Towards Non-Targeted Screening of Lipid Biomarkers for Improved Equine Anti-Doping
Abstract
1. Biomarkers for Equine Anti-Doping
2. Current Challenges for Targeted Screening
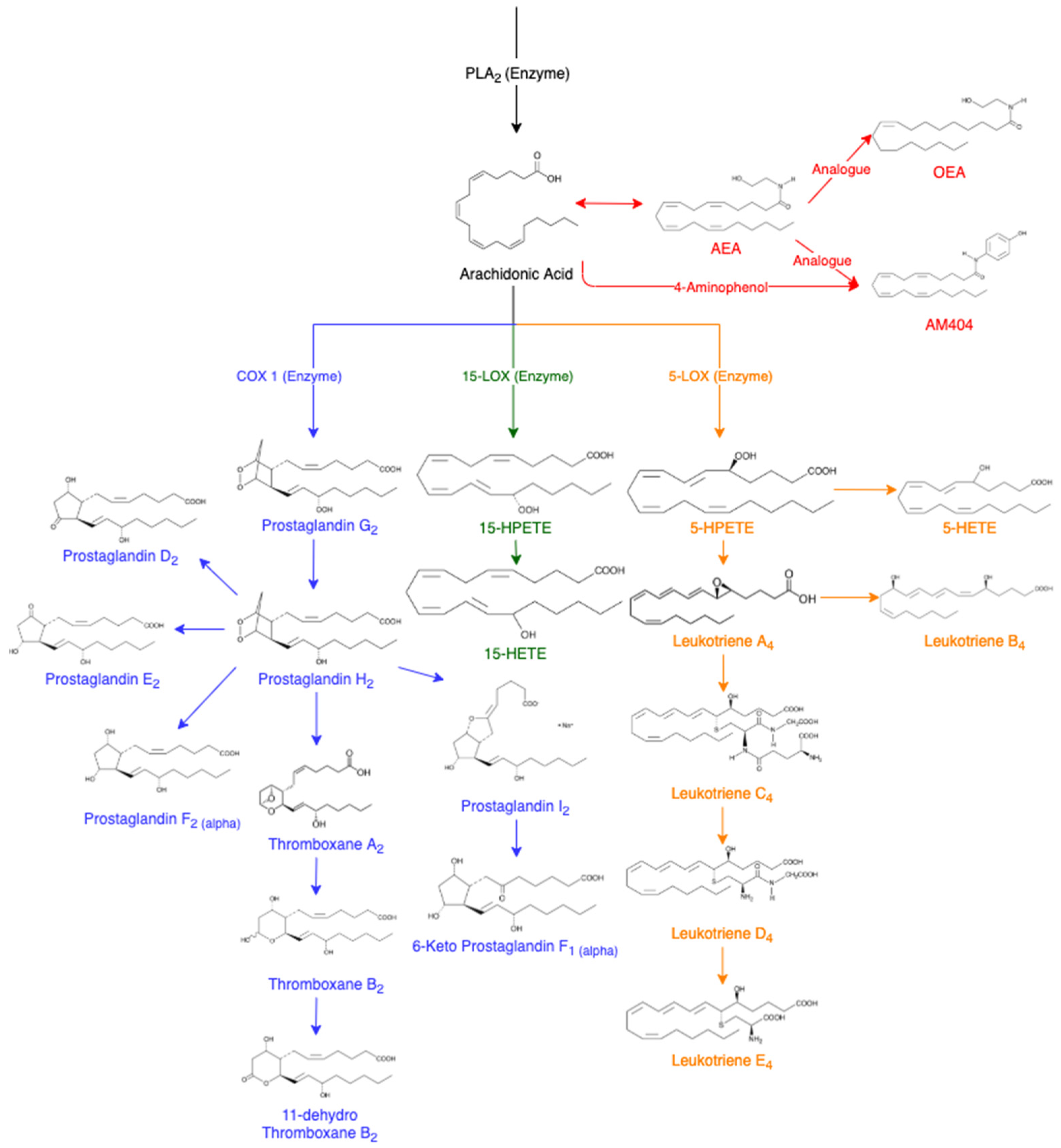
3. Lipids
4. Analytical Techniques for Lipidomics
4.1. Sample Preparation
4.2. Analysis
4.3. Data Acquisition Modes
5. Statistical Analysis
6. Studies Monitoring Eicosanoids for Equine Anti-Doping Screening
6.1. Lipid Screening in Equine Samples
6.2. Lipid Inflammatory Markers and Their Effect from a Corticosteroid Administration
6.3. Lipid Inflammatory Markers and Their Effect from an NSAID Administration
6.4. Lipid Inflammatory Markers and Their Effect from a Cannabidiol Administration
7. Potential for Lipidomics to Contribute Longitudinal Assessments for Equine Anti-Doping
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fragkaki, A.G.; Kioukia-Fougia, N.; Kiousi, P.; Kioussi, M.; Tsivou, M. Challenges in detecting substances for equine anti-doping. Drug Test. Anal. 2017, 9, 1291–1303. [Google Scholar] [CrossRef] [PubMed]
- Teale, P.; Barton, C.; Driver, P.M.; Kay, R.G. Biomarkers: Unrealized potential in sports doping analysis. Bioanalysis 2009, 1, 1103–1118. [Google Scholar] [CrossRef] [PubMed]
- Narduzzi, L.; Dervilly, G.; Audran, M.; Le Bizec, B.; Buisson, C. A role for metabolomics in the antidoping toolbox? Drug Test. Anal. 2020, 12, 677–690. [Google Scholar] [CrossRef] [PubMed]
- Marclay, F.; Mangin, P.; Margot, P.; Saugy, M. Perspectives for Forensic Intelligence in anti-doping: Thinking outside of the box. Forensic Sci. Int. 2013, 229, 133–144. [Google Scholar] [CrossRef] [PubMed]
- Stojiljkovic, N.; Paris, A.; Garcia, P.; Popot, M.-A.; Bonnaire, Y.; Tabet, J.-C.; Junot, C. Evaluation of horse urine sample preparation methods for metabolomics using LC coupled to HRMS. Bioanalysis 2014, 6, 785–803. [Google Scholar] [CrossRef]
- Reichel, C. OMICS-strategies and methods in the fight against doping. Forensic Sci. Int. 2011, 213, 20. [Google Scholar] [CrossRef]
- Mangal, D.; Uboh, C.E.; Soma, L.R. Analysis of bioactive eicosanoids in equine plasma by stable isotope dilution reversed-phase liquid chromatography/multiple reaction monitoring mass spectrometry. Rapid Commun. Mass Spectrom. 2011, 25, 585–598. [Google Scholar] [CrossRef]
- Dass, C. Characterization of Lipids. In Fundamentals of Contemporary Mass Spectrometry; John Wiley & Sons: New York, NY, USA, 2007; pp. 423–451. [Google Scholar] [CrossRef]
- López-Bascón, M.A.; Calderón-Santiago, M.; Díaz-Lozano, A.; Camargo, A.; López-Miranda, J.; Priego-Capote, F. Development of a qualitative/quantitative strategy for comprehensive determination of polar lipids by LC–MS/MS in human plasma. Anal. Bioanal. Chem. 2020, 412, 489–498. [Google Scholar] [CrossRef]
- Koelmel, J.P.; Li, X.; Stow, S.M.; Sartain, M.J.; Murali, A.; Kemperman, R.; Tsugawa, H.; Takahashi, M.; Vasiliou, V.; Bowden, J.A.; et al. Lipid Annotator: Towards Accurate Annotation in Non-Targeted Liquid Chromatography High-Resolution Tandem Mass Spectrometry (LC-HRMS/MS) Lipidomics Using A Rapid and User-Friendly Software. Metabolites 2020, 10, 101. [Google Scholar] [CrossRef]
- Nolazco Sassot, L.; Villarino, N.F.; Dasgupta, N.; Morrison, J.J.; Bayly, W.M.; Gang, D.; Sanz, M.G. The lipidome of Thoroughbred racehorses before and after supramaximal exercise. Equine Vet. J. 2019, 51, 696–700. [Google Scholar] [CrossRef]
- Harwood, J.L.; Frayn, K.N.; Murphy, D.J.; Michell, R.H.; Gurr, M.I. Lipids: Biochemistry, Biotechnology and Health; John Wiley & Sons, Incorporated: Hoboken, UK, 2016. [Google Scholar]
- Fahy, E.; Cotter, D.; Sud, M.; Subramaniam, S. Lipid classification, structures and tools. Biochim. Et Biophys. Acta. Mol. Cell Biol. Lipids 2011, 1811, 637–647. [Google Scholar] [CrossRef] [PubMed]
- Granström, E. The arachidonic acid cascade. Inflammation 1984, 8, S15–S25. [Google Scholar] [CrossRef] [PubMed]
- Thakare, R.; Chhonker, Y.S.; Gautam, N.; Nelson, A.; Casaburi, R.; Criner, G.; Dransfield, M.T.; Make, B.; Schmid, K.K.; Rennard, S.I.; et al. Simultaneous LC–MS/MS analysis of eicosanoids and related metabolites in human serum, sputum and BALF. Biomed. Chromatogr. 2018, 32, e4102. [Google Scholar] [CrossRef] [PubMed]
- Toewe, A.; Balas, L.; Durand, T.; Geisslinger, G.; Ferreirós, N. Simultaneous determination of PUFA-derived pro-resolving metabolites and pathway markers using chiral chromatography and tandem mass spectrometry. Anal. Chim. Acta 2018, 1031, 185–194. [Google Scholar] [CrossRef] [PubMed]
- Giles, H.; Leff, P. The biology and pharmacology of PGD2. Prostaglandins 1988, 35, 277–300. [Google Scholar] [CrossRef]
- Jackson, C.A.; Colahan, P.T.; Rice, B. Use of a Commercially Available Prostaglandin E2 Enzyme-Linked Immunosorbent Assay for Non-Steroidal Anti-Inflammatory Drug Screening. In Proceedings of the 16th International Conference of Racing Analysts and Veterinarians, Tokyo, Japan, 21–27 October 2006; Volume 16, pp. 477–482. [Google Scholar]
- Lees, P.; Ewins, C.P.; Taylor, J.B.O.; Sedgwick, A.D. Serum thromboxane in the horse and its inhibition by aspirin, phenylbutazone and flunixin. Br. Vet. J. 1987, 143, 462–476. [Google Scholar] [CrossRef]
- Lopez, L.R.; Guyer, K.E.; Torre, I.G.D.L.; Pitts, K.R.; Matsuura, E.; Ames, P.R. Platelet thromboxane (11-dehydro-Thromboxane B2) and aspirin response in patients with diabetes and coronary artery disease. World J. Diabetes 2014, 5, 115–127. [Google Scholar] [CrossRef]
- Johnson, R.A.; Morton, D.R.; Kinner, J.H.; Gorman, R.R.; McGuire, J.C.; Sun, F.F.; Whittaker, N.; Bunting, S.; Salmon, J.; Moncada, S.; et al. The chemical structure of prostaglandin X (prostacyclin). Prostaglandins 1976, 12, 915–928. [Google Scholar] [CrossRef]
- Connolly, P.J.; Wetter, S.K.; Beers, K.N.; Hamel, S.C.; Chen, R.H.K.; Wachter, M.P.; Ansell, J.; Singer, M.M.; Steber, M.; Ritchie, D.M.; et al. N-Hydroxyurea and hydroxamic acid inhibitors of cyclooxygenase and 5-lipoxygenase. Bioorganic Med. Chem. Letters 1999, 9, 979–984. [Google Scholar] [CrossRef]
- Samuelsson, B.; Dahlen, S.-E.; Lindgren, J.A.; Rouzer, C.A.; Serhan, C.N. Leukotrienes and lipoxins: Structures, biosynthesis, and biological effects. Science 1987, 237, 1171–1176. [Google Scholar] [CrossRef]
- McMillan, R.M.; Foster, S.J. Leukotriene B4 and inflammatory disease. Agents Actions 1988, 24, 114–119. [Google Scholar] [CrossRef] [PubMed]
- Goodman, L.; Coles, T.B.; Budsberg, S. Leukotriene inhibition in small animal medicine. J. Vet. Pharmacol. Ther. 2008, 31, 387–398. [Google Scholar] [CrossRef] [PubMed]
- Felder, C.C.; Briley, E.M.; Axelrod, J.; Simpson, J.T.; Mackie, K.; Devane, W.A. Anandamide, an endogenous cannabimimetic eicosanoid, binds to the cloned human cannabinoid receptor and stimulates receptor-mediated signal transduction. Proc. Natl. Acad. Sci. USA 1993, 90, 7656–7660. [Google Scholar] [CrossRef]
- Beltramo, M.; Stella, N.; Calignano, A.; Lin, S.Y.; Makriyannis, A.; Piomelli, D. Functional role of high-affinity anandamide transport, as revealed by selective inhibition. Science 1997, 277, 1094–1097. [Google Scholar] [CrossRef]
- Sharma, C.V.; Long, J.H.; Shah, S.; Rahman, J.; Perrett, D.; Ayoub, S.S.; Mehta, V. First evidence of the conversion of paracetamol to AM404 in human cerebrospinal fluid. J. Pain Res. 2017, 10, 2703–2709. [Google Scholar] [CrossRef] [PubMed]
- Högestätt, E.D.; Jönsson, B.A.G.; Ermund, A.; Andersson, D.A.; Björk, H.; Alexander, J.P.; Cravatt, B.F.; Basbaum, A.I.; Zygmunt, P.M. Conversion of Acetaminophen to the Bioactive N-Acylphenolamine AM404 via Fatty Acid Amide Hydrolase-dependent Arachidonic Acid Conjugation in the Nervous System. J. Biol. Chem. 2005, 280, 31405–31412. [Google Scholar] [CrossRef] [PubMed]
- Ricciotti, E.; FitzGerald, G.A. Prostaglandins and Inflammation. Arterioscler. Thromb. Vasc. Biol. 2011, 31, 986–1000. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Wu, L.; Chen, J.; Dong, L.; Chen, C.; Wen, Z.; Hu, J.; Fleming, I.; Wang, D.W. Metabolism pathways of arachidonic acids: Mechanisms and potential therapeutic targets. Signal Transduct. Target. Ther. 2021, 6, 94. [Google Scholar] [CrossRef]
- Chambers, E.; Wagrowski-Diehl, D.M.; Lu, Z.; Mazzeo, J.R. Systematic and comprehensive strategy for reducing matrix effects in LC/MS/MS analyses. J. Chromatogr. B 2007, 852, 22–34. [Google Scholar] [CrossRef]
- Sarafian, M.H.; Gaudin, M.; Lewis, M.R.; Martin, F.-P.; Holmes, E.; Nicholson, J.K.; Dumas, M.-E. Objective Set of Criteria for Optimization of Sample Preparation Procedures for Ultra-High Throughput Untargeted Blood Plasma Lipid Profiling by Ultra Performance Liquid Chromatography–Mass Spectrometry. Anal. Chem. 2014, 86, 5766–5774. [Google Scholar] [CrossRef]
- Cajka, T.; Fiehn, O. Toward Merging Untargeted and Targeted Methods in Mass Spectrometry-Based Metabolomics and Lipidomics. Anal. Chem. 2016, 88, 524–545. [Google Scholar] [CrossRef] [PubMed]
- Zahra, P.W.; Simpson, N.J.K. Application of Alternative SPE Sorbents and Formats for Extraction of Urine and Plasma. In Proceedings of the 20th International Conference of Racing Analysts and Veterinarians, Mauritius, 20–27 September 2015; Volume 20, pp. 324–330. [Google Scholar]
- Bruce, S.J.; Tavazzi, I.; Parisod, V.; Rezzi, S.; Kochhar, S.; Guy, P.A. Investigation of Human Blood Plasma Sample Preparation for Performing Metabolomics Using Ultrahigh Performance Liquid Chromatography/Mass Spectrometry. Anal. Chem. 2009, 81, 3285–3296. [Google Scholar] [CrossRef]
- Carrier, D.J.; Bogri, T.; Cosentino, G.P.; Guse, I.; Rakhit, S.; Singh, K. HPLC studies on leukotriene A4 obtained from the hydrolysis of its methyl ester. Prostaglandins Leukot. Essent. Fat. Acids 1988, 34, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Terragno, A.; Rydzik, R.; Terragno, N.A. High performance liquid chromatography and UV detection for the separation and quantitation of prostaglandins. Prostaglandins 1981, 21, 101–112. [Google Scholar] [CrossRef] [PubMed]
- Gandhi, A.S.; Budac, D.; Khayrullina, T.; Staal, R.; Chandrasena, G. Quantitative analysis of lipids: A higher-throughput LC-MS/MS-based method and its comparison to ELISA. Future Sci. OA 2017, 3, FSO157. [Google Scholar] [CrossRef] [PubMed]
- Tsikas, D.; Zoerner, A.A. Analysis of eicosanoids by LC-MS/MS and GC-MS/MS: A historical retrospect and a discussion. J. Chromatogr. B 2014, 964, 79–88. [Google Scholar] [CrossRef]
- Reis, A.; Rudnitskaya, A.; Blackburn, G.J.; Mohd Fauzi, N.; Pitt, A.R.; Spickett, C.M. A comparison of five lipid extraction solvent systems for lipidomic studies of human LDL. J. Lipid Res. 2013, 54, 1812–1824. [Google Scholar] [CrossRef]
- Lu, X.; Zhao, X.; Bai, C.; Zhao, C.; Lu, G.; Xu, G. LC–MS-based metabonomics analysis. J. Chromatogr. B 2008, 866, 64–76. [Google Scholar] [CrossRef]
- Kinyua, J.; Negreira, N.; Ibáñez, M.; Bijlsma, L.; Hernández, F.; Covaci, A.; van Nuijs, A.L.N. A data-independent acquisition workflow for qualitative screening of new psychoactive substances in biological samples. Anal. Bioanal. Chem. 2015, 407, 8773–8785. [Google Scholar] [CrossRef]
- Chernushevich, I.V.; Loboda, A.V.; Thomson, B.A. An introduction to quadrupole–time-of-flight mass spectrometry. J. Mass Spectrom. 2001, 36, 849–865. [Google Scholar] [CrossRef]
- Dass, C. Mass Analysis and Ion Detection. In Fundamentals of Contemporary Mass Spectrometry; John Wiley & Sons: New York, NY, USA, 2007; pp. 67–117. [Google Scholar] [CrossRef]
- Hopfgartner, G. Can MS fully exploit the benefits of fast chromatography? Bioanalysis 2011, 3, 121–123. [Google Scholar] [CrossRef] [PubMed]
- Fenaille, F.; Barbier Saint-Hilaire, P.; Rousseau, K.; Junot, C. Data acquisition workflows in liquid chromatography coupled to high resolution mass spectrometry-based metabolomics: Where do we stand? J. Chromatogr. A 2017, 1526, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Costa, C.; Martínez-Bartolomé, S.; McClatchy, D.B.; Saviola, A.J.; Yu, N.-K.; Yates, J.R. Impact of the Identification Strategy on the Reproducibility of the DDA and DIA Results. J. Proteome Res. 2020, 19, 3153–3161. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Xu, Z.; Strashnov, I.; Liao, X. Use of information dependent acquisition mass spectra and sequential window acquisition of all theoretical fragment-ion mass spectra for fruit juices metabolomics and authentication. Metabolomics 2020, 16, 81. [Google Scholar] [CrossRef] [PubMed]
- Drotleff, B.; Calderon Castro, C.; Cebo, M.; Dittrich, K.; Fu, X.; Laemmerhofer, M. Untargeted LC-MS Lipidomics with Data Independent Acquisition using Sequential Window Acquisition of All Theoretical Fragment-Ion Spectra. LC GC Eur. 2021, 34, 7–20. [Google Scholar]
- Doerr, A. DIA mass spectrometry. Nat. Methods 2015, 12, 35. [Google Scholar] [CrossRef]
- Raetz, M.; Bonner, R.; Hopfgartner, G. SWATH-MS for metabolomics and lipidomics: Critical aspects of qualitative and quantitative analysis. Metabolomics 2020, 16, 71. [Google Scholar] [CrossRef]
- Wrona, M.; Mauriala, T.; Bateman, K.P.; Mortishire-Smith, R.J.; O’Connor, D. ‘All-in-One’ analysis for metabolite identification using liquid chromatography/hybrid quadrupole time-of-flight mass spectrometry with collision energy switching. Rapid Commun. Mass Spectrom. 2005, 19, 2597–2602. [Google Scholar] [CrossRef]
- Plumb, R.S.; Johnson, K.A.; Rainville, P.; Smith, B.W.; Wilson, I.D.; Castro-Perez, J.M.; Nicholson, J.K. UPLC/MSE; a new approach for generating molecular fragment information for biomarker structure elucidation. Rapid Commun. Mass Spectrom. 2006, 20, 1989–1994. [Google Scholar] [CrossRef]
- Koopmans, F.; Ho, J.T.C.; Smit, A.B.; Li, K.W. Comparative Analyses of Data Independent Acquisition Mass Spectrometric Approaches: DIA, WiSIM-DIA, and Untargeted DIA. Proteomics 2018, 18, 1700304. [Google Scholar] [CrossRef]
- Tsugawa, H.; Hanada, A.; Ikeda, K.; Isobe, Y.; Senoo, Y.; Senoo, M. High-Throughput Lipid Profiling with SWATH Aquisition and MS-Dial; SCIEX: Framingham, MA, USA, 2019; pp. 1–5. [Google Scholar]
- Ren, S.; Hinzman, A.A.; Kang, E.L.; Szczesniak, R.D.; Lu, L.J. Computational and statistical analysis of metabolomics data. Metabolomics 2015, 11, 1492–1513. [Google Scholar] [CrossRef]
- Saccenti, E.; Hoefsloot, H.C.; Smilde, A.K.; Westerhuis, J.A.; Hendriks, M.M. Reflections on univariate and multivariate analysis of metabolomics data. Metabolomics 2014, 10, 361–374. [Google Scholar] [CrossRef]
- Tsugawa, H.; Ikeda, K.; Takahashi, M.; Satoh, A.; Mori, Y.; Uchino, H.; Okahashi, N.; Yamada, Y.; Tada, I.; Bonini, P.; et al. A lipidome atlas in MS-DIAL 4. Nat. Biotechnol. 2020, 38, 1159–1163. [Google Scholar] [CrossRef] [PubMed]
- Courant, F.; Antignac, J.-P.; Dervilly-Pinel, G.; Le Bizec, B. Basics of mass spectrometry based metabolomics. Proteomics 2014, 14, 2369–2388. [Google Scholar] [CrossRef]
- Schymanski, E.L.; Jeon, J.; Gulde, R.; Fenner, K.; Ruff, M.; Singer, H.P.; Hollender, J. Identifying Small Molecules via High Resolution Mass Spectrometry: Communicating Confidence. Environ. Sci. Technol. 2014, 48, 2097–2098. [Google Scholar] [CrossRef]
- Mangal, D.; Uboh, C.E.; Soma, L.R.; Liu, Y. Inhibitory effect of triamcinolone acetonide on synthesis of inflammatory mediators in the equine. Eur. J. Pharmacol. 2014, 736, 1–9. [Google Scholar] [CrossRef]
- Knych, H.K.; Arthur, R.M.; McKemie, D.S.; Seminoff, K.; Hamamoto-Hardman, B.; Kass, P.H. Phenylbutazone blood and urine concentrations, pharmacokinetics, and effects on biomarkers of inflammation in horses following intravenous and oral administration of clinical doses. Drug Test. Anal. 2019, 11, 792–803. [Google Scholar] [CrossRef]
- Knych, H.K.; Weiner, D.; Arthur, R.M.; Baden, R.; McKemie, D.S.; Kass, P.H. Serum concentrations, pharmacokinetic/pharmacodynamic modeling, and effects of dexamethasone on inflammatory mediators following intravenous and oral administration to exercised horses. Drug Test. Anal. 2020, 12, 1087–1101. [Google Scholar] [CrossRef]
- Cuniberti, B.; Odore, R.; Barbero, R.; Cagnardi, P.; Badino, P.; Girardi, C.; Re, G. In vitro and ex vivo pharmacodynamics of selected non-steroidal anti-inflammatory drugs in equine whole blood. Vet. J. 2012, 191, 327–333. [Google Scholar] [CrossRef]
- Slifer, P. A Review of Therapeutic Drugs Used for Doping of Race Horses: NSAIDs, Acepromazine, and Furosemide; Components, C., Ed.; Iowa State University: Ames, IA, USA, 2018; Volume 105. [Google Scholar]
- Lees, P.; Landoni, M.F.; Giraudel, J.; Toutain, P.L. Pharmacodynamics and pharmacokinetics of nonsteroidal anti-inflammatory drugs in species of veterinary interest. J. Vet. Pharmacol. Ther. 2004, 27, 479–490. [Google Scholar] [CrossRef]
- Murakami, M.; Naraba, H.; Tanioka, T.; Semmyo, N.; Nakatani, Y.; Kojima, F.; Ikeda, T.; Fueki, M.; Ueno, A.; Oh-ishi, S.; et al. Regulation of Prostaglandin E2 Biosynthesis by Inducible Membrane-associated Prostaglandin E2 Synthase That Acts in Concert with Cyclooxygenase-2. J. Biol. Chem. 2000, 275, 32783–32792. [Google Scholar] [CrossRef] [PubMed]
- Knych, H.K.; Arthur, R.M.; McKemie, D.S.; Baden, R.; Oldberg, N.; Kass, P.H. Pharmacokinetics of intravenous flumetasone and effects on plasma hydrocortisone concentrations and inflammatory mediators in the horse. Equine Vet. J. 2019, 51, 238–245. [Google Scholar] [CrossRef] [PubMed]
- Ryan, D.; McKemie, D.S.; Kass, P.H.; Puschner, B.; Knych, H.K. Pharmacokinetics and effects on arachidonic acid metabolism of low doses of cannabidiol following oral administration to horses. Drug Test. Anal. 2021, 13, 1305–1317. [Google Scholar] [CrossRef] [PubMed]
- Cawley, A.T.; Keledjian, J. Intelligence-based anti-doping from an equine biological passport. Drug Test. Anal. 2017, 9, 1441–1447. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tou, K.; Cawley, A.; Bowen, C.; Bishop, D.P.; Fu, S. Towards Non-Targeted Screening of Lipid Biomarkers for Improved Equine Anti-Doping. Molecules 2023, 28, 312. https://doi.org/10.3390/molecules28010312
Tou K, Cawley A, Bowen C, Bishop DP, Fu S. Towards Non-Targeted Screening of Lipid Biomarkers for Improved Equine Anti-Doping. Molecules. 2023; 28(1):312. https://doi.org/10.3390/molecules28010312
Chicago/Turabian StyleTou, Kathy, Adam Cawley, Christopher Bowen, David P. Bishop, and Shanlin Fu. 2023. "Towards Non-Targeted Screening of Lipid Biomarkers for Improved Equine Anti-Doping" Molecules 28, no. 1: 312. https://doi.org/10.3390/molecules28010312
APA StyleTou, K., Cawley, A., Bowen, C., Bishop, D. P., & Fu, S. (2023). Towards Non-Targeted Screening of Lipid Biomarkers for Improved Equine Anti-Doping. Molecules, 28(1), 312. https://doi.org/10.3390/molecules28010312





