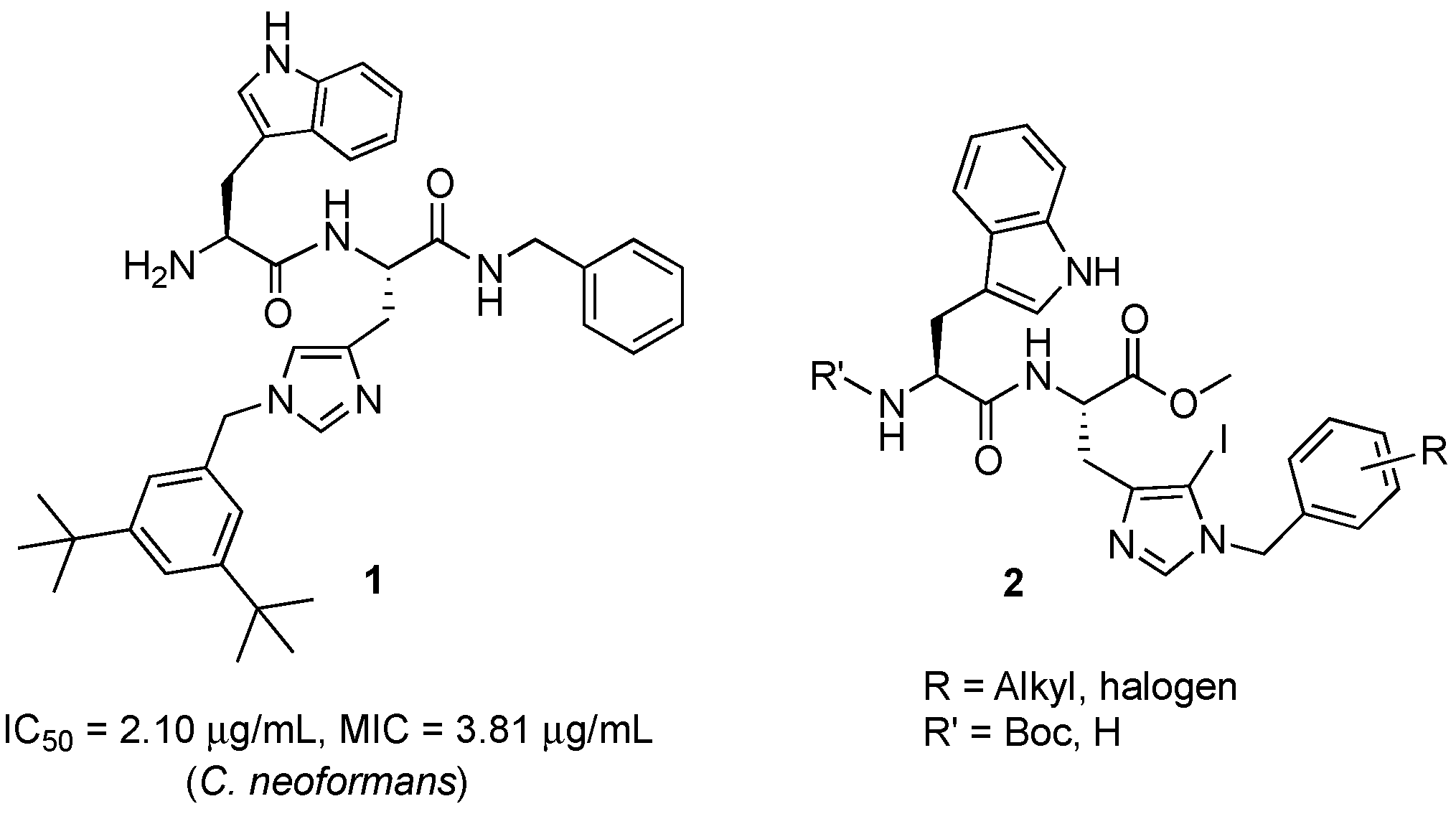
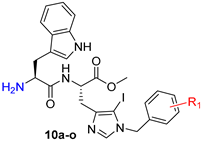
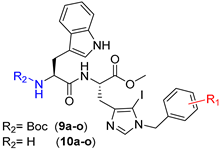
Herein, we modulated the activity of a previously reported dipeptide 1 (Figure 1) [26] by incorporating an iodo group. In the previously reported series of Trp-His class of dipeptides, introduction of various substituted benzyl groups at the N-1 position led to the identification of an anticryptococcal peptide (1). The peptide 1 possesses a 3,5-di-tert-butylbenzyl group at the N-1 position of histidine and NHBzl group at the C-terminal providing hydrophobic character in the peptide. In the current study, we replaced the NHBzl with a methyl ester group and in an attempt to keep the hydrophobicity intact or balanced, an iodo group was introduced at the C-5 position of the histidine ring (2). In our recent work, we observed that the most active dipeptides possessed an NHBzl group at the C-terminal and free N-terminal. Further replacement of the C-terminal NHBzl with OMe and introduction of Boc at the N-terminal led to peptides with similar activities to that of the most active peptides. The slight difference in activities was dependent on the substitutions at the N-1 position of histidine [27]. Therefore, in the present work, a less bulky C-terminal, i.e., OMe was chosen, as a bulky halogen group was introduced at the C-5 position of histidine.
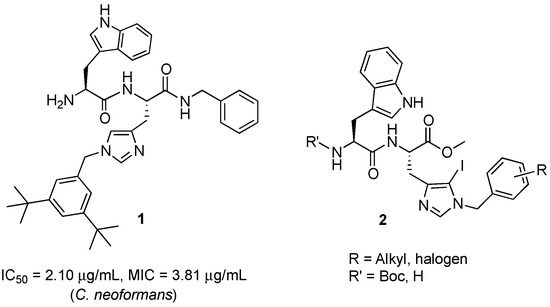
Figure 1.
Structures of dipeptides.
A series of dipeptides 2 (Figure 1) were synthesized by utilizing the backbone of peptide 1 and introducing an iodo group at the C-5 position of the histidine along with speculating the effect of various electron donating and electron withdrawing groups on the N-1 benzyl group in the modulation of antimicrobial activities.
2. Results and Discussion
2.1. Synthesis
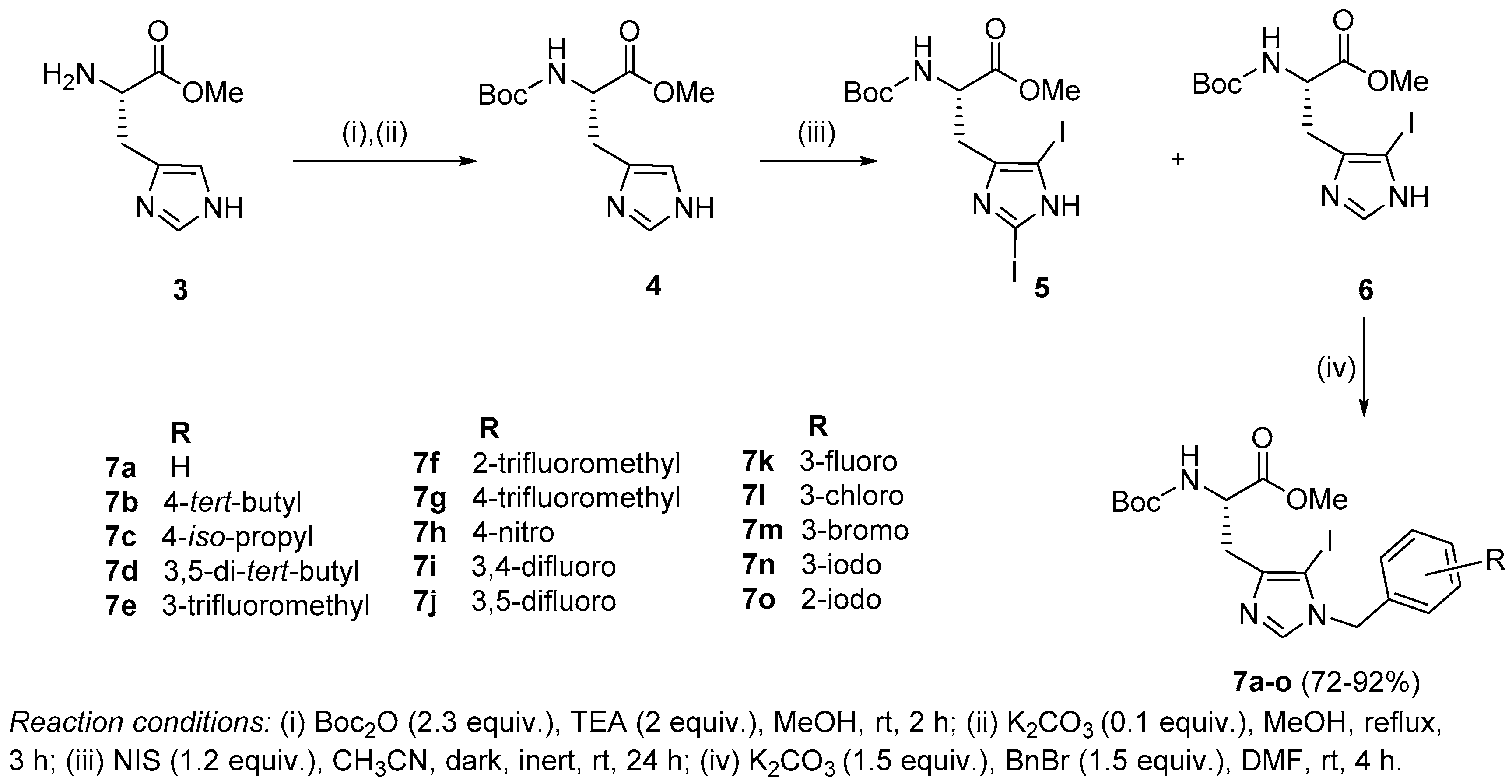
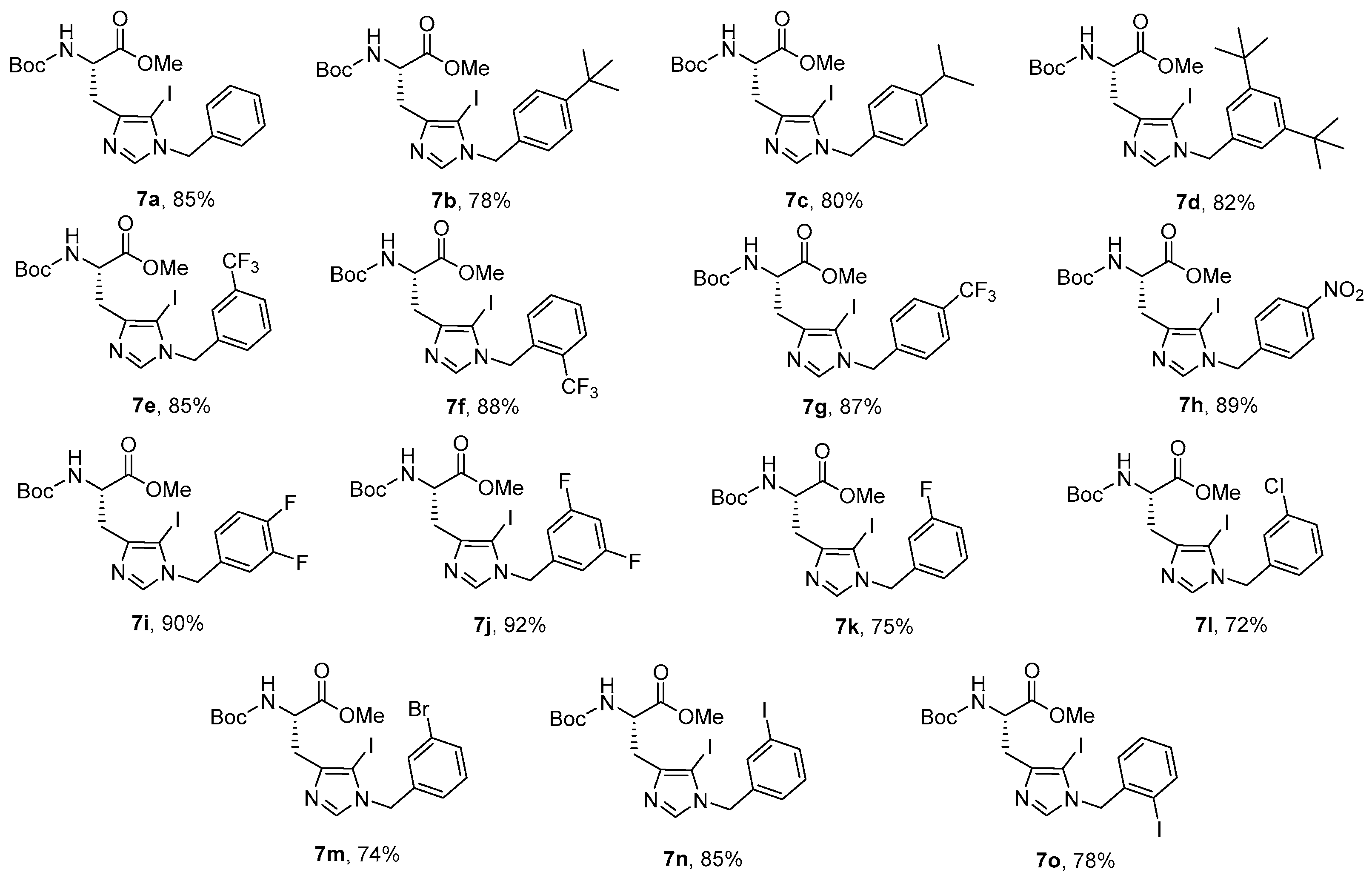
Synthesis of Boc-His (1-Bzl-5-iodo)-OMe: The N-terminal of His-OMe (3) was protected with a Boc group in the presence of di-tert-butyl dicarbonate (Boc2O) to obtain Boc-His-OMe (4, Scheme 1) which was further subjected to iodination by direct electrophilic halogenation in the presence of N-iodosuccinimide at ambient temperatures under dark and inert conditions [28]. Under these conditions, both C-2,5-diiodo (5) and C-5-iodo (6) products were obtained. The crude products were purified using a silica gel column with hexane:ethyl acetate as a solvent system. Boc-His-5-iodo-OMe (6) was then benzylated at the N-1 position using various substituted benzyl bromides and the final derivatives (7a–o) were isolated in 72–92% yield (Figure 2).
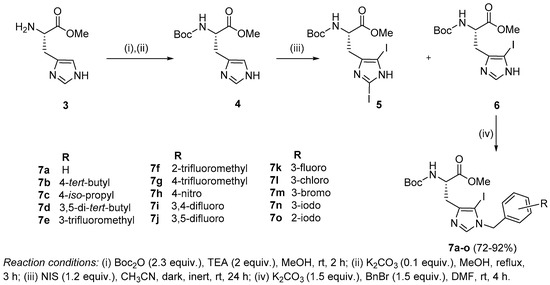
Scheme 1.
Synthesis of N-benzylated 5-iodohistidine derivatives.
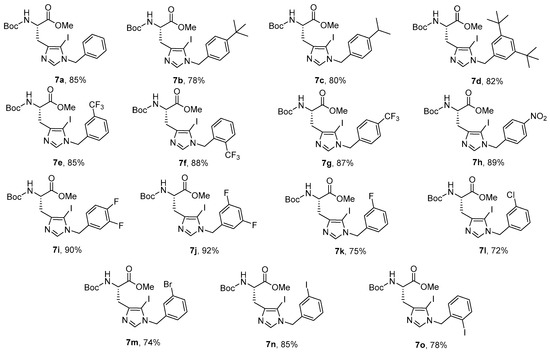
Figure 2.
N-benzylated 5-iodohistidine derivatives.
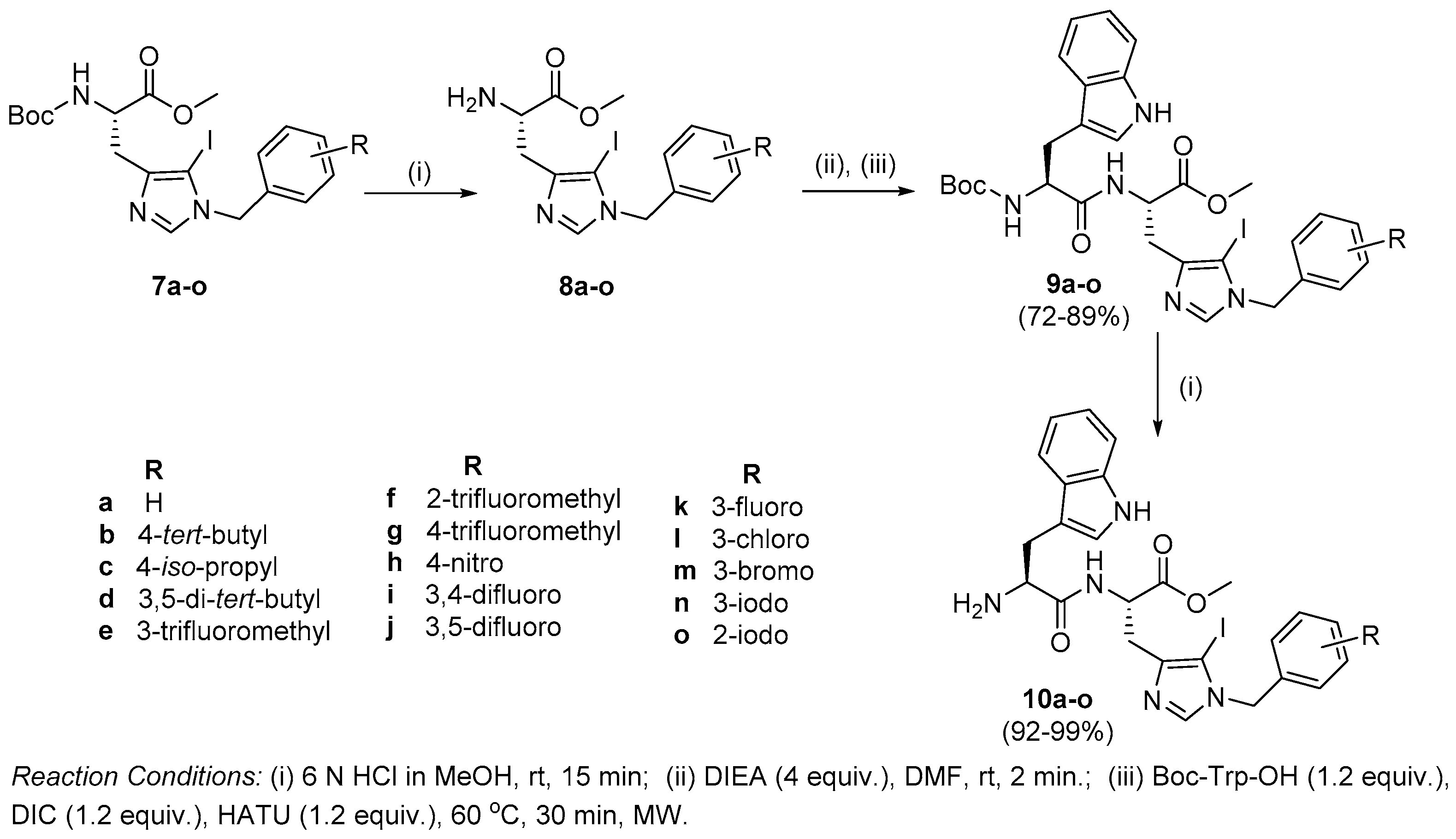
Synthesis of peptides (Series 1–2): Synthesis of the Trp-His(1-Bzl-5-iodo)-OMe class of peptides was carried out from the C- to the N-terminus by sequential deprotection and coupling reactions (Scheme 2). Acidolysis of 7a–o was accomplished with 6 M HCl to obtain 8a–o, and then neutralized with DIEA.
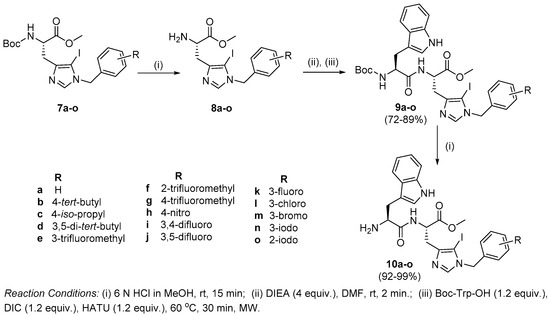
Scheme 2.
Synthesis of Trp-His (1-Bzl)-NHBzl class of dipeptides.
The coupling of 8a–o with Boc-Trp-OH was executed in the presence of a coupling cocktail combination of HOAt and DIC in DMF under microwave irradiation. The crude peptides were purified on a fully automated flash column chromatography system with solvent system of CH2Cl2:MeOH. Deprotection at the N-terminal of purified dipeptides (9a–o) was performed with methanolic HCl to obtain dipeptides 10a–o. The purity of all the synthesized peptides was analyzed by HPLC on a LC-18 column with a run time of 40 min with a flow of 1 mL/min using a gradient system of 95–5% (A:B) where buffer A and B were 0.1% CF3COOH in H2O and CH3CN (see SI).
2.2. Antimicrobial Susceptibility Testing
Peptides 9a–o and 10a–o were tested against selected strains of fungi and bacteria and the results are summarized in Table 1, Table 2 and Table 3. In Series 1, with all Boc-protected peptides 9a–o (Table 1), moderate to low activities were observed against C. neoformans. Most of the peptides with moderately bulky groups such as 9a (R = benzyl), 9c (R = 4-iso-propyl), 9e (R = 3-trifluoromethyl) and with halogenated benzyl groups 9i–9n showed weak fungicidal effects (IC50 values ranging from 17.62 to 23.58 μg/mL) in comparison to the reference standard amphotericin B. The peptides 9a–o were found to be inactive against all other fungal strains.

Table 1.
In vitro antifungal screening of peptides (Series 1).

Table 2.
In vitro antifungal screening of peptides (Series 2).

Table 3.
In vitro antibacterial screening of peptides (Series 1–2).
In Series 2, removal of a Boc group (Table 2) led to a significant increase in the anticryptococcal activity of the peptides. With an overall increase in the bulk from a benzyl group (10a, IC50 = 17.68 μg/mL) to 3,5-di-tert-butylbenzyl (10d, IC50 = 2.20 μg/mL, MIC = 4.01 μg/mL), a substantial increase in bioactivity was observed. Other peptides with electron donating groups such as 10b (R = 4-tert-butyl) and 10c (R = 4-iso-propyl) exhibited moderate activity with IC50 values of 5.15 μg/mL (MIC = 9.36 μg/mL) and 8.81 μg/mL (MIC = 16.02 μg/mL), respectively. Hence, it is concluded that in peptides with electron donating side chains, the activity decreased with the decrease in bulk in the order: 3,5-di-tert-butylbenzyl ˃ 4-tert-butyl ˃ 4-iso-propyl ˃ benzyl.
However, when the methyl group of the tert-butyl moiety was replaced with the less hydrophobic and highly electron withdrawing fluorine atom 10g (IC50 = 13.53 μg/mL, MIC = 24.6 μg/mL), the activity was reduced by 2.6-fold. The position of the trifluoromethyl group on the benzyl ring also affected the activity, such that 3-CF3 (10e) was found to be moderately active with an IC50 value of 4.87 μg/mL (MIC = 8.86 μg/mL) whereas the 2-CF3 group containing peptide, 10f, exhibited a much lower activity with an IC50 value of 17.70 μg/mL (MIC = 32.18 μg/mL). Hence, the activity decreased in order 3-CF3 ˃ 4-CF3 ˃ 2-CF3. The effect of the fluorine group was observed by direct introduction at different positions on the benzyl ring. The derivatives of 3,4-difluorobenzyl (10i) and 3,5-difluorobenzyl (10j) exhibited moderate activities with IC50 values of 10.28 μg/mL (MIC = 18.7 μg/mL) and 6.09 μg/mL (MIC = 11.08 μg/mL), respectively. However, the 3-fluorobenzyl derivative (10k) displayed a significant increase in activity with an IC50 value of 2.78 μg/mL (MIC = 4.64 μg/mL). This prompted us to evaluate various other 3-halobenzyl substituted derivatives. Peptides 10l, 10m and 10n containing 3-chloro, 3-bromo, 3-iodo groups, respectively, were moderately active with IC50 values of 17.6, 9.23 and 17.6 μg/mL, respectively. However, when iodo was placed at the ortho position of the benzyl ring, a significant increase in activity was observed with an IC50 value of 2.59 μg/mL (MIC = 4.59 μg/mL). Furthermore, the incorporation of an electron withdrawing nitro group (10h) at the fourth position of the benzyl ring drastically reduced the activity of the peptide (IC50 < 20 μg/mL). In a nutshell, with an optimum balance of hydrophobicity, electronegativity and polarity, peptides tend to show enhanced activity. All the peptides were found to be inactive against C. albicans, C. glabrata, C. parapsilosis and C. krusei at the highest tested concentrations. Peptides with IC50 values less than 20 μg/mL from both series were further tested against bacterial strains (Table 3) but none showed very significant activity.
To summarize, peptides 10d (R = 3,5-di-tert-butylbenzyl) and 10o (R = 2-iodobenzyl) showed the best activities of the two series with IC50 values of 2.20 μg/mL (MIC = 4.01 μg/mL) and 2.59 μg/mL (MIC = 4.59 μg/mL), respectively. The presence of an iodo group at the C-5 position of the histidine in this series of dipeptides imparts the quintessential hydrophobicity, therefore aiding the fungicidal activity of the peptides. The presence of different halogen groups (Br, Cl and F) at C-5 and their impact on the biological activities will be further explored in future.
2.3. Cytotoxicity Assay
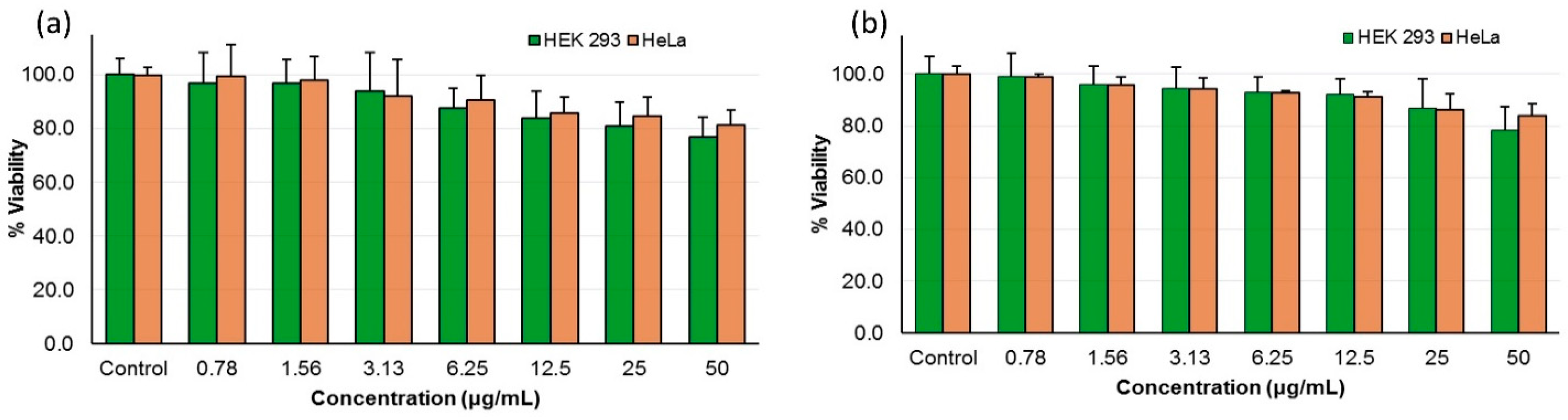
Cytotoxicity assays were carried out to determine the selectivity of the most active peptides 10d and 10o towards fungal cells and their toxicity towards mammalian cells. The peptides showed a non-cytotoxic nature against human cancer cells line (HeLa) and a non-cancerous mammalian cell line (HEK 293) at their MIC concentration and higher (Figure 3, Table 4).
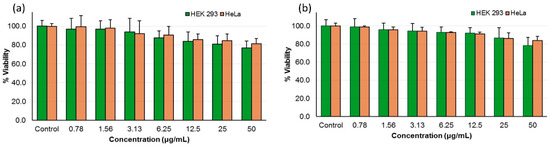
Figure 3.
Mammalian cell cytotoxicity of peptide (a) 10d and (b) 10o as percentage viability of HEK-293 and HeLa cells at different concentrations.

Table 4.
Cytotoxicity and hemolytic activities of the active peptides.
2.4. Hemolytic Assay
Non-selective peptides do not differentiate between fungal cells and human erythrocytes and thereby interact with both, causing hemolysis. A hemolytic study was carried out to demonstrate the selectivity of the synthesized peptides towards fungal cells. Peptides 10d and 10o exhibited non-hemolytic behavior at their MIC values and above, as evident from the HC10 and HC50 values (Table 4); therefore, they show high selectivity towards C. neoformans cells.
2.5. Time Kill Kinetics
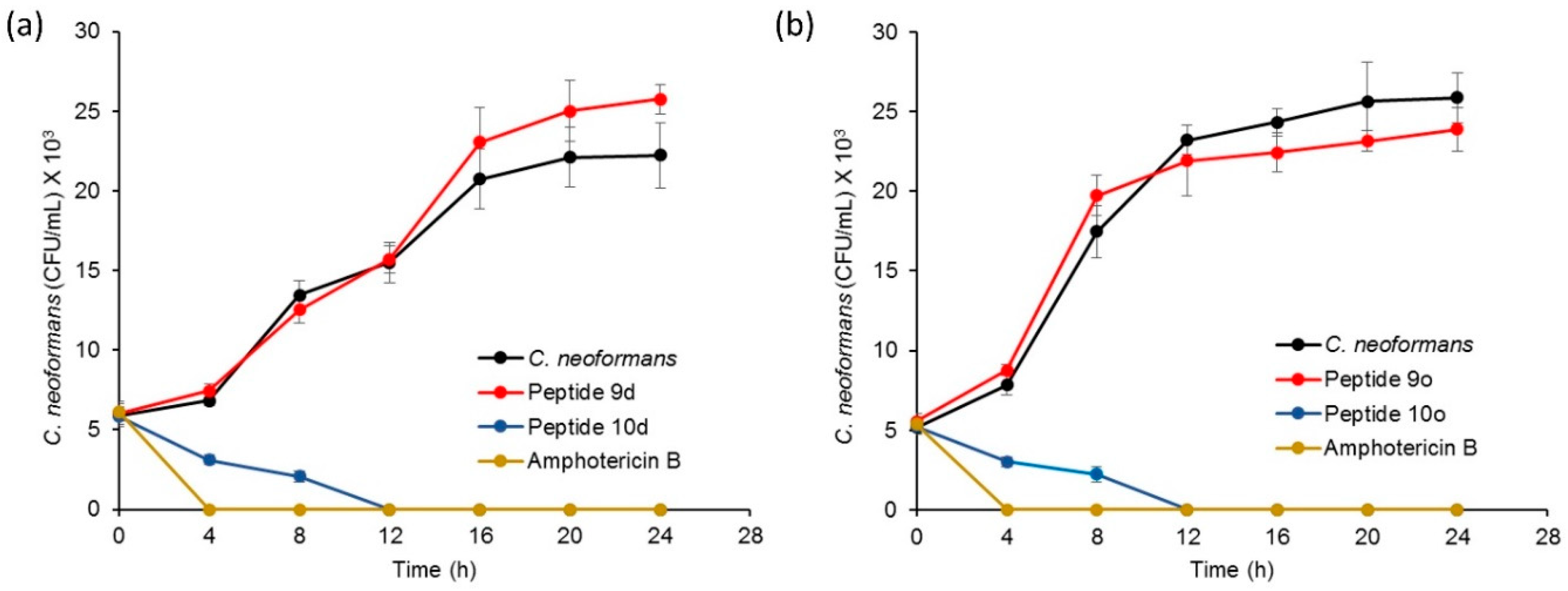
To determine the time-dependent fungicidal action of peptides 10d and 10o, their time–kill profile was studied. The assay was carried out with the active peptides 10d and 10o-treated cryptococcal cells while corresponding inactive counterparts 9d and 9o-treated cells along with untreated cells were taken as negative controls. Amphotericin B-treated cells were used as the positive control. The growth profile of fungal cells was determined for each sample over a period of 24 h.
The growth pattern of amphotericin B-treated cryptococcal cells showed a significant drop in colony forming units (CFUs) after 4 h. Peptide 10d displayed significant fungicidal activity and inhibited the growth of cryptococcal cells after 12 h (Figure 4a). However, inactive peptide 9d-treated cells did not show any growth inhibition and exhibited the growth curve similar to that of a standard curve shown by untreated cells. Furthermore, time–kill kinetics of the active peptide 10o revealed a gradual decrease followed by complete inhibition of CFUs after 12 h (Figure 4b). However, negative peptide 9o-treated and untreated cells showed similar growth curves. The results confirmed the fungicidal action of the tested peptides.
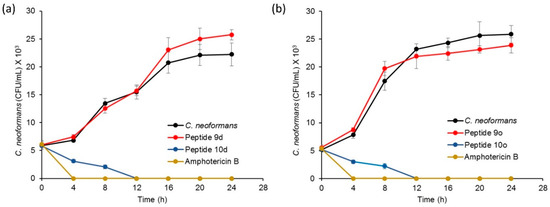
Figure 4.
Time–kill kinetics profile for (a) peptide 10d, with amphotericin B as positive control, while C. neoformans cells and peptide 9d were negative controls; (b) peptide 10o, with amphotericin B as positive control while C. neoformans cells and peptide 9o were negative controls.
2.6. Drug Combination Study
The fungicidal nature of the active peptides was further evaluated when given in combination with clinical standards that are used for the treatment of invasive cryptococcal infections. Cryptococcal cells were treated with different concentrations of peptides 10d and 10o in combination with varying concentrations of amphotericin or fluconazole and the fractional inhibitory concentration (FIC) index was calculated (Table 5) [29].

Table 5.
Evaluation of combinative effect of bioactive peptides with amphotericin B (Amp B) and fluconazole (Flu).
FICI ≤ 0.5: synergistic; FICI > 4: antagonistic; and FICI 0.6–4.0: additive or indifferent.
Peptide 10d, when used in combination with amphotericin B, exhibited a significant synergistic action with an FICI value of 0.18. The MIC value of peptide 10d showed a 16-fold decrease from 4.01 to 0.25 μg/mL, whereas that of amphotericin B showed an 8-fold decrease in the MIC value. Furthermore, 10d also exhibited synergism with fluconazole, showing an FICI value of 0.19. An approx. 8-fold decrease in the MIC of 10d (from 4.01 to 0.501 μg/mL) was observed and fluconazole showed an approx. 16-fold decrease in the MIC value (from 10 to 0.625 μg/mL). Peptide 10o, at a non-cidal concentration, resulted in the 4-fold potentiation of amphotericin B in combination, while the MIC of peptide 10o was lowered by around 16-fold (MIC from 4.59 to 0.285 μg/mL). The FICI value of this combination was found to be 0.31, indicating synergistic action. However, with fluconazole, peptide 10o exhibited an additive effect with an FICI value of 1.0.
2.7. Mechanistic Investigations
The mechanism of action of the bioactive peptides was determined by analyzing the cell death pattern, by establishing the morphological changes after the treatment using electron microscopy and by identifying the broad intracellular targets using confocal imaging.
2.7.1. Cell Death Pattern Analysis Using Flow Cytometry
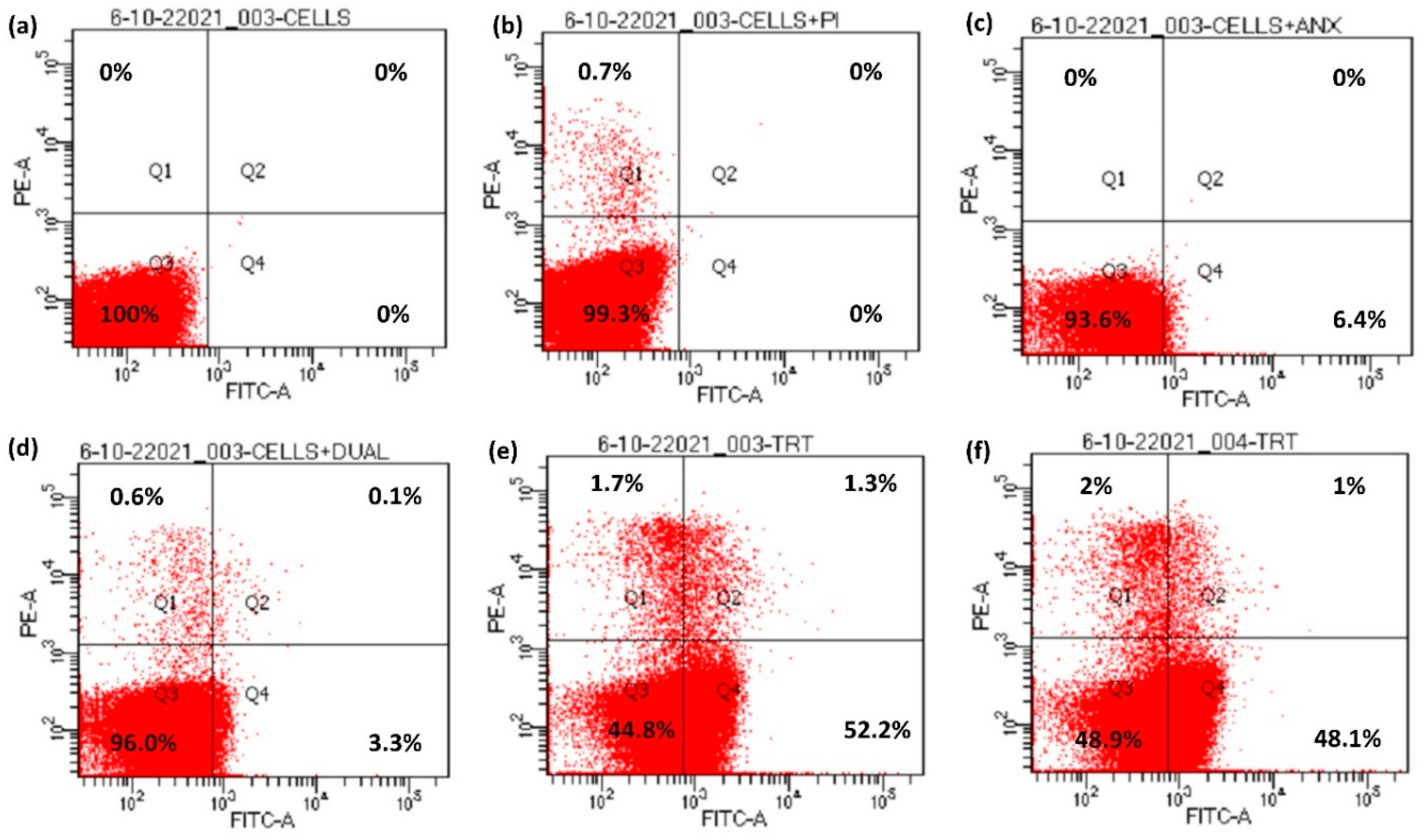
Apoptosis includes a series of events occurring inside the cell, causing changes in plasma membrane, genetic material and various internal proteins, therefore leading to programmed cell death. One of the early markers of apoptosis is the translocation of a membrane phospholipid, phosphatidylserine, from the inner to the outer leaflet of the plasma membrane without altering the integrity of the plasma membrane [30]. Annexin V-FITC interacts with the externalized phosphatidylserine, therefore showing the cell change towards apoptosis which can be analyzed using flow cytometry [31,32]. The four quadrants in flow cytometry indicate different stages of the cells. The percentage depicts the population of the cells in a specific stage. Q1 corresponds to the percentage of cells undergoing necrosis, Q2 depicts the early apoptotic stage of the cells whereas Q4 corresponds to the cells in late-stage apoptosis and Q3 depicts the cells in the native state [33].
The control samples for peptide 10o showed cryptococcal cells alone, exhibiting 100% population in native state Q3 (Figure 5a), whereas PI incubated samples (Figure 5b), showed 99.3% of cell population in the native state and 0.7% were in Q1. After incubation with annexin V-FITC (Figure 5c), the population of cryptococcal cells scattered more towards Q4 (6.4%), while 93.6% were in native quadrant Q3. Further incubation of cells with both PI and annexin V-FITC (Figure 5d) did not have a significant effect on the redistribution of the cell population over all quadrants, as more than 95% cells were in native state Q3, Q1 had 0.6% cells, Q2 showed 0.1% and Q4 constituted 3.3% of total cell population. However, when peptide 10d-treated fungal cells were incubated with both PI and annexin V-FITC (Figure 5e), a marked variation in the pattern of distribution of cells was observed over the four quadrants, as 52.2% of cell population was pushed towards the Q4 quadrant showing late-stage apoptosis, while 1.7% cells exhibited necrosis (Q1), 1.3% were in Q2 and 44.8% of cell population remained in native state Q3. On a similar note, when cells were treated with 10o and incubated with PI and annexin V-FITC (Figure 5f), 2% necrosis was observed (Q1), 48.1% exhibited late stage apoptosis and only 1% were in Q2, whereas 48.9% were in the native state Q3.
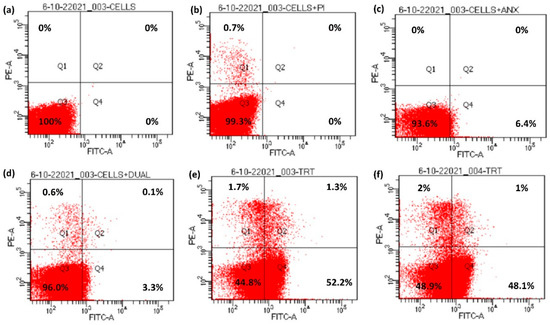
Figure 5.
Externalization of phosphatidylserine in bioactive peptide-treated cells. (a) Unstained cells, (b) cells + PI, (c) cells + Annexin V-FITC, (d) cells with Annexin V-FITC + PI, (e) cells treated with Annexin V-FITC + PI + peptide 10d and (f) cells treated with Annexin V-FITC + PI + peptide 10o.
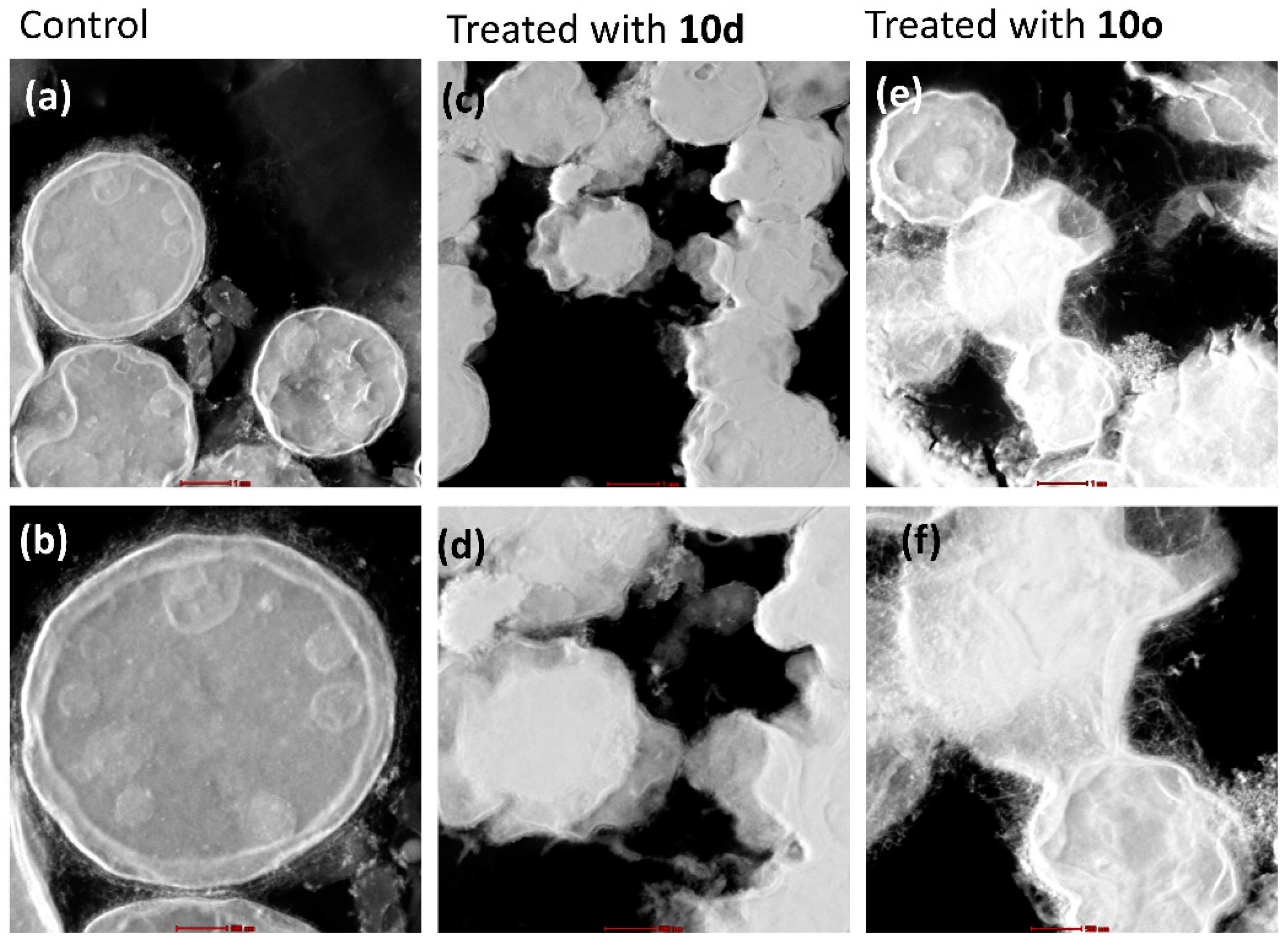
2.7.2. Surface Morphology Analysis Using Scanning Electron Microscopy (SEM)
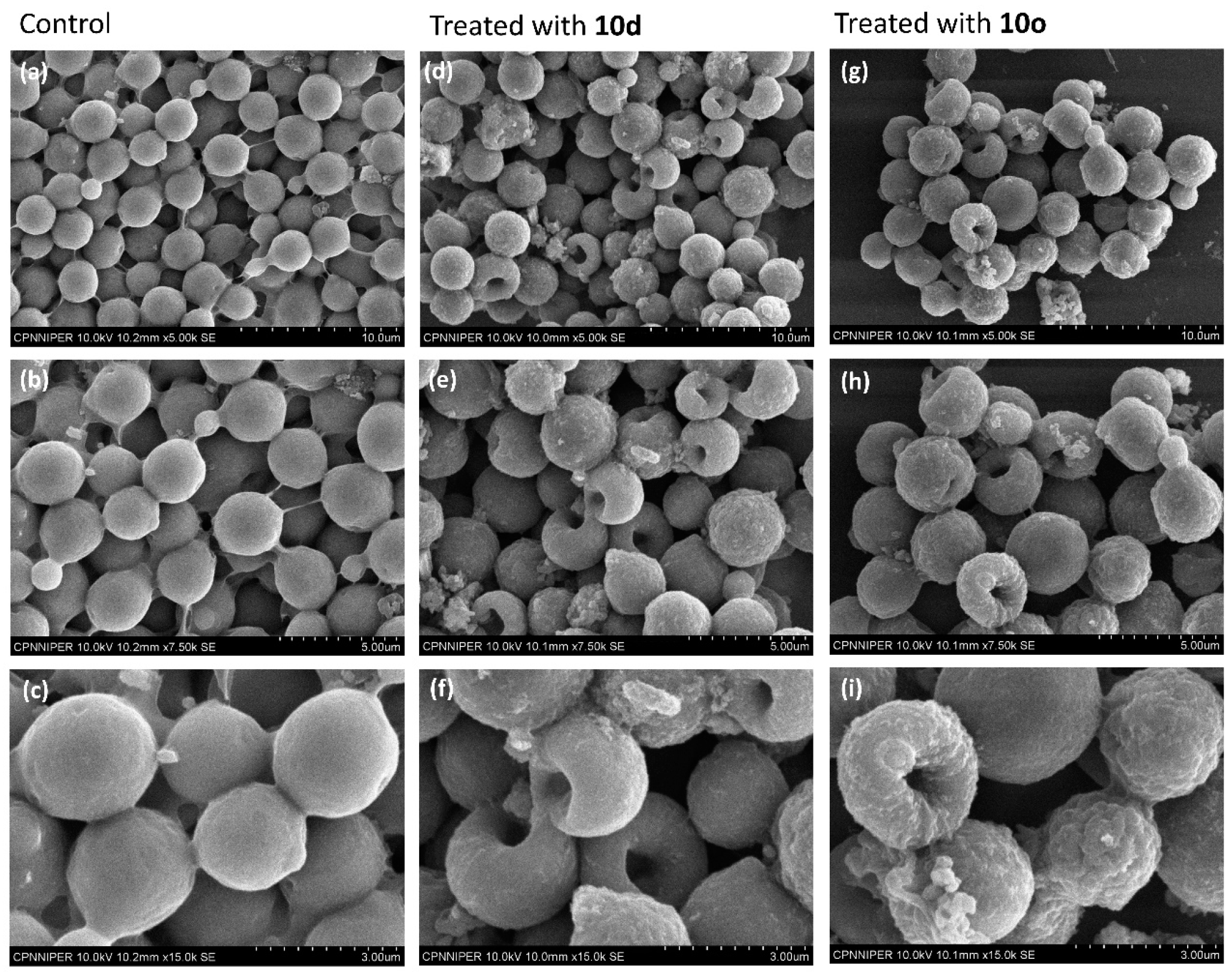
A comparative analysis of the surface morphologies of treated and untreated cells was carried out using scanning electron microscopy (SEM). The untreated cells appeared as oval-shaped with smooth surfaces and intact cell membranes (Figure 6a–c) when observed in SEM. However, cells treated with peptide 10d showed large pores in the middle of the cells (Figure 6d–f) giving a doughnut-shaped appearance. The cell surface comparatively became highly wrinkled and a blatant change in morphology of the cells was observed in comparison to the untreated cells. Likewise, cryptococcal cells treated with peptide 10o showed a highly wrinkled cell surface with pores in the middle making their overall appearance doughnut-like (Figure 6g–i) when compared to the untreated cells. The cells were highly deformed and the intracellular contents appear to have come outside the cell, thereby causing cell death.
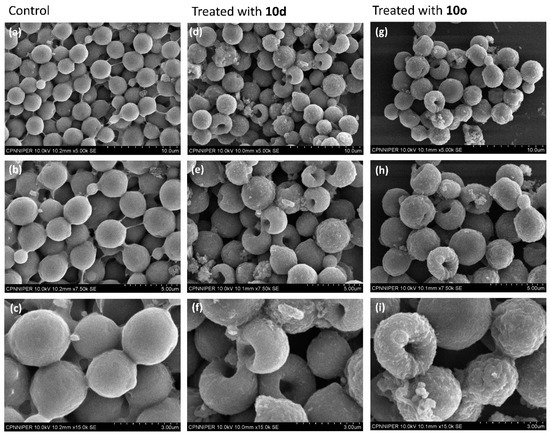
Figure 6.
SEM-based surface morphological analysis of 10d and 10o treated and untreated C. neoformans cells. Images (a–c) represent the morphology of untreated cells; images (d–f) represent the morphology of cells treated with 10d; images (g–i) represent the morphology of cells treated with 10o. The scale bar for images (a,d,g) is 10 μm, for images (b,e,h) it is 5 μm and for images (c,f,i) it is 3 μm.
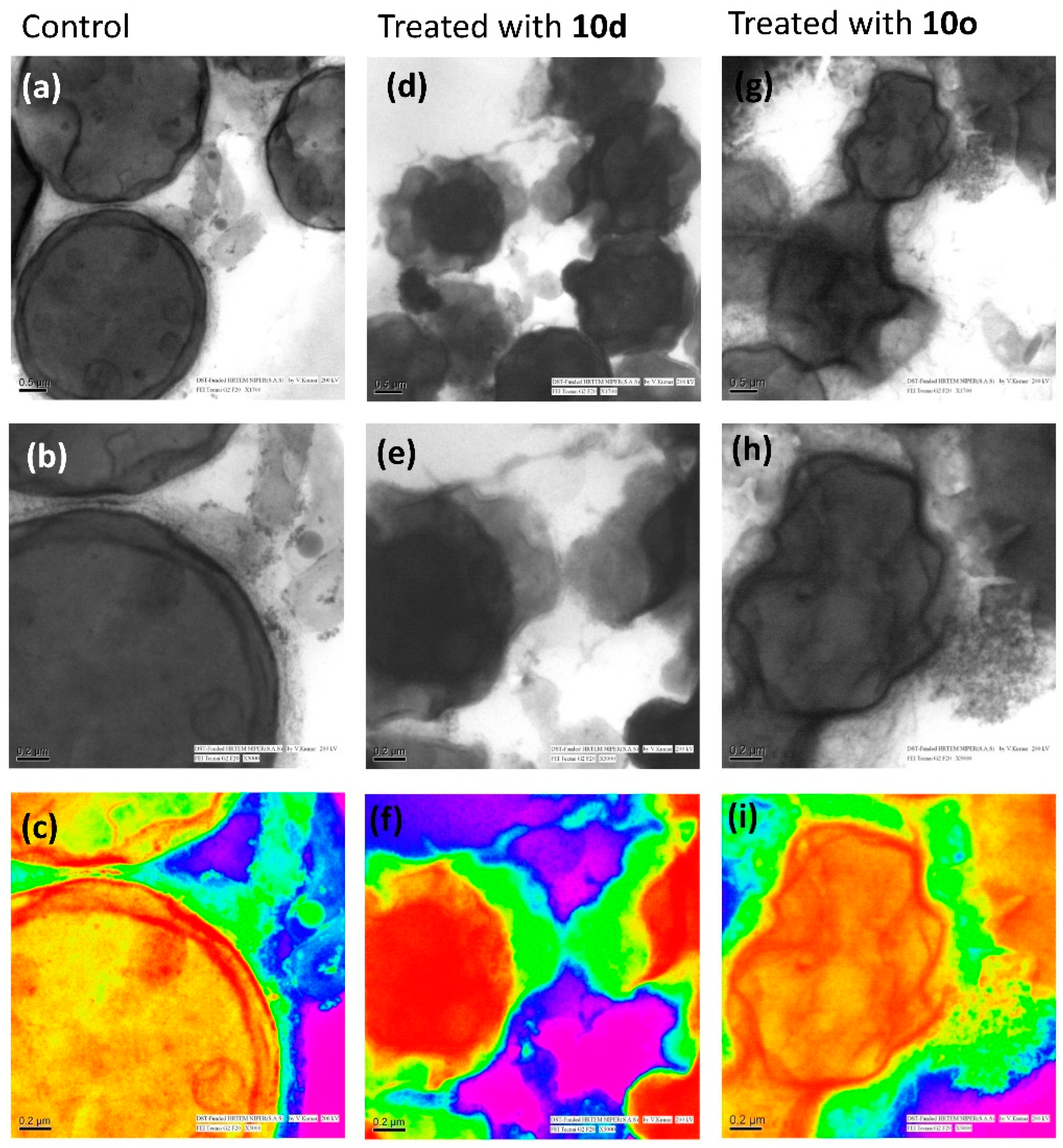
2.7.3. Internal Cell Morphology Analysis Using Transmission Electron Microscopy (TEM)
High-resolution transmission electron microscopy (HRTEM) provided an explicit view of the structural details of cells along with intricate changes that occur during the cell wall disruption. The cryptococcal cells in Figure 7a–c display a distinct inner plasma membrane covered with a layer of outer membrane followed by a faded capsule. The cell organelles can also be distinguished faintly from the images. However, the cells treated with peptide 10d (Figure 7d–f) show complete lysis of the cell membrane. The outer membrane and capsule are not clearly distinguishable. The intracellular content of cells have oozed out and the cells appear as a stack of non-distinguishable structures lying one over the other. However, the level of cell membrane disruption was much more severe in the case of the 10o-treated cells (Figure 7g–i). The cells lost their shape completely and appear to have fused intracellular material. The cells were highly shrunken and the cell membrane was completely damaged as compared to the untreated cells.
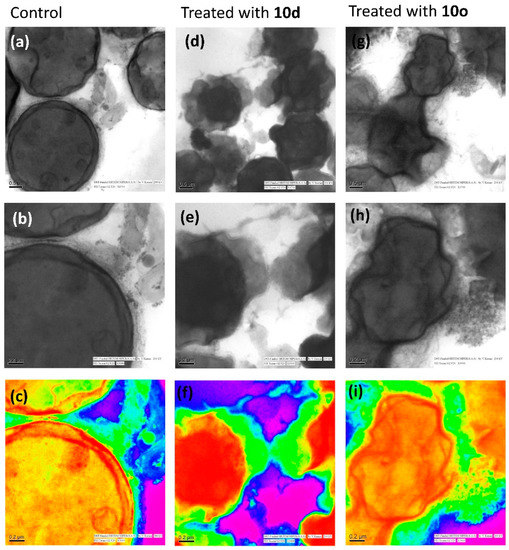
Figure 7.
Morphological study of C. neoformans cells and cells treated with bioactive peptides by high-resolution transmission electron microscopy. Images (a–c) represent the morphology of untreated/healthy cells; images (d–f) represent the cells treated with 10d and images (g–i) represent the cells treated with 10o; images (c,f,i) are the rainbow form of images (b,e,h), respectively. The scale bar for images (a,d,e) is 0.5 μm and for images (b,c,e,f,h,i) it is 0.2 μm.
Scanning transmission electron microscopy (STEM) analysis provides high contrast images of the cells and the results were in coherence with the HRTEM. The STEM images of untreated cells (Figure 8a,b) depict smooth walled, rounded cells with cell organelles intact, whereas the images of cells treated with peptide 10d (Figure 8c,d) show highly shrunken cells with irregular cell membranes. The intracellular material has oozed out and a clear distinct cell was not observed. The deformed cells have merged to appear similar to a mass of highly irregular structure. Similarly, cryptococcal cells treated with 10o (Figure 8e,f) were highly deformed and shrunken. The membrane separating the cells seemed to have disappeared and merged to form a cluster of deformed cells.
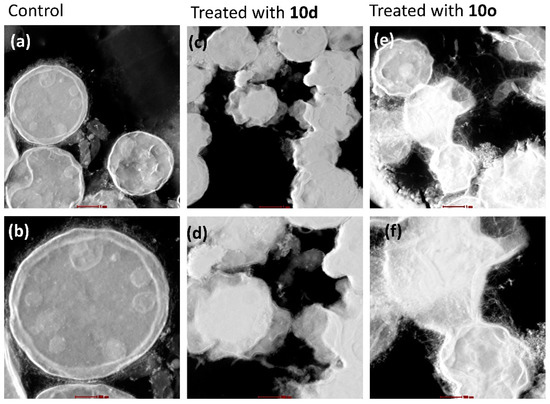
Figure 8.
Morphological study of C. neoformans cells and cells treated with bioactive peptides by STEM. Images (a,b) represent the morphology of untreated/healthy cells; images (c,d) represent the cells treated with 10d; and images (e,f) represent the cells treated with 10o. The scale bar for images (a,c,e) is 1 μm and for images (b,d,f) it is 0.5 μm.
2.7.4. Confocal Laser Scanning Microscopy (CLSM)
- (a)
- Membrane permeabilization and localization of FITC-labeled peptide
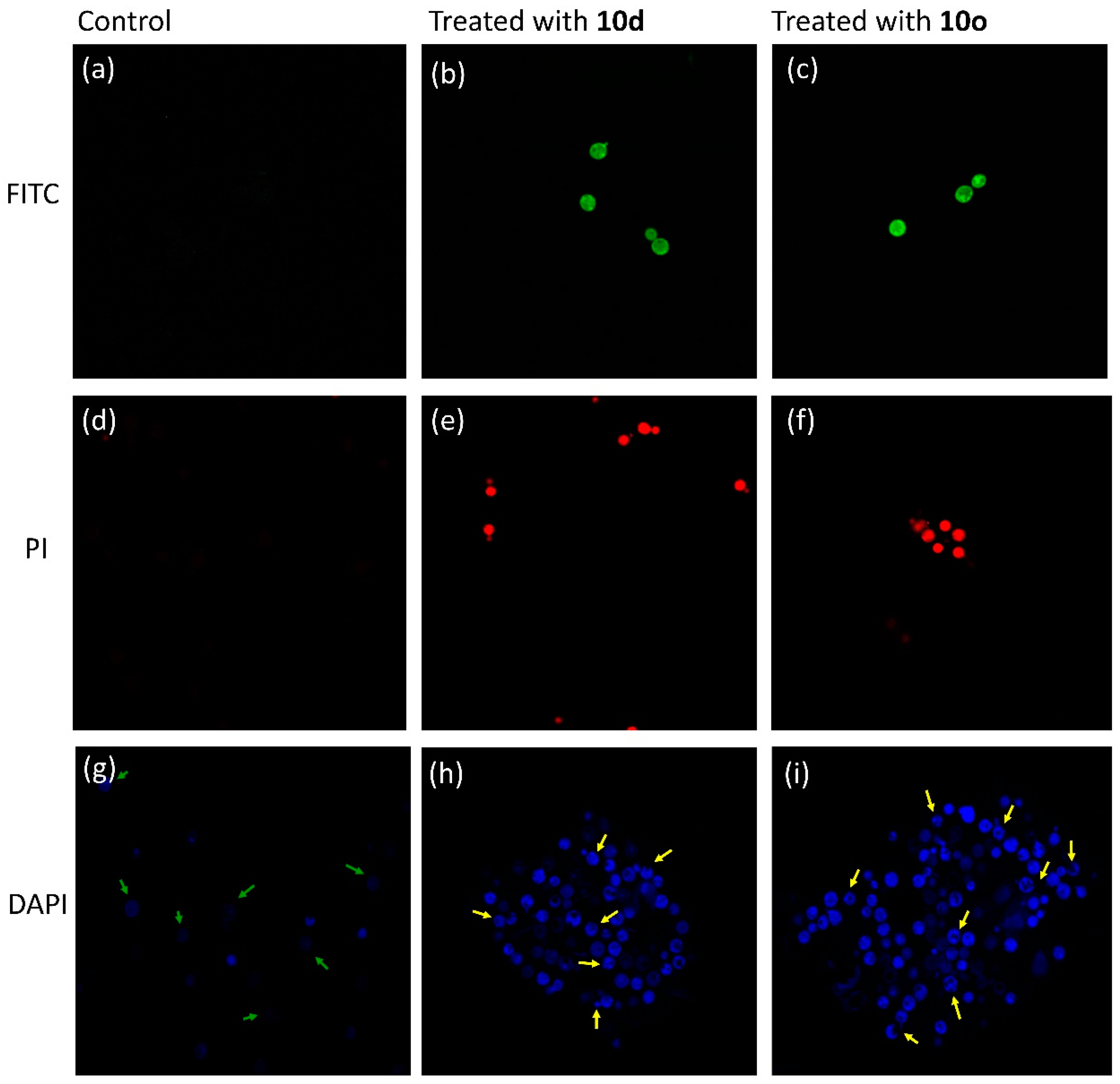
The bioactive peptides 10d and 10o along with an inactive peptide 10h were labeled with FITC (fluorescein isothiocyanate, a green, fluorescent dye, with excitation at 480 nm). The permeabilization of peptides inside the cell was analyzed by incubating the FITC-labeled peptides with C. neoformans cells and observing the incubated slides under CLSM. The FITC-tagged inactive peptide, on incubation with cryptococcal cells, did not show any fluorescence when observed under CLSM (Figure 9a), depicting their inability to cross the cell membrane. However, the images of cells incubated with FITC-tagged peptides 10d (Figure 9b) and 10o (Figure 9c) showed prominent green fluorescence. Hence, it was concluded that peptides act via membrane permeabilization; therefore, the membrane integrity of the cells is lost leading to permeabilization of the FITC-peptide inside the cells followed by localization, providing a bright green fluorescence.
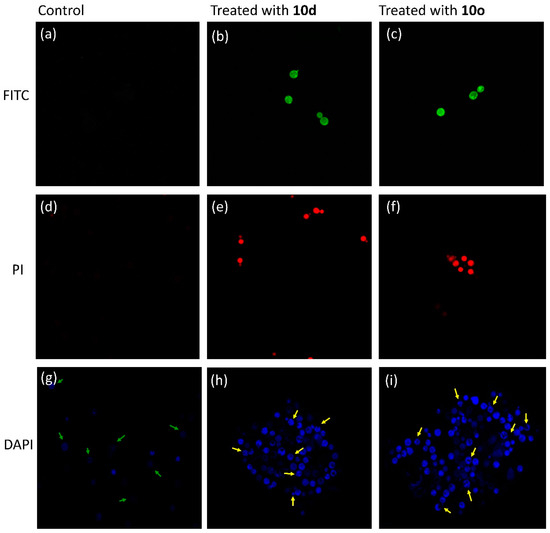
Figure 9.
Confocal micrographs. Image (a) was obtained after incubating cells with inactive FITC-labeled peptide 10h, while image (b) was obtained after treatment with active peptide 10d tagged with FITC and image (c) was obtained after treatment with active peptide 10o tagged with FITC. PI uptake assay: image (d) was obtained after incubating cells with PI, while image (e) was obtained after treatment with active peptide 10d and image (f) was obtained after treatment with active peptide 10o. DAPI uptake assay: image (g) was obtained after incubating cells with DAPI, while image (h) was obtained after incubating cells treated with 10d with DAPI and image (i) was obtained after incubating cells treated with 10o with DAPI.
- (b)
- Membrane disruption and DNA interaction analysis by propidium iodide (PI) uptake
Propidium iodide (PI) dye cannot permeate the walls of healthy cells; hence, when cryptococcal cells were incubated with PI and observed under CLSM (excitation at 480 nm), they did not show any fluorescence (Figure 9d). However, when the cells were treated with peptide 10d (Figure 9e) and 10o (Figure 9f) and then incubated with PI, a prominent red fluorescence was observed. The results indicated that the peptides acted by lysis of the cell wall, which allowed the dye to penetrate the cell and intercalate with the DNA giving the whole cell a bright red fluorescence.
- (c)
- Detection of nuclear fragmentation
The molecule 4′,6-diamidino-2-phenylindole (DAPI) easily permeates inside cells and interacts with the DNA giving a pinpoint blue fluorescence when observed under CLSM. It selectively binds to the minor groove of the DNA; therefore, any change in the fluorescence intensity pattern reveals the degree of damage to DNA. Untreated cells, when incubated with DAPI, showed faint blue pinpoint fluorescence (Figure 9g) indicating staining of the nucleus (marked by green arrows). However, cells treated with 10d (Figure 9h) and 10o (Figure 9i) exhibited fragmented bright blue fluorescence showing destroyed cells (marked by yellow arrows). Therefore, it can be concluded that after entering the cells, the peptides interact with the genetic material, triggering the event of cell death.
2.7.5. Possible Mechanism
Primarily, the negatively charged surface of fungi demands a positively charged moiety for easy interaction; hence, an overall positive charge was introduced in the peptides while designing them by incorporating charged amino acids. Upon interaction with the fungal cell membrane, the hydrophobicity of peptides plays a crucial role and helps the peptide to permeate inside the lipophilic membrane of the fungal cell. During permeation, the peptide induces morphological changes in the otherwise intact fungal cell membrane leading to the appearance of wrinkles on the cell surface, as evident from SEM. The peptide then interacts with various intracellular components inside the cell leading to depletion of cytoplasm (TEM), and triggering various apoptotic pathways as confirmed by flow cytometry. The depletion leads to an altered morphology of the cell, making it appear doughnut-shaped (SEM). Consequently, the intact membrane become indistinguishable from cytoplasm (TEM). The collective effect of different interactions result in distortion of the cell surface integrity, membrane permeabilization, pore formation, cell lysis and death.
3. Conclusions and Summary
In a series of N-substituted histidine-containing His-Trp class of dipeptides, we found that an additional substitution at the C-5 position with an iodo group influenced the biological activity against C. neoformans. In vitro antifungal susceptibility assays revealed that peptides Trp-His[1-(3,5-di-tert-butylbenzyl)-5-iodo]-OMe (10d) and Trp-His[1-(2-iodophenyl)-5-iodo)]-OMe (10o) exhibited the most significant antifungal activities against C. neoformans among the whole series and showed high selectivity and safety profiles. The mechanistic studies revealed that the peptides are membrane active in nature as evident from SEM and TEM analyses, which demonstrated the contrasting morphologies of healthy and bioactive peptide-treated cells. Perfectly intact oval-shaped cryptococcal cells attained a highly deformed structure with a large pore in the middle which gave it a doughnut-like appearance. Furthermore, CLSM analysis showed that the peptides easily moved across the cell membrane by increasing fluidity, thereby causing membrane lysis and disruption and interaction with various intracellular targets expediting the cell death. Furthermore, flow cytometry showed that the cell death was hastened by the induction of apoptotic pathways.
4. Material and Methods
Microbial strains: Microbial strains were procured from National Culture Collection of Pathogenic Fungi (NCCPF) at the Postgraduate Institute of Medical Education & Research (PGIMER), Chandigarh and Microbial Type Culture Collection and Gene bank (MTCC) at the Institute of Microbial Technology (IMTECH), Chandigarh, India. Peptides were screened against five different strains of fungi: viz. Candida albicans (NCCPF400034), Candida glabrata (MTCC 3019), Candida parapsilosis (NCCPF 440002), Candida krusei (NCCPF 440002) and Cryptococcus neoformans (NCCPF250316), and five strains of bacteria: viz. Escherichia coli (MTCC 2961), Staphylococcus aureus (MTCC 3160), Enterococcus faecalis (MTCC 439), Streptococcus pyogenes (MTCC 442) and Pseudomonas aeruginosa (MTCC 3542).
Growth media: Antifungal susceptibility testing was carried out using standard broth microdilution assays as per the Clinical and Laboratory Standards Institute (CLSI) guidelines for fungi [34,35]. RPMI 1640 (Sigma), yeast extract, peptone and dextrose (YPED, Himedia) were used as the incubation broths. The cells were diluted to a cell density of 103 colony forming unit (CFU/mL) and incubated at 30 °C for 48 h. Furthermore, Muller Hinton (Himedia) media was utilized for bacterial strains and antibacterial activity was evaluated per CLSI guidelines for bacteria [36]. Bacterial cells were diluted to a cell density of 105 CFU/mL and incubated at 37 °C for 24 h. Amphotericin B (Himedia, India) and rifampicin (Himedia, India) were used as standard drugs for the evaluation of activity against all the fungal and bacterial strains, respectively.
For the detailed synthetic procedures and characterization data please refer to supplementary information.
4.1. Broth Microdilution Assay
The stock solutions of peptides (2 mg/mL) were prepared in DMSO. Specific amounts of peptide from stock solution were added to a 96-well microtiter plate containing 100 μL of growth media and then serially diluted two-fold. A further 100 μL of growth media containing microbial cells were added to each well and incubated at 30–37 °C for 24–48 h. The optical density (OD) was measured at 600 nm using a BioStack Ready (BioTek Instruments, Winooski, VT, USA) microplate reader to analyze microbial growth inhibition, and IC50 and MIC values were calculated. To visualize the growth and inhibition pattern, a solution of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) in water was added to the plate and incubated for 2–3 h. MTT reacts with the mitochondrial dehydrogenases, which are present only in a viable cell, to give blue colored formazan crystals. Therefore, a darker color indicates a high number of viable cells; however, non-viable cells appear pale or colorless.
4.2. Cytotoxicity Assay
The cytotoxicity studies of peptides 10d and 10o were carried out in standard 96-well cell culture plates against HEK-293 and HeLa cells. The cells (50,000 cells/well) were seeded to the wells of the plate in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum at 37 °C overnight and incubated for 24 h to achieve confluency. Peptides at different concentrations of 50, 25, 12.5, 6.25, 3.12, 1.56 and 0.78 μg/mL were added to the cells in separate wells and incubated at 37 °C for 18 h. A further 20 µL MTT solution (5 mg/mL stock conc. in PBS) was added and the plate was incubated for another 3–4 h. To detect the viable cells that react with MTT to make formazan crystals, 120 μL of supernatant was removed and 100 μL DMSO was added to dissolve the crystals. Untreated cells and DMSO (10%) were used as negative and positive controls, respectively [37]. The percent viability of cells was calculated using equation:
4.3. Hemolytic Assay
Human blood in 10% citrate phosphate dextrose was obtained from Government Hospital, Chandigarh, India. Red blood cells (RBCs) were harvested by spinning at 1000× g for 5 min followed by washing with PBS 3–5 times. The packed cell volume was obtained and utilized to make a 0.8% (v/v) suspension of RBCs in PBS. The suspension (100 μL) was transferred to each well of a 96-well microtiter plate and mixed with peptides 10d and 10o (100 μL) at concentrations of 50, 25, 12.5, 6.25, 3.12, 1.56 and 0.78 μg/mL. The plate was incubated at 37 °C for 60 min followed by centrifugation at 1000× g for 5 min at 25 °C. The supernatant (100 μL) was transferred to a fresh microtiter plate and absorbance was recorded at 414 nm (Varioskan LUX multimode microplate reader, Thermo Scientific, Waltham, MS, USA) to analyze RBC lysis. Untreated RBCs in PBS and lysed RBCs in 0.1% Triton X-100 were used as negative and positive controls, respectively [29]. Percent hemolysis was calculated using following equation [38].
4.4. Time Kill Assay
C. neoformans cells (~1 × 103 CFU/mL) were inoculated in media and four samples for each test peptide were prepared, where the cryptococcal cells were either (i) incubated with test peptides (10d or 10o), (ii) negative peptides (9d or 9o), (iii) amphotericin B or (iv) left untreated in separate tubes with incubation broth. The tubes were then incubated for 24 h at 30 °C with 150× g in a shaking incubator. Aliquots of 100 μL were removed from each sample at predetermined time intervals of 0, 4, 8, 12, 16, 20 and 24 h. Aliquots of 10 μL were serially diluted (10-fold) in saline and plated on nutrient agar plates. The plates were then incubated at 30 °C for 24 h and the colony forming units (CFUs) of C. neoformans cells were counted [29].
4.5. Drug Combination Study
Various dilutions of standard drugs, i.e., amphotericin B, fluconazole and bioactive peptides (10d and 10o) were prepared. Incubation broth was added to 96-well microplates, followed by addition of different concentrations of the standard drugs across the rows, and different concentrations of bioactive peptides across the columns of the microtiter plates. Hence, a unique combination of concentrations of standard drug and test peptide were obtained in each well. Inocula were added to the microplate by correcting the OD600 of fungal suspension in incubation broth. The plate was then incubated for 48–72 h at 30 °C. The optical density of each well of the plate was recorded on an ELISA plate reader to calculate the FIC index.
4.6. Flow Cytometry
C. neoformans cells were treated with the bioactive peptides (10d or 10o) at MIC value and incubated at 30 °C at 200× g for 12 h. The treated cells along with a batch of untreated cells were centrifuged at 2000× g for 10 min at 4 °C and suspended in cold phosphate-buffered saline (PBS), followed by 2–3 washings with PBS. The harvested treated and untreated cells were then incubated with zymolyase for 1 h at ambient temperature to obtain cell protoplasm. The staining of obtained protoplasm was done using an Annexin V-FITC detection kit (Sigma-Aldrich, Saint Louis, DE, USA). The protoplasm of untreated cells was divided into 4 tubes (2 mL) and into 3 tubes a different staining agent was added while one was kept unstained. Hence, four samples of untreated cells were prepared with (i) unstained cells alone, (ii) cells with propidium iodide (PI), (iii) cells with annexin V-FITC and (iv) cells with both PI and annexin V-FITC. Furthermore, two more samples were prepared where protoplasm of 10d- and 10o-treated cells were stained with both PI and annexin V-FITC. All 6 samples were then analyzed by using an FACS Calibur (Becton-Dickinson, San Jose, CA, USA) at excitation of 488 nm (for FITC) and 560 nm (for PI) band filter. Approximately 10,000 cells were used for counting [29].
4.7. SEM
The bioactive peptides (10d or 10o) were added at their MIC value to a suspension of C. neoformans cells (~103) in an incubation broth at 30 °C for 12 h. Then, the treated, as well as untreated, cells were centrifuged separately at 2000× g for 10 min at 4 °C and suspended in cold PBS and followed by 2–3 washings with PBS. The cells were fixed with 2% glutaraldehyde in 0.1% phosphate buffer for 1 h at ambient temperature and washed 2–3 times with PBS. An amount of 1% osmium tetroxide in 0.1 M PBS was added to the cells to further fix and stain them. The cells were incubated with OsO4 for 1 h at 4 °C. The samples were then dehydrated by incubating them with increasing gradients of 10%, 30%, 50% and 70% of ethanol in water followed by 100% ethanol. The samples were placed on a round glass cover slip previously attached to the aluminum stub by adhesive carbon tape. The samples were dried using a vacuum freeze-drier HITACHI 2030 at ambient temperature, then sputter-coated with gold using a HITACHI E1010 and observed under the SEM (Hitachi S-3400N, Tokyo, Japan) [39].
4.8. TEM
The samples were prepared in a similar manner to as mentioned in the procedure for SEM sample preparation, up until the dehydration of cells with increasing gradients of ethanol. The dehydrated samples were then embedded in resin which was prepared from an epoxy embedding medium kit (Sigma, 45359-1EA-F). Increasing gradients (10%, 20%, 30%, 50%, 70% and 90%) of resin were added to the samples followed by incubation for 1–2 h at room temperature after addition of each gradient. Undiluted resin (100%) was added to the sample and incubated for 24 h at 70 °C. Semi-thin and ultrathin sections were cut with an ultramicrotome (Ultramicotome Lecia EM UC6). The sections of samples were placed on a 3.05 mm diameter and 200 mesh copper grid and stained with uranyl acetate or lead acetate, before observation under the HRTEM (FEI Technai G2 F20, Eindhoven, The Netherlands) and analysis at 120 KV.
4.9. CLSM
- (a)
- FITC-labeled peptide uptake assay
Active and inactive peptides tagged with fluorescein isothiocyanate were incubated with C. neoformans cells (~1 × 103 CFU/mL) at their MIC for 12 h. After incubation, cells were harvested by centrifugation at 2000× g for 10 min at 4 °C and suspended in PBS and mounted on a slide for visualization under a confocal microscope with excitation and emission wavelengths of 488 nm and 515 nm, respectively.
- (b)
- PI uptake assay
The samples for PI were prepared by incubating C. neoformans (ATCC350) cells (~1 × 103 CFU/mL) with test peptides (10d and 10o) for 12 h at their MIC values. After incubation, treated and untreated cells were centrifuged at 2000× g for 10 min at 4 °C and suspended in cold PBS and followed by 2–3 washings with PBS. The cells were then incubated with PI (1.49 μM) for 1 h at room temperature with constant shaking (150× g rpm), and later harvested by centrifugation and suspended in PBS. Then, the cells were examined using confocal microscopy (Olympus Fluoview™ FV1000 SPD; Olympus, Tokyo, Japan) with a wavelength of >560 nm for PI. C. neoformans cells without treatment of active peptides served as a control.
- (c)
- DAPI uptake assay
C. neoformans (ATCC350) cells (~1 × 103 CFU/mL) were incubated with test peptides for 12 h at their MIC values. After incubation, treated and untreated cells were centrifuged at 2000× g for 10 min at 4 °C and suspended in cold PBS and followed by 2–3 washings with PBS. The cells were then incubated with DAPI (3.01 μM) for 10 min at room temperature with constant shaking, harvested by centrifugation and suspended in PBS. Then, the fungal cells were examined using confocal microscopy (Olympus Fluoview™ FV1000 SPD; Olympus, Tokyo, Japan) with excitation at 350 nm and emission at 470 nm [40].
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/molecules28010087/s1. General procedure for the synthesis of histidine derivatives and peptides, characterization data, 1H and 13C-NMR spectra, HRMS data and analytical HPLC of peptides are included in the supplementary information [41,42].
Author Contributions
Investigation, K.S., S.A., I.K.M., S.S., S.M.R., V.K. and K.T.; Data curation, K.S. and R.J.; Writing—original draft, K.S.; Supervision, R.J.; Project administration, R.J. All authors have read and agreed to the published version of the manuscript.
Funding
Doctoral fellowship and research facilities were provided by National Institute of Pharmaceutical Education and Research, S.A.S. Nagar, India funded by Ministry of Chemicals and Fertilizers, Government of India.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Informed consent was obtained for hemolytic study.
Data Availability Statement
The data will be made available by the corresponding author on reasonable request.
Acknowledgments
Komal Sharma thanks the National Institute of Pharmaceutical Education and Research, S.A.S. Nagar, India, for providing the doctoral fellowship and all research facilities.
Conflicts of Interest
The authors declare no competing financial interests.
References
- Rajasingham, R.; Govender, N.P.; Jordan, A.; Loyse, A.; Shroufi, A.; Denning, D.W.; Meya, D.B.; Chiller, T.M.; Boulware, D.R. The global burden of HIV-associated cryptococcal infection in adults in 2020: A modelling analysis. Lancet Infect. Dis. 2022, 22, 1748–1755. [Google Scholar] [CrossRef] [PubMed]
- Ngan, N.T.T.; Flower, B.; Day, J.N. Treatment of Cryptococcal Meningitis: How Have We Got Here and Where are We Going? Drugs 2022, 82, 1237–1249. [Google Scholar] [CrossRef] [PubMed]
- Jarvis, J.N.; Lawrence, D.S.; Meya, D.B.; Kagimu, E.; Kasibante, J.; Mpoza, E.; Rutakingirwa, M.K.; Ssebambulidde, K.; Tugume, L.; Rhein, J. Single-Dose Liposomal Amphotericin B Treatment for Cryptococcal Meningitis. N. Engl. J. Med. 2022, 386, 1109–1120. [Google Scholar] [CrossRef] [PubMed]
- Muhaj, F.F.; George, S.J.; Nguyen, C.D.; Tyring, S.K. Antimicrobials and resistance part II: Antifungals, antivirals, and antiparasitics. J. Am. Acad. Dermatol. 2022, 86, 1207–1226. [Google Scholar] [CrossRef] [PubMed]
- Magana, M.; Pushpanathan, M.; Santos, A.L.; Leanse, L.; Fernandez, M.; Ioannidis, A.; Giulianotti, M.A.; Apidianakis, Y.; Bradfute, S.; Ferguson, A.L.; et al. The value of antimicrobial peptides in the age of resistance. Lancet Infect. Dis. 2020, 20, e216–e230. [Google Scholar] [CrossRef] [PubMed]
- Mahindra, A.; Sharma, K.K.; Rathore, D.; Khan, S.I.; Jacob, M.R.; Jain, R. Synthesis and antimicrobial activities of His(2-aryl)-Arg and Trp-His(2-aryl) classes of dipeptidomimetics. MedChemComm 2014, 5, 671–676. [Google Scholar] [CrossRef] [PubMed]
- Iyer, K.R.; Revie, N.M.; Fu, C.; Robbins, N.; Cowen, L.E. Treatment strategies for cryptococcal infection: Challenges, advances and future outlook. Nat. Rev. Microbiol. 2021, 19, 454–466. [Google Scholar] [CrossRef]
- Struyfs, C.; Cammue, B.P.A.; Thevissen, K. Membrane-interacting antifungal peptides. Front. Cell Dev. Biol. 2021, 9, 649875. [Google Scholar] [CrossRef]
- Sharma, K.K.; Maurya, I.K.; Khan, S.I.; Jacob, M.R.; Kumar, V.; Tikoo, K.; Jain, R. Discovery of a membrane-active, ring-modified histidine containing ultrashort amphiphilic peptide that exhibits potent inhibition of Cryptococcus neoformans. J. Med. Chem. 2017, 60, 6607–6621. [Google Scholar] [CrossRef]
- Sharma, K.K.; Ravi, R.; Maurya, I.K.; Kapadia, A.; Khan, S.I.; Kumar, V.; Tikoo, K.; Jain, R. Modified histidine containing amphipathic ultrashort antifungal peptide, His-[2-p-(n-butyl) phenyl]-Trp-Arg-OMe exhibits potent anticryptococcal activity. Eur. J. Med. Chem. 2021, 223, 113635. [Google Scholar] [CrossRef]
- Mahindra, A.; Bagra, N.; Wangoo, N.; Jain, R.; Khan, S.I.; Jacob, M.R.; Jain, R. Synthetically modified L-histidine-rich peptidomimetics exhibit potent activity against Cryptococcus neoformans. Bioorg. Med. Chem. Lett. 2014, 24, 3150–3154. [Google Scholar] [CrossRef]
- Mahindra, A.; Bagra, N.; Wangoo, N.; Khan, S.I.; Jacob, M.R.; Jain, R. Discovery of short peptides exhibiting high potency against Cryptococcus neoformans. ACS Med. Chem. Lett. 2014, 5, 315–320. [Google Scholar] [CrossRef] [PubMed]
- Sharma, K.K.; Sharma, K.; Kudwal, A.; Khan, S.I.; Jain, R. Peptide-Heterocycle Conjugates as Antifungals against Cryptococcosis. Asian J. Org. Chem. 2022, 11, e202200196. [Google Scholar] [CrossRef]
- Greco, I.; Molchanova, N.; Holmedal, E.; Jenssen, H.; Hummel, B.D.; Watts, J.L.; Håkansson, J.; Hansen, P.R.; Svenson, J. Correlation between hemolytic activity, cytotoxicity and systemic in vivo toxicity of synthetic antimicrobial peptides. Sci. Rep. 2020, 10, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Sharma, K.; Sharma, K.K.; Sharma, A.; Jain, R. Peptide-based drug discovery: Current status and recent advances. Drug Discov. Today 2022, 103464. [Google Scholar] [CrossRef] [PubMed]
- Mahindra, A.; Bagra, N.; Jain, R. Palladium-catalyzed regioselective C-5 arylation of protected L-histidine: Microwave-assisted C–H activation adjacent to donor arm. J. Org. Chem. 2013, 78, 10954–10959. [Google Scholar] [CrossRef] [PubMed]
- Sharma, K.K.; Mandloi, M.; Jain, R. Regioselective copper-catalyzed N(1)-(hetero)arylation of protected histidine. Org. Biomol. Chem. 2016, 14, 8937–8941. [Google Scholar] [CrossRef]
- Sharma, K.K.; Mandloi, M.; Jain, R. Regioselective access to 1,2-diarylhistidines through the copper-catalyzed n1-arylation of 2-arylhistidines. Eur. J. Org. Chem. 2017, 2017, 984–988. [Google Scholar] [CrossRef]
- Song, L.; Ojeda-Carralero, G.M.; Parmar, D.; González-Martínez, D.A.; Van Meervelt, L.; Van der Eycken, J.; Goeman, J.; Rivera, D.G.; Van der Eycken, E.V. Chemoselective Peptide Backbone Diversification and Bioorthogonal Ligation by Ruthenium-Catalyzed C−H Activation/Annulation. Adv. Synth. Catal. 2021, 363, 3297–3304. [Google Scholar] [CrossRef]
- O’Brien, E.A.; Sharma, K.K.; Byerly-Duke, J.; Camacho, L.A., III; VanVeller, B. A General Strategy to Install Amidine Functional Groups Along the Peptide Backbone. J. Am. Chem. Soc. 2022, 144, 22397–22402. [Google Scholar] [CrossRef]
- Jia, F.; Zhang, Y.; Wang, J.; Peng, J.; Zhao, P.; Zhang, L.; Yao, H.; Ni, J.; Wang, K. The effect of halogenation on the antimicrobial activity, antibiofilm activity, cytotoxicity and proteolytic stability of the antimicrobial peptide Jelleine-I. Peptides 2019, 112, 56–66. [Google Scholar] [CrossRef] [PubMed]
- Arias, M.; Hoffarth, E.R.; Ishida, H.; Aramini, J.M.; Vogel, H.J. Recombinant expression, antimicrobial activity and mechanism of action of tritrpticin analogs containing fluoro-tryptophan residues. Biochim. Biophys. Acta Biomembr. 2016, 1858, 1012–1023. [Google Scholar] [CrossRef] [PubMed]
- Cruz, J.o.C.S.; Iorio, M.; Monciardini, P.; Simone, M.; Brunati, C.; Gaspari, E.; Maffioli, S.I.; Wellington, E.; Sosio, M.; Donadio, S. Brominated variant of the lantibiotic NAI-107 with enhanced antibacterial potency. J. Nat. Prod. 2015, 78, 2642–2647. [Google Scholar] [CrossRef] [PubMed]
- Molchanova, N.; Nielsen, J.E.; Sørensen, K.B.; Prabhala, B.K.; Hansen, P.R.; Lund, R.; Barron, A.E.; Jenssen, H. Halogenation as a tool to tune antimicrobial activity of peptoids. Sci. Rep. 2020, 10, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Gottler, L.M.; de la Salud Bea, R.; Shelburne, C.E.; Ramamoorthy, A.; Marsh, E.N.G. Using Fluorous Amino Acids to Probe the Effects of Changing Hydrophobicity on the Physical and Biological Properties of the β-Hairpin Antimicrobial Peptide Protegrin-1. Biochemistry 2008, 47, 9243–9250. [Google Scholar] [CrossRef]
- Sharma, K.; Aaghaz, S.; Maurya, I.K.; Rudramurthy, S.M.; Singh, S.; Kumar, V.; Tikoo, K.; Jain, R. Antifungal evaluation and mechanistic investigations of membrane active short synthetic peptides-based amphiphiles. Bioorg. Chem. 2022, 127, 106002. [Google Scholar] [CrossRef]
- Sharma, K.; Aaghaz, S.; Maurya, I.K.; Sharma, K.K.; Singh, S.; Rudramurthy, S.M.; Kumar, V.; Tikoo, K.; Jain, R. Synthetic amino acids-derived peptides target Cryptococcus neoformans by inducing cell membrane disruption. Bioorg. Chem. 2023, 130, 106252. [Google Scholar] [CrossRef]
- Jain, R.; Avramovitch, B.; Cohen, L.A. Synthesis of ring-halogenated histidines and histamines. Tetrahedron 1998, 54, 3235–3242. [Google Scholar] [CrossRef]
- Maurya, I.K.; Thota, C.K.; Sharma, J.; Tupe, S.G.; Chaudhary, P.; Singh, M.K.; Thakur, I.S.; Deshpande, M.; Prasad, R.; Chauhan, V.S. Mechanism of action of novel synthetic dodecapeptides against Candida albicans. Biochim. Biophys. Acta 2013, 1830, 5193–5203. [Google Scholar] [CrossRef]
- Vermes, I.; Haanen, C.; Steffens-Nakken, H.; Reutelingsperger, C. Flow cytometric detection of phosphatidylserine expression on early apoptotic cells using fluorescein labelled Annexin V. J. Immunol. Methods 1995, 184, 39–51. [Google Scholar] [CrossRef]
- Adan, A.; Alizada, G.; Kiraz, Y.; Baran, Y.; Nalbant, A. Flow cytometry: Basic principles and applications. Crit. Rev. Biotechnol. 2017, 37, 163–176. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Zhu, W.; Yang, X.; Chen, L.; Yang, R.; Hua, Z.; Li, G. Detection of apoptosis based on the interaction between annexin V and phosphatidylserine. Anal. Chem. 2009, 81, 2410–2413. [Google Scholar] [CrossRef]
- Henry, C.M.; Hollville, E.; Martin, S.J. Measuring apoptosis by microscopy and flow cytometry. Methods 2013, 61, 90–97. [Google Scholar] [CrossRef] [PubMed]
- CLSI. Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts, CLSI Supplement M27; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2017. [Google Scholar]
- CLSI. Reference Method for Broth Dilution Antifungal Susceptibility Testing of Filamentous Fungi, CLSI Supplement M38; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2017. [Google Scholar]
- CLSI. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically, CLSI Supplement M07; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018. [Google Scholar]
- Sharma, A.; Singh, S.; Tewari, R.; Bhatt, V.P.; Sharma, J.; Maurya, I.K. Phytochemical analysis and mode of action against Candida glabrata of Paeonia emodi extracts. J. Mycol. Med. 2018, 28, 443–451. [Google Scholar] [CrossRef] [PubMed]
- Green, R.M.; Bicker, K.L. Evaluation of peptoid mimics of short, lipophilic peptide antimicrobials. Int. J. Antimicrob. Agents 2020, 56, 106048. [Google Scholar] [CrossRef]
- Mares, D. Electron microscopy of Microsporum cookei after ’in vitro’ treatment with protoanemonin: A combined SEM and TEM study. Mycopathologia 1989, 108, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Park, C.; Lee, D.G. Melittin induces apoptotic features in Candida albicans. Biochem. Biophys. Res. Commun. 2010, 394, 170–172. [Google Scholar] [CrossRef]
- Abdo, M.-R.; Joseph, P.; Boigegrain, R.-A.; Liautard, J.-P.; Montero, J.-L.; Köhler, S.; Winum, J.-Y. Brucella suis histidinol dehydrogenase: Synthesis and inhibition studies of a series of substituted benzylic ketones derived from histidine. Bioorg. Med. Chem. 2007, 15, 4427–4433. [Google Scholar] [CrossRef]
- Meena, C.L.; Thakur, A.; Nandekar, P.P.; Sangamwar, A.T.; Sharma, S.S.; Jain, R. Synthesis of CNS active thyrotropin-releasing hormone (TRH)-like peptides: Biological evaluation and effect on cognitive impairment induced by cerebral ischemia in mice. Bioorg. Med. Chem. 2015, 23, 5641–5653. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).