Triterpenic Acid Amides as Potential Inhibitors of the SARS-CoV-2 Main Protease
Abstract
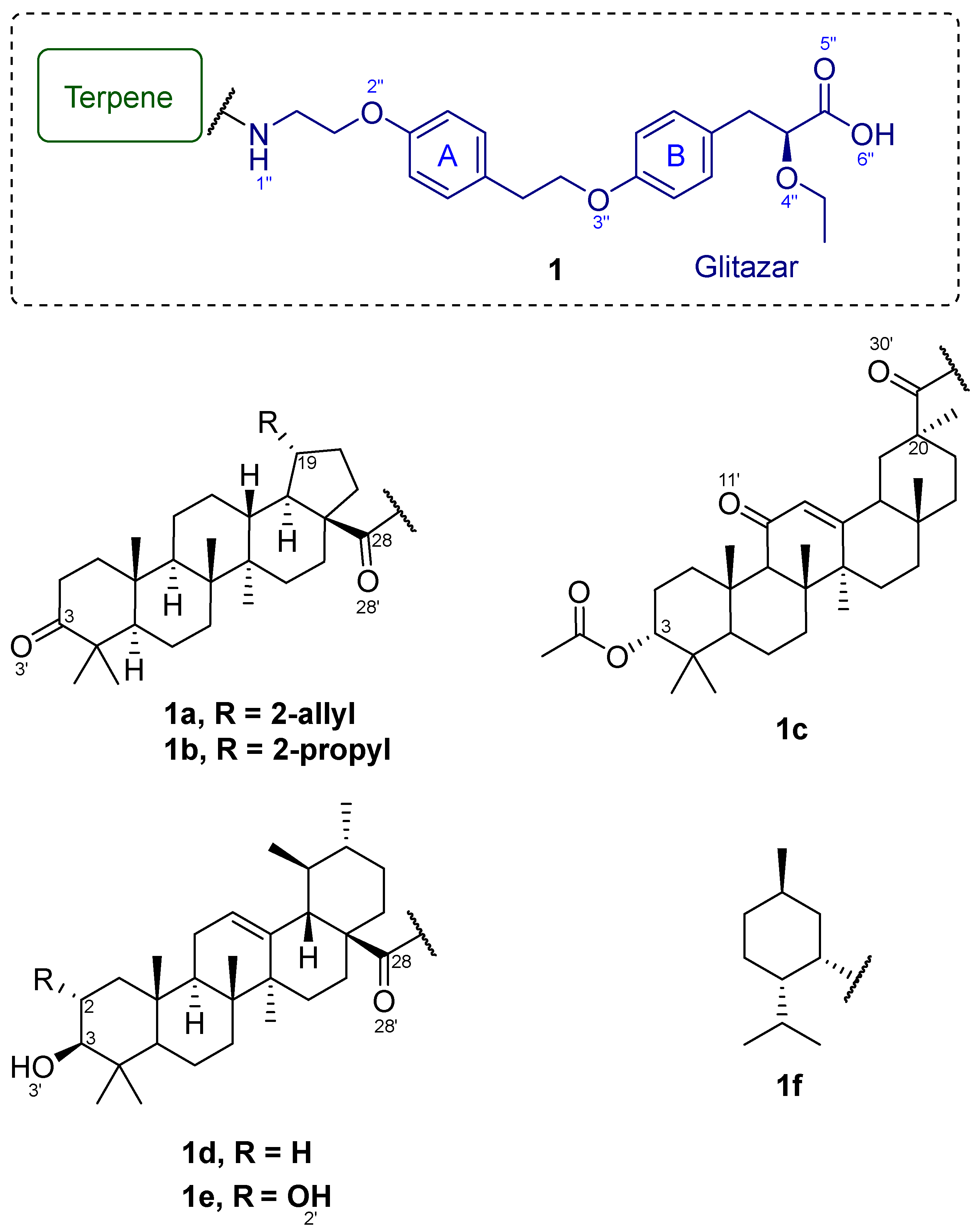
1. Introduction
2. Results
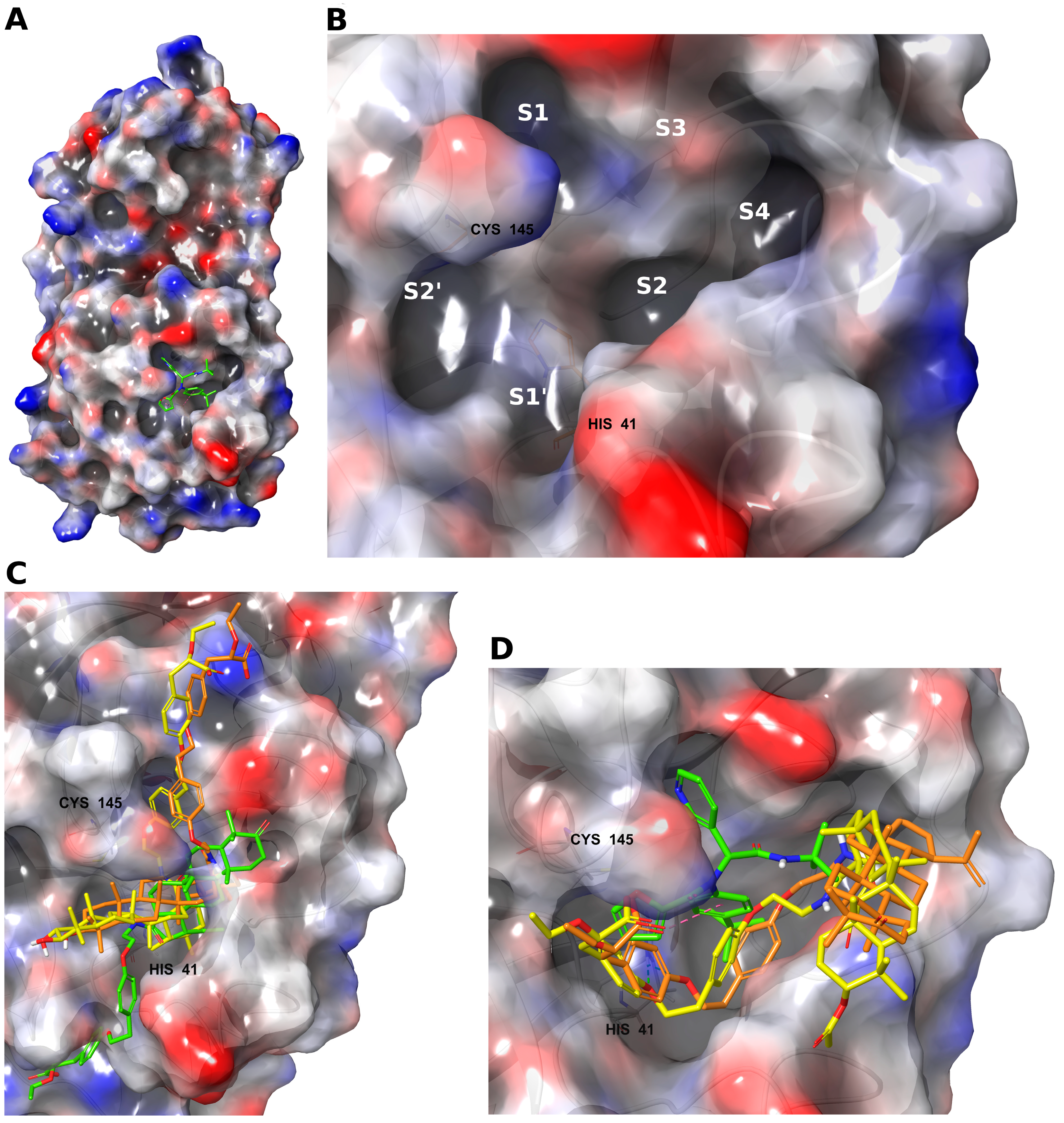
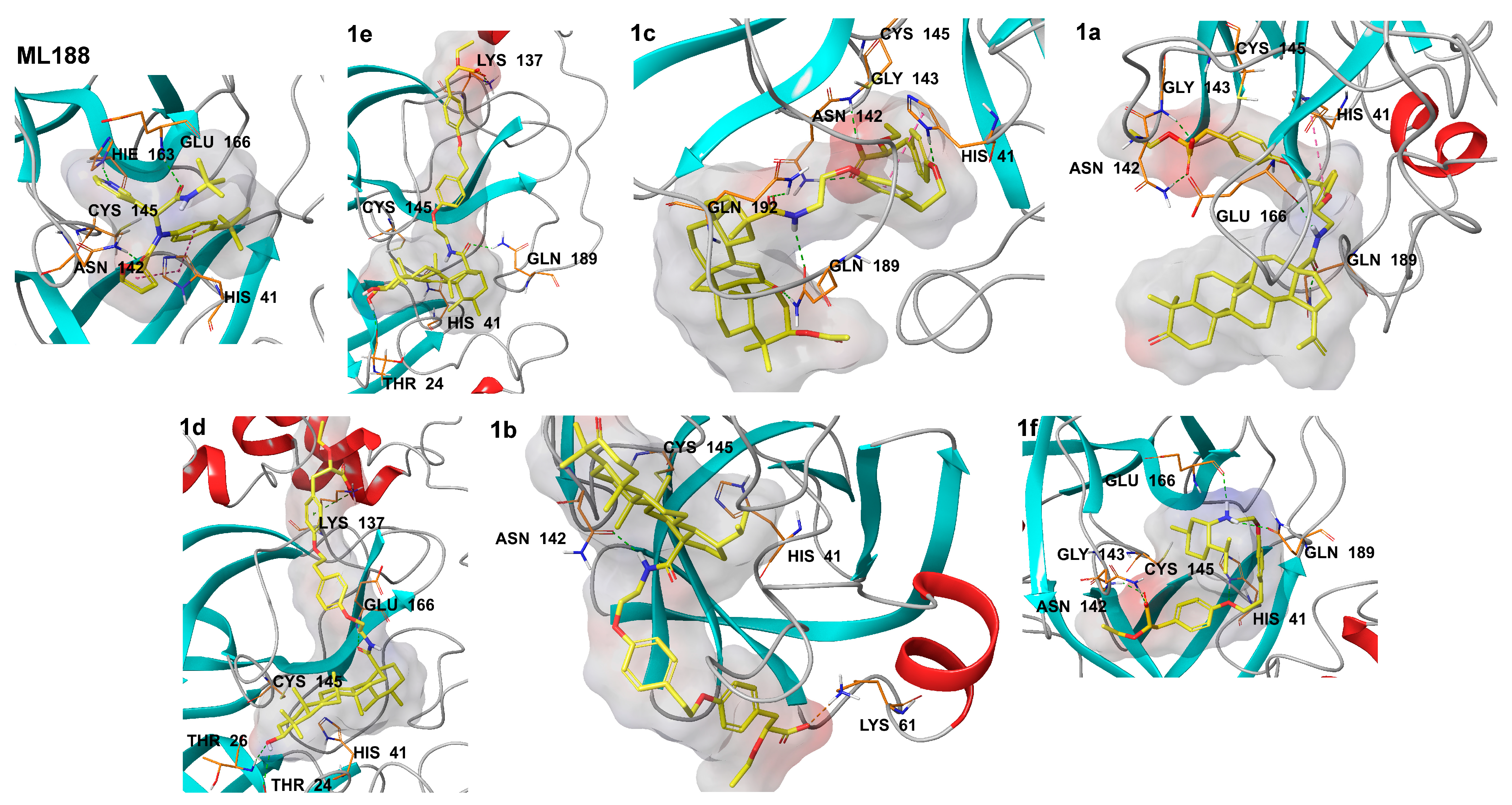
2.1. Molecular Modeling
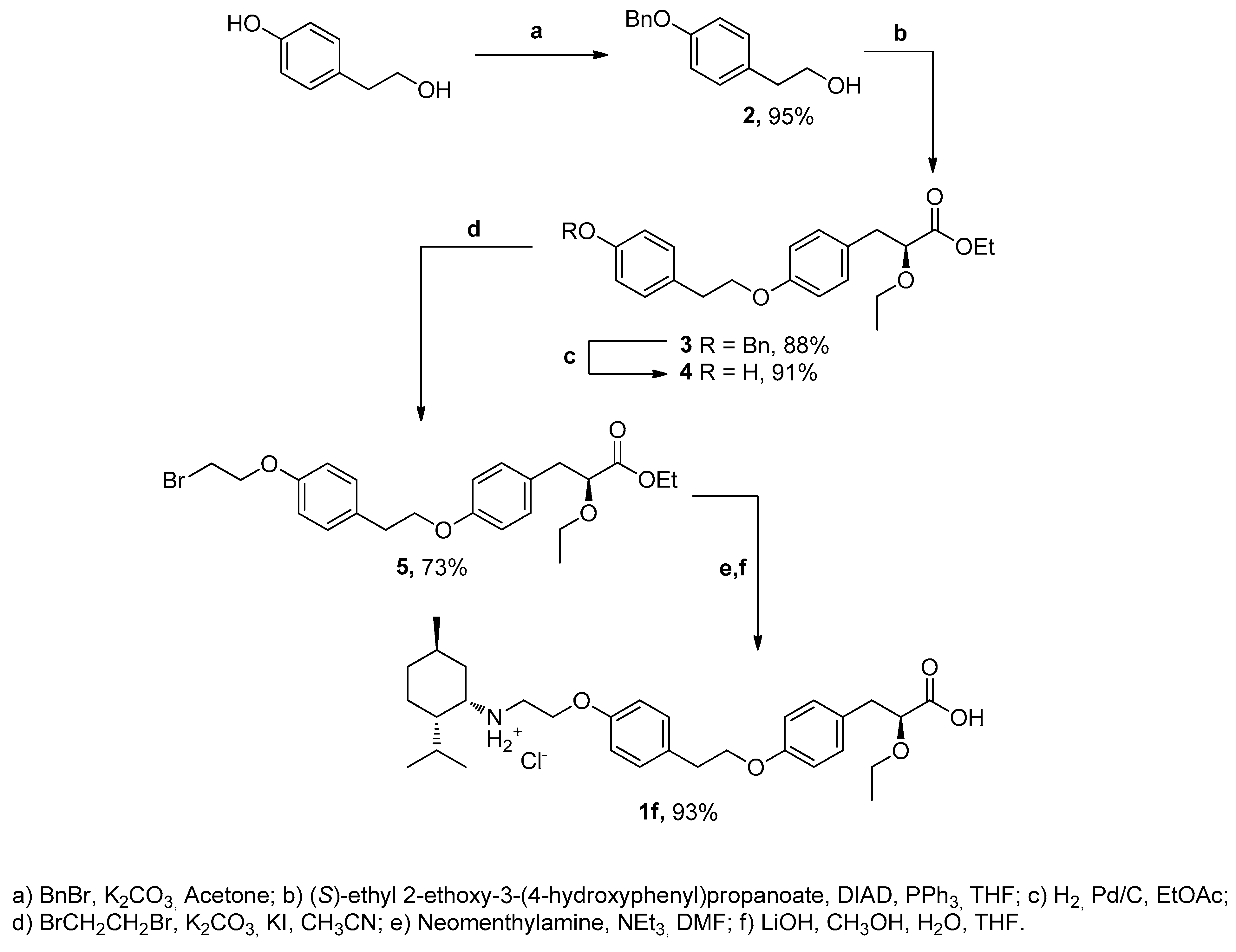
2.2. Chemistry
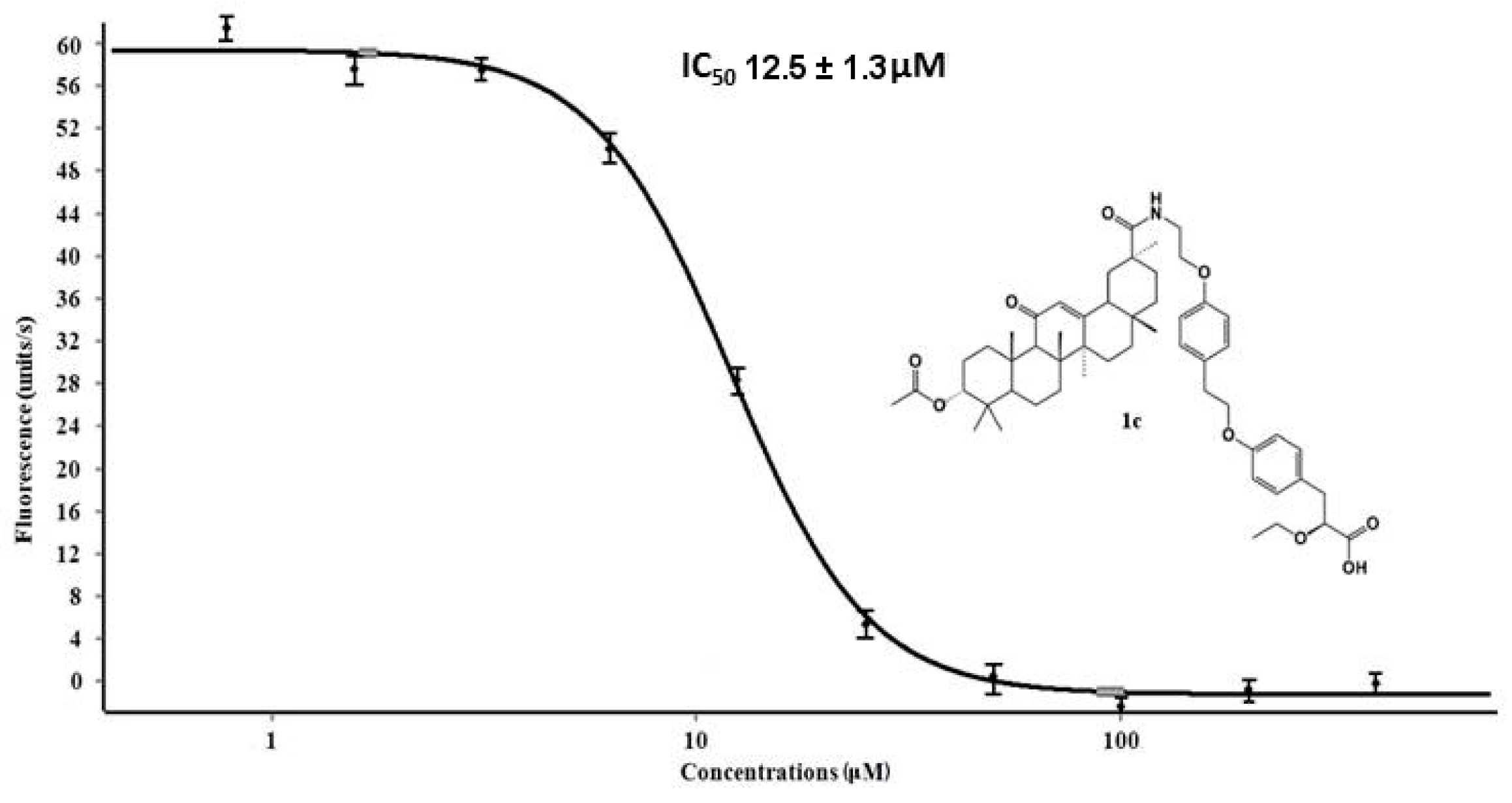
2.3. SARS-CoV-2 Mpro Inhibiting Activity
3. Discussion
4. Materials and Methods
4.1. Molecular Modeling
4.2. Chemistry
- 2-(4-(benzyloxy)-phenyl) ethanol (2)
- (S)-ethyl 3-(4-(4-(4-(benzyloxy)-phenethoxy)-phenyl)-2-ethoxypropanoate (3)
- (S)-ethyl-2-ethoxy-3-(4-(4-hydroxyphenethoxy)-phenyl) propanoate (4)
- Ethyl 3-(4-(4-(4-(2-bromoethoxy)-phenethoxy)-phenyl)-2-ethoxy propanoate (5)
- (S)-ethyl 2-ethoxy-3-(4-(4-(2-(mentylamino)-ethoxy)-phenethoxy)-phenyl) propanoate (6)
- N-(2-(4-(2-(4-((S)-2-carboxy-2-ethoxyethyl)phenoxy)ethyl)phenoxy)ethyl)-2-isopropyl-5-methylcyclohexanaminium chloride (1f)
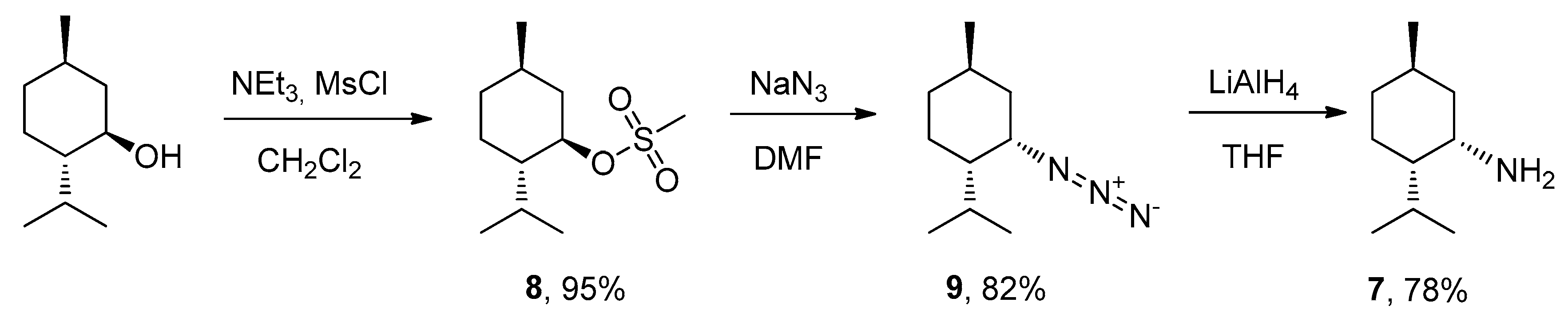
- (1R,2S,5R)-2-isopropyl-5-methylcyclohexyl methanesulfonate (8)
- (1S,2S,4R)-2-azido-1-isopropyl-4-methylcyclohexane (9)
- (1S,2S,4R)-neomenthylamine (7)
4.3. Evaluation of Inhibitory Activity against the Main Viral Protease
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Ghosh, A.K.; Mishevich, J.L.; Mesecar, A.; Mitsuya, H. Recent Drug Development and Medicinal Chemistry Approaches for the Treatment of SARS-CoV-2 Infection and COVID-19. Chem. Med. Chem. 2022, 17, e202200440. [Google Scholar] [CrossRef] [PubMed]
- Ullrich, S.; Nitsche, C. The SARS-CoV-2 main protease as drug target. Bioorg. Med. Chem. Lett. 2020, 30, 127377. [Google Scholar] [CrossRef] [PubMed]
- Jin, Z.; Du, X.; Xu, Y.; Deng, Y.; Liu, M.; Zhao, Y.; Zhang, B.; Li, X.; Zhang, L.; Peng, C.; et al. Structure of Mpro from COVID-19 virus and discovery of its inhibitors. Nature 2020, 582, 289–293. [Google Scholar] [CrossRef] [PubMed]
- Xiao, S.; Tian, Z.; Wang, Y.; Si, L.; Zhang, L.; Zhou, D. Recent progress in the antiviral activity and mechanism study of pentacyclic triterpenoids and their derivatives. Med. Res. Rev. 2018, 38, 951–976. [Google Scholar] [CrossRef] [PubMed]
- Murck, H. Symptomatic Protective Action of Glycyrrhizin (Licorice) in COVID-19 Infection? Front. Immunol. 2020, 11, 1239. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Chan, K.H.; Jiang, Y.; Kao, R.Y.T.; Lu, H.T.; Fan, K.W.; Cheng, V.C.C.; Tsui, W.H.W.; Hung, I.F.N.; Lee, T.S.W.; et al. In vitro susceptibility of 10 clinical isolates of SARS coronavirus to selected antiviral compounds. J. Clin. Virol. 2004, 31, 69–75. [Google Scholar] [CrossRef]
- da Silva, E.F.; Antunes Fernandes, K.H.; Diedrich, D.; Gotardi, J.; Freire Franco, M.S.; Tomich de Paula da Silva, C.H.; Duarte de Souza, A.P.; Baggio Gnoatto, S.C.; Beilstein, J. New triazole-substituted triterpene derivatives exhibiting anti-RSV activity: Synthesis, biological evaluation, and molecular modeling. Org. Chem. 2022, 18, 1524–1531. [Google Scholar] [CrossRef]
- Liu, Y.; Yang, L.; Wang, H.; Xiong, Y. Recent Advances in Antiviral Activities of Triterpenoids. Pharmaceuticals 2022, 15, 1169. [Google Scholar] [CrossRef]
- Ma, C.; Nakamura, N.; Hattori, M.; Kakuda, H.; Qiao, J.; Yu, H. Inhibitory Effects on HIV-1 Protease of Constituents from the Wood of Xanthocerassorbifolia. J. Nat. Prod. 2000, 63, 238–242. [Google Scholar] [CrossRef]
- Ma, C.; Nakamura, N.; Miyashiro, H.; Hattori, M.; Shimotohno, K. Inhibitory Effects of Constituents from Cynomorium songaricum and Related Triterpene Derivatives on HIV-1 Protease. Chem. Pharm. Bull. 1999, 47, 141–145. [Google Scholar] [CrossRef]
- Ma, C.-M.; Wu, X.-H.; Hattori, M.; Wang, X.-J.; Kano, Y. HCV Protease Inhibitory, Cytotoxic and Apoptosis-Inducing Effects of Oleanolic Acid Derivatives. J. Pharm. Pharm. Sci. 2009, 12, 243. [Google Scholar] [CrossRef]
- Flekhter, O.B.; Boreko, E.I.; Nigmatullina, L.R.; Tret’yakova, E.V.; Pavlova, N.I.; Baltina, L.A.; Nikolaeva, S.N.; Savinova, O.V.; Eremin, V.F.; Galin, F.Z.; et al. Synthesis and Antiviral Activity of Betulonic Acid Amides and Conjugates with Amino Acids. Russ. J. Bioorg. Chem. 2004, 30, 80–88. [Google Scholar] [CrossRef] [PubMed]
- Baltina, L.A.; Flekhter, O.B.; Nigmatullina, L.R.; Boreko, E.I.; Pavlova, N.I.; Nikolaeva, S.N.; Savinova, O.V.; Tolstikov, G.A. Lupane triterpenes and derivatives with antiviral activity. Bioorg. Med. Chem. Lett. 2003, 13, 3549–3552. [Google Scholar] [CrossRef] [PubMed]
- Fomenko, V.; Blokhin, M.; Kuranov, S.; Khvostov, M.; Baev, D.; Borisova, M.S.; Luzina, O.; Tolstikova, T.G.; Salakhutdinov, N.F. Triterpenic Acid Amides as a Promising Agent for Treatment of Metabolic Syndrome. Sci. Pharm. 2021, 89, 4. [Google Scholar] [CrossRef]
- Vuong, W.; Khan, M.B.; Fischer, C.; Arutyunova, E.; Lamer, T.; Shields, J.; Saffran, H.A.; McKay, R.T.; van Belkum, M.J.; Joyce, M.A.; et al. Feline coronavirus drug inhibits the main protease of SARS-CoV-2 and blocks virus replication. Nat. Commun. 2020, 11, 4282. [Google Scholar] [CrossRef] [PubMed]
- Simón, L.; Goodman, J.M. Enzyme catalysis by hydrogen bonds: The balance between transition State binding and substrate binding in oxyanion holes. J. Org. Chem. 2010, 75, 1831–1840. [Google Scholar] [CrossRef]
- Zhang, L.; Lin, D.; Sun, X.; Curth, U.; Drosten, C.; Sauerhering, L.; Becker, S.; Rox, K.; Hilgenfeld, R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science 2020, 368, 409–412. [Google Scholar] [CrossRef]
- Shcherbakov, D.; Baev, D.; Kalinin, M.; Dalinger, A.; Chirkova, V.; Belenkaya, S.; Khvostov, A.; Krut’ko, D.; Medved’ko, A.; Volosnikova, E.; et al. Design and Evaluation of Bispidine-Based SARS-CoV-2 Main Protease Inhibitors. ACS Med. Chem. Lett. 2022, 13, 140–147. [Google Scholar] [CrossRef]
- Filimonov, A.; Yarovaya, O.; Zaykovskaya, A.; Rudometova, N.; Shcherbakov, D.; Chirkova, V.; Baev, D.; Borisevich, S.; Luzina, O.; Pyankov, O.; et al. (+)-Usnic Acid and Its Derivatives as Inhibitors of a Wide Spectrum of SARS-CoV-2 Viruses. Viruses 2022, 14, 2154. [Google Scholar] [CrossRef]
- Alhadrami, H.A.; Sayed, A.M.; Sharif, A.M.; Azhar, E.I.; Rateb, M.E. Olive-Derived Triterpenes Suppress SARS CoV-2 Main Protease: A Promising Scaffold for Future Therapeutics. Molecules 2021, 26, 2654. [Google Scholar] [CrossRef]
- Yi, Y.; Li, J.; Lai, X.; Zhang, M.; Kuang, Y.; Bao, Y.; Yu, R.; Hong, W.; Muturi, E.; Xue, H.; et al. Natural triterpenoids from licorice potently inhibit SARS-CoV-2 infection. J. Adv. Res. 2022, 36, 201–210. [Google Scholar] [CrossRef] [PubMed]
- Schrodinger LLC. Small Molecule Drug Discovery Suite; Schrödinger LLC: New York, NY, USA, 2020. [Google Scholar]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
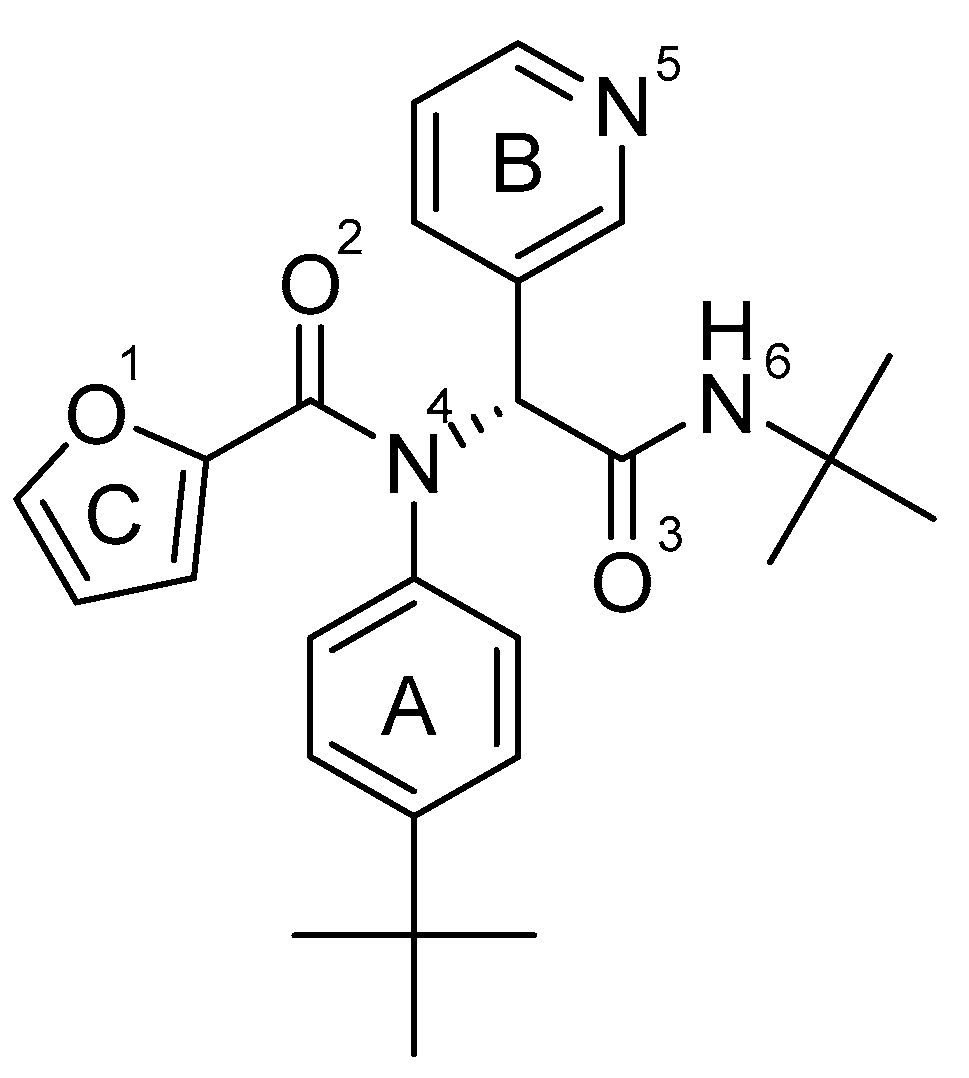
- Lockbaum, G.J.; Reyes, A.C.; Lee, J.M.; Tilvawala, R.; Nalivaika, E.A.; Ali, A.; Kurt Yilmaz, N.; Thompson, P.R.; Schiffer, C.A. Crystal Structure of SARS-CoV-2 Main Protease in Complex with the Non-Covalent Inhibitor ML188. Viruses 2021, 13, 174. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Wu, C.; Ghoreishi, D.; Chen, W.; Wang, L.; Damm, W.; Ross, G.A.; Dahlgren, M.K.; Russell, E.; Von Bargen, C.D. OPLS4: Improving force field accuracy on challenging regimes of chemical space. J. Chem. Theory Comput. 2021, 17, 4291–4300. [Google Scholar] [CrossRef] [PubMed]
- Sherman, W.; Day, T.; Jacobson, M.P.; Friesner, R.A.; Farid, R. Novel procedure for modeling ligand/receptor induced fit effects. J. Med. Chem. 2006, 49, 534–553. [Google Scholar] [CrossRef]
- Friesner, R.A.; Murphy, R.B.; Repasky, M.P.; Frye, L.L.; Greenwood, J.R.; Halgren, T.A.; Sanschagrin, P.C.; Mainz, D.T. Extra Precision Glide: Docking and Scoring Incorporating a Model of Hydrophobic Enclosure for Protein−Ligand Complexes. J. Med. Chem. 2006, 49, 6177–6196. [Google Scholar] [CrossRef]
- Jacobson, M.P.; Pincus, D.L.; Rapp, C.S.; Day, T.J.F.; Honig, B.; Shaw, D.E.; Friesner, R.A. A Hierarchical Approach to All-Atom Protein Loop Prediction. Proteins Struct. Funct. Genet. 2004, 55, 351–367. [Google Scholar] [CrossRef]
- Wen, X.; Sun, H.; Liu, J.; Cheng, K.; Zhang, P.; Zhang, L.; Hao, J.; Zhang, L.; Ni, P.; Zographos, S.; et al. Naturally Occurring Pentacyclic Triterpenes as Inhibitors of Glycogen Phosphorylase: Synthesis, Structure Activity Relationships, and X-ray Crystallographic Studies. J. Med. Chem. 2008, 51, 3540–3554. [Google Scholar] [CrossRef]
- Cai, Z.; Liu, Q.; Li, P.; Guo, Z.; Shen, Z. Synthesis and anti-diabetic activity of (RS)-2-ethoxy-3-{4-[2-(4-trifluoro-8-methanesulfonyloxy-phenyl)-ethoxy]-phenyl}-propionic acid. Acta Pharmacol. Sin. 2006, 27, 597–602. [Google Scholar] [CrossRef][Green Version]
- Wappes, E.A.; Fosu, S.C.; Chopko, T.C.; Nagib, D.A. Triiodide-Mediated δ-Amination of Secondary C−H Bonds. Angew. Chem. Int. Ed. 2016, 55, 9974–9978. [Google Scholar] [CrossRef]
- Waldvogel, S.; Welschoff, N. Practical Synthesis of Optically Pure Menthylamines Starting from Racemic Neomenthol. Synthesis 2010, 2010, 3596–3601. [Google Scholar] [CrossRef]
- Barton, D.H.R.; Morgan, L.R. Photochemical transformations. Part XII. Photolysis Azides. J. Chem. Soc. 1962, 117, 622. [Google Scholar] [CrossRef]
- Ma, C.; Hu, Y.; Townsend, J.A.; Lagarias, P.I.; Marty, M.T.; Kolocouris, A.; Wang, J. Ebselen, Disulfiram, Carmofur, PX-12, Tideglusib, and Shikonin Are Nonspecific Promiscuous SARS-CoV-2 Main Protease Inhibitors. ACS Pharmacol. Transl. Sci. 2020, 3, 1265–1277. [Google Scholar] [CrossRef] [PubMed]
| Ligand | Docking Parameters, kcal/mol | |||
|---|---|---|---|---|
| Docking Score | LE | Emodel | IFD Score | |
| 1e | −11.784 | −0.196 | −101.329 | −670.17 |
| 1c | −10.532 | −0.167 | −118.702 | −667.78 |
| 1a | −10.479 | −0.178 | −104.657 | −665.62 |
| 1d | −9.843 | −0.167 | −93.581 | −666.84 |
| 1b | −8.672 | −0.147 | −102.7 | −666.49 |
| ML188 | −8.379 | −0.262 | −83.299 | −666.082 |
| 1f | −7.39 | −0.2 | −86.486 | −663.78 |
| Ligand | Interactions | Distance (Å) | Bonding | Bonding Type |
|---|---|---|---|---|
| 1e | Lys137:NZ — 1e:O6’’ | 4.97 | Ionic Bond | Salt Bridge |
| Lys137:HZ2 — 1e:O5’’ | 1.84 | Hydrogen Bond | Conventional | |
| Gln189:HE22 — 1e:O28’ | 2.15 | Hydrogen Bond | Conventional | |
| 1e:H2’ — Thr24:O | 1.86 | Hydrogen Bond | Conventional | |
| 1c | His41:HD1 — 1c:O3’’ | 1.89 | Hydrogen Bond | Conventional |
| Asn142:HD22 — 1c:O5’’ | 1.81 | Hydrogen Bond | Conventional | |
| Gly143:HN — 1c:O6’’ | 1.91 | Hydrogen Bond | Conventional | |
| Gln189:HE21 — 1c:O11’ | 2.03 | Hydrogen Bond | Conventional | |
| Gln192:HE22 — 1c:O28’ | 2.14 | Hydrogen Bond | Conventional | |
| 1c:H1’’ — Gln189:OE1 | 2.16 | Hydrogen Bond | Conventional | |
| His41:Imidazol ring — 1c:Ring A | 4.29 | Hydrophobic | Pi-Pi Stacked | |
| 1a | Asn142:HD22 — 1a:O5’’ | 1.67 | Hydrogen Bond | Conventional |
| Gly143:HN — 1a:O6’’ | 1.67 | Hydrogen Bond | Conventional | |
| Gln189:HE22 — 1a:O28’ | 1.73 | Hydrogen Bond | Conventional | |
| 1a:H1’’ — Glu166:O | 1.99 | Hydrogen Bond | Conventional | |
| His41:Imidazol ring — 1c:Ring A | 5.49 | Hydrophobic | Pi-Pi T-shaped | |
| 1d | Lys137:NZ — 1d:O6’’ | 2.82 | Ionic Bond | Salt Bridge |
| Lys137:NZ — 1d:Ring B | 5.26 | Ionic Bond | Pi-Cation | |
| Thr26:HN — 1d:O3’ | 2.12 | Hydrogen Bond | Conventional | |
| Glu166:HN — 1d:O28’ | 2.07 | Hydrogen Bond | Conventional | |
| 1d:H3’ — Thr24:O | 2.69 | Hydrogen Bond | Conventional | |
| 1b | 1b:H1’’ — Asn142:OD1 | 2.00 | Hydrogen Bond | Conventional |
| Lys61:NZ — 1b:O6’’ | 4.44 | Ionic Bond | Salt Bridge | |
| ML188 | Asn142:HD22 — ML188:O2 | 1.71 | Hydrogen Bond | Conventional |
| His163:HE2 — ML188:N5 | 2.08 | Hydrogen Bond | Conventional | |
| Glu166:HN — ML188:O3 | 2.03 | Hydrogen Bond | Conventional | |
| His41:Imidazol ring — ML188:Ring C | 5.18 | Hydrophobic | Pi-Pi T-shaped | |
| His41:Imidazol ring — ML188:Ring A | 5.27 | Hydrophobic | Pi-Pi T-shaped | |
| 1f | His41:HD1 — 1f:O3’’ | 2.04 | Hydrogen Bond | Conventional |
| Asn142:HD22 — 1f:O6’’ | 1.67 | Hydrogen Bond | Conventional | |
| Gly143:HN — 1f:O5’’ | 1.84 | Hydrogen Bond | Conventional | |
| 1f:H1’’ — Gln189:OE1 | 2.42 | Hydrogen Bond | Conventional | |
| 1f:H1’’* — Glu166:O | 2.35 | Hydrogen Bond | Conventional |
| Ligand | IC50, μM |
|---|---|
| 1e | 25 ± 3 |
| 1c | 125 ± 13 |
| 1a | 100 ± 5 |
| 1d | 100 ± 5 |
| 1b | 100 ± 5 |
| ML188 | 1.6 ± 0.6 |
| 1f | <400 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baev, D.S.; Blokhin, M.E.; Chirkova, V.Y.; Belenkaya, S.V.; Luzina, O.A.; Yarovaya, O.I.; Salakhutdinov, N.F.; Shcherbakov, D.N. Triterpenic Acid Amides as Potential Inhibitors of the SARS-CoV-2 Main Protease. Molecules 2023, 28, 303. https://doi.org/10.3390/molecules28010303
Baev DS, Blokhin ME, Chirkova VY, Belenkaya SV, Luzina OA, Yarovaya OI, Salakhutdinov NF, Shcherbakov DN. Triterpenic Acid Amides as Potential Inhibitors of the SARS-CoV-2 Main Protease. Molecules. 2023; 28(1):303. https://doi.org/10.3390/molecules28010303
Chicago/Turabian StyleBaev, Dmitry S., Mikhail E. Blokhin, Varvara Yu. Chirkova, Svetlana V. Belenkaya, Olga A. Luzina, Olga I. Yarovaya, Nariman F. Salakhutdinov, and Dmitry N. Shcherbakov. 2023. "Triterpenic Acid Amides as Potential Inhibitors of the SARS-CoV-2 Main Protease" Molecules 28, no. 1: 303. https://doi.org/10.3390/molecules28010303
APA StyleBaev, D. S., Blokhin, M. E., Chirkova, V. Y., Belenkaya, S. V., Luzina, O. A., Yarovaya, O. I., Salakhutdinov, N. F., & Shcherbakov, D. N. (2023). Triterpenic Acid Amides as Potential Inhibitors of the SARS-CoV-2 Main Protease. Molecules, 28(1), 303. https://doi.org/10.3390/molecules28010303







