Pyridinium Salts of Dehydrated Lanthanide Polychlorides
Abstract
1. Introduction
2. Results and Discussion
2.1. Synthesis
2.2. Characterization
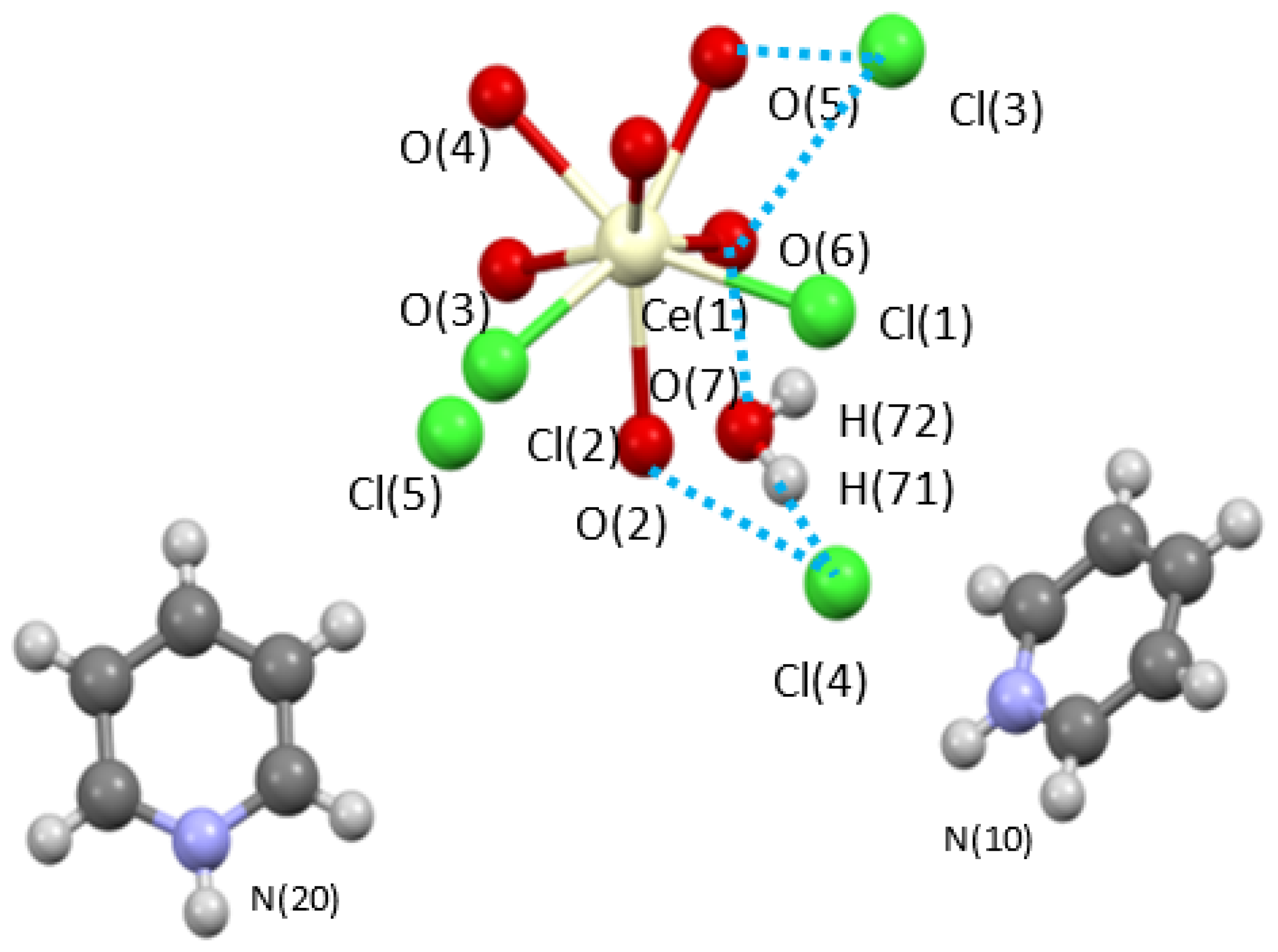
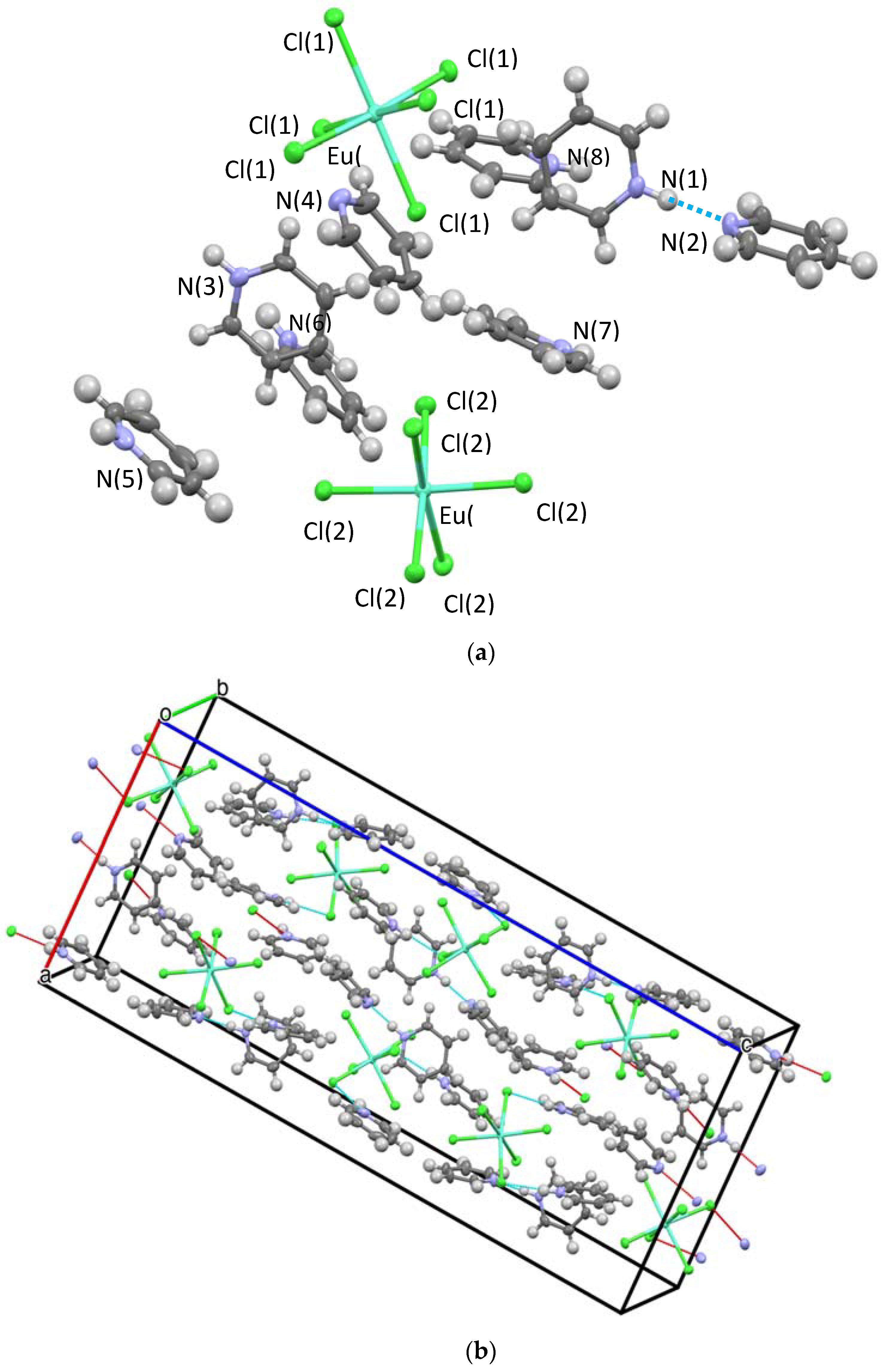
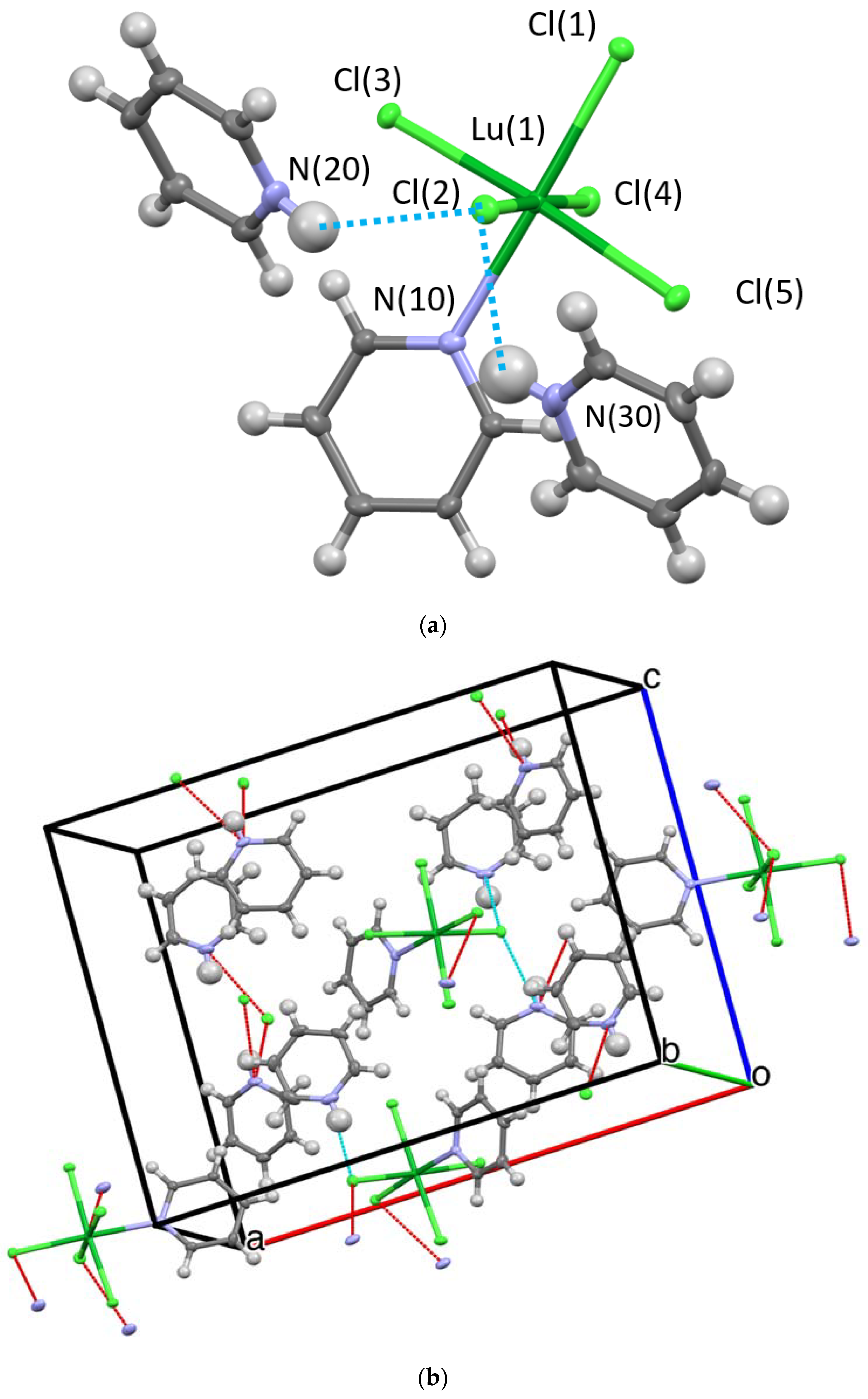
2.2.1. Crystal Structures
Hydrogen Bonding
Bonding Energy
Lattice Energy
Crystal Stress Analysis
Stress Summary
3. Materials and Methods
3.1. Ln-6 General Synthesis
3.2. Ln-5 General Syntheses
4. Analytical Analyses
4.1. Infrared Spectroscopy
4.2. X-ray Crystal Structure Information
5. Summary and Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Boyle, T.J.; Guerrero, F.; Cramer, R.E.; Boye, D.A.; Brooks, H.; Reuel, P.C. Synthesis and Characterization of Solvated Lanthanide Tris(trimethylsilyl)siloxides. Inorg. Chem. 2022, 61, 5048–5059. [Google Scholar] [CrossRef] [PubMed]
- Lanthanides Isolation and Production; Web Solutions, LLC.: Waukesha, WI, USA, 2022; Available online: https://science.jrank.org/pages/3825/Lanthanides-Isolation-production.html (accessed on 20 September 2022).
- Boyle, T.J.; Ottley, L.A.M. Lanthanide halides. In The Rare Earth Elements: Fundamentals and Applications; Atwood, D., Ed.; Wiley & Sons Ltd.: Hoboken, NJ, USA, 2012. [Google Scholar]
- Gruber, V.; Carsky, M. New technology for lanthanide recovery from spent Nd-Fe-B magnets. S. Afr. J. Chem. Eng. 2020, 33, 35–38. [Google Scholar] [CrossRef]
- Mitchem, S. Scientists Gain Insight into Recycling Processes for Nuclear and Electronic Waste. Argonne National Laboratory, 2012. Available online: https://www.anl.gov/article/scientists-gain-insight-into-recycling-processes-for-nuclear-and-electronic-waste (accessed on 20 September 2022).
- Taylor, M.D. Preparation of Anhydrous Lanthanon Halides. Chem. Rev. 1962, 62, 503–511. [Google Scholar] [CrossRef]
- Boyle, T.J.; Cramer, R.E.; Fasulo, F.; Padilla, N. Solvation coordination compounds of scandium chloride from the dehydration of scandium chloride hexahydrate. Polyhedron 2021, 208, 115437. [Google Scholar] [CrossRef]
- Li, J.-S.; Neumuller, B.; Dehnicke, K. Pyridinium-Chlorometallate von Lanthanoid-Elementen. Die Kristallstrukturen von [HPy]2[LnCl5(Py)] mit Ln = Eu, Er, Yb und von [H(Py)2][YbCl4(Py)2] · Py. Z. Anorg. Allg. Chem. 2002, 628, 2785–2789. [Google Scholar] [CrossRef]
- Cook, D. Vibrational Spectra of pyridinium salts. Can. J. Chem. 1961, 39, 2009–2024. [Google Scholar] [CrossRef]
- Groom, C.R.; Bruno, I.J.; Lightfoot, M.P.; Ward, S.C. The Cambridge Structural Database. Acta Crystallogr. Sec. E 2016, B72, 171–179. [Google Scholar] [CrossRef]
- Cramer, R.E.; Rimsza, J.M.; Boyle, T.J. Lanthanide Contraction is a variable. Inorg. Chem. 2021, 61, 6120–6127. [Google Scholar] [CrossRef]
- Szalewicz, K. Hydrogen Bond Encyclopedia of Physical Science and Technology, 3rd ed.; Academic Press: Cambridge, MA, USA, 2003; pp. 505–538. [Google Scholar]
- Löble, M.W.; Keith, J.M.; Altman, A.B.; Stieber, S.C.E.; Batista, E.R.; Boland, K.S.; Conradson, S.D.; Clark, D.L.; Pacheco, J.L.; Kozimor, S.A.; et al. Covalency in Lanthanides. An X-ray Absorption Spectroscopy and Density Functional Theory Study of LnCl6x– (x = 3, 2). J. Am. Chem. Soc. 2015, 137, 2506–2523. [Google Scholar] [CrossRef]
- Becker, A.; Urland, W. Crystal structures and magnetic behaviour of new complex lanthanide chlorides with organic cations. J. Alloys Compd. 1998, 24, 62–66. [Google Scholar] [CrossRef]
- Petricek, S. Syntheses and Crystal Structures of Anionic Lanthanide Chloride Complexes [(CH3)2NH2][LnCl4(HMPA)2] (Ln = La, Nd) and [(CH3)2NH2]4[LnCl6]Cl (Ln = Nd, Sm, Eu). Acta Chim. Slov. 2005, 52, 398–403. [Google Scholar]
- Czjek, M.; Fuess, H. Crystal structure of tetra(monomethylammonium) hexachloroytterbatochloride (CH3NH3)4YbCl7. Z. Krist. Cryst. Mater. 1987, 179, 49. [Google Scholar]
- Czjek, M.; Fuess, H.; Pabst, I. Crystal structure and magnetic properties of tetra(monomethylammonium) hexachloroytterbatochloride ( CH3NH3)4YbCl7. Z. Anorg. Allg. Chem. 1992, 617, 105–109. [Google Scholar] [CrossRef]
- Diamantopoulou, E.; Papefstathiou, G.S.; Terzis, A.; PRaptopoulou, C.P.; Desseyn, H.O.; Perlepes, S.P. Hydrogen bonded networks based on lanthanide(III) complexes of N,N′-dimethylurea (DMU): Preparation, characterisation, and crystal structures of [Nd(DMU)(6)][NdCl6] and [Nd(NO3)(3)(DMU)(3)]. Polyhedron 2003, 22, 825. [Google Scholar] [CrossRef]
- Runge, P.; Shulze, M.; Urland, W. Darstellung und Kristallstruktur von (CH3NH3)8[NdCl6][NdCl4(H2O)2]2Cl3. Z. Anorg. Allg. Chem. 1991, 592, 115–120. [Google Scholar] [CrossRef]
- Hallfeldt, J.; Urland, W. Synthese, Kristallstruktur und magnetisches Verhalten von (2,4,6-Trimethylpyridinium)–[ErCl6][ErCl5(H2O)]2Cl3. Z. Anorg. Allg. Chem. 2002, 628, 2661–2664. [Google Scholar] [CrossRef]
- Rogers, R.D.; Rollins, A.N.; Henry, R.F.; Murdoch, J.S.; Etzenhouser, R.D.; Huggins, S.E.; Nunez, L. Direct comparison of the preparation and structural features of crown ether and polyethylene glycol complexes of neodymium trichloride hexahydrate. Inorg. Chem. 1991, 30, 4945–4954. [Google Scholar] [CrossRef]
- Izgorodina, E.I.; Bernard, U.L.; Dean, P.M.; Pringle, J.M.; MacFarlane, D.R. The Madelung Constant of Organic Salts. Cryst. Growth Des. 2009, 11, 4834–4839. [Google Scholar] [CrossRef]
- The SHELEX Suite of Package of Programs—Saint, SADABS, Apex3; Bruker AXS: Madison, WI, USA, 2012.
- Sheldrick, G.M. A short history of SHELX. Acta Crystallogr. Sect. A 2008, 64, 112–122. [Google Scholar] [CrossRef]
- Sheldrick, G.M. Crystal structure refinement with SHELXL. Acta Crystallogr. Sect. C 2015, C17, 3–8. [Google Scholar]
- Dolomanov, O.V.; Bourhis, L.J.; Gildea, R.J.; Howard, J.A.K.; Pushmann, H. OLEX2: A complete structure solution, refinement and analysis program. J. Appl. Cryst. 2009, 42, 339–341. [Google Scholar] [CrossRef]
- Downward, L.; Booth, C.H.; Lukens, W.W.; Bridges, F. A Variation of the F-Test for Determining Statistical Relevance of Particular Parameters in EXAFS Fits. AIP Conf. Proc. 2007, 882, 129–131. [Google Scholar]
| Compound | La-6 | Ce-6 | Pr-6 |
|---|---|---|---|
| Chem. Form | C20H23Cl6N4La | C20H23CeCl6N4 | C20H23Cl6N4Pr |
| Form. weight | 335.02 | 672.24 | 673.03 |
| temp (K) | 100 (2) | 100 (2) | 100 (2) |
| space group | Orthorhombic P 21 21 21 | Orthorhombic P 21 21 21 | Monoclinic P21/c |
| a (Å) | 9.7104(6) | 9.6382(3) | 15.6181(11) |
| b (Å) | 15.6614(10 | 15.6027(4) | 9.5868(8) |
| c (Å) | 18.1015(12) | 18.0596(6) | 36.244(3) |
| b (deg) | 89.999(3) | ||
| V (Å3) | 2752.8(3) | 2715.84(14) | 5426.8(7) |
| Z | 4 | 4 | 8 |
| Dcalcd (Mg/m3) | 1.619 | 1.644 | 1.648 |
| m (Mo, Ka) (mm−1) | 2.149 | 2.282 | 2.402 |
| Flack Parameter | 0.500(6) | 0.50(1) | NA c |
| R1 a (%) (all data) | 2.25 (2.65) | 2.22 (2.22) | 5.55 (10.16) |
| wR2 b (%) (all data) | 4.86 (5.18) | 6.14 (6.15) | 10.15 (12.71) |
| Compound | Nd-6 | Sm-6 | Eu-6 |
| Chem. Form | C20H23Cl6N4Nd | C20H23Cl6N4Sm | C20H23Cl6EuN4 |
| Form. weight | 676.36 | 682.47 | 684.08 |
| temp (K) | 100 (2) | 100 (2) | 100 (2) |
| space group | Monoclinic P21/c | Monoclinic P21/c | Monoclinic P21/c |
| a (Å) | 15.561(2) | 15.5230(14) | 15.534(3) |
| b (Å) | 9.5667(15) | 9.5386(9) | 9.5290(19) |
| c (Å) | 36.204(5) | 36.265(4) | 36.287(7) |
| b (deg) | 90.306(5) | 90.244(4) | 90.091(7) |
| V (Å3) | 5389.3(14) | 5369.6(9) | 5371.5(18) |
| Z | 8 | 8 | 8 |
| Dcalcd(Mg/m3) | 1.667 | 1.688 | 1.692 |
| m (Mo, Ka) (mm−1) | 2.537 | 2.800 | 2.948 |
| Flack Parameter | NA | NA | NA |
| R1 a (%) (all data) | 8.14 (13.45) | 4.45 (6.04) | 4.81 (9.68) |
| wR2 b (%) (all data) | 16.45 (19.59) | 9.48 (10.78) | 11.22 (15.11) |
| Compound | Gd-6 | Tb-5 | Dy-5 |
| Chem. Form | C20H23Cl6GdN4 | C15H17Cl5N3Tb | C15H17Cl5DyN3 |
| Form. weight | 689.37 | 572.48 | 579.06 |
| temp (K) | 100 (2) | 100 (2) | 100(2) |
| space group | Monoclinic P21/c | Orthorhombic Pna21 | Orthorhombic Pna21 |
| a (Å) | 15.5351(13) | 15.561(2) | 18.693(3) |
| b (Å) | 9.5079(8) | 9.5667(15) | 7.3078(10) |
| c (Å) | 36.279(3) | 36.204(5) | 14.789(2) |
| b (deg) | 90.055(3) | ||
| V (Å3) | 5358.6(8) | 2028.3(3) | 2020.3(5) |
| Z | 8 | 4 | 4 |
| Dcalcd(Mg/m3) | 1.709 | 1.885 | 1.904 |
| m (Mo, Ka) (mm−1) | 3.089 | 4.148 | 4.362 |
| Flack Parameter | NA | 0.500(9) | 0.459(6) |
| R1 a (%) (all data) | 6.38 (7.36) | 1.78 (1.98) | 1.37 (1.40) |
| wR2 b (%) (all data) | 13.85 (14.77) | 3.65 (3.71) | 3.25 (3.30) |
| Compound | Ho-5 | Er-5 | Tm-5 |
| Chem. Form | C15H17Cl5HoN3 | C15H17Cl5ErN3 | C15H17Cl5N3Tm |
| Form. weight | 581.49 | 583.82 | 585.49 |
| temp (K) | 100 (2) | 100 (2) | 100 (2) |
| space group | Orthorhombic Pna21 | Orthorhombic Pna21 | Orthorhombic Pna21 |
| a (Å) | 18.6738(17) | 18.6315(7) | 18.6333(11) |
| b (Å) | 7.2976(6) | 7.2898(3) | 7.2815(4) |
| c (Å) | 14.7913(13) | 14.7705(5) | 14.7648(9) |
| V (Å3) | 2015.7(3) | 2006.13(13) | 2003.3(2) |
| Z | 4 | 4 | 4 |
| Dcalcd(Mg/m3) | 1.916 | 1.933 | 1.941 |
| m (Mo, Ka) (mm−1) | 4.590 | 4.852 | 5.098 |
| Flack Parameter | 0.465(5) | 0.486(5) | 0.350(8) |
| R1 a (%) (all data) | 1.65 (1.71) | 1.05 (1.06) | 2.27 (2.76) |
| wR2 b (%) (all data) | 3.78 (3.79) | 2.67 (2.68) | 4.92 (5.28) |
| Compound | Yb-5 | Lu-5 | Ce-H2O/py-H |
| Chem. Form | C15H17Cl5N3Yb | C15H17Cl5N3Lu | C10H26CeCl5N2O7 |
| Form. weight | 589.60 | 591.54 | 603.70 |
| temp (K) | 100 (2) | 100 (2) | 100(2) |
| space group | Orthorhombic Pna21 | Orthorhombic Pna21 | Monoclinic P21/n |
| a (Å) | 18.6135(11) | 18.8935(3) | 8.4460(8) |
| b (Å) | 7.2804(4) | 7.27030(10) | 17.5550(18) |
| c (Å) | 147520(9)) | 14.7377(3) | 15.5427(18) |
| b(deg) | 91.534(3) | ||
| V (Å3) | 1999.1(2) | 1992.25(6) | 2303.7(4) |
| Z | 4 | 4 | 4 |
| Dcalcd(Mg/m3) | 1.959 | 1.972 | 1.741 |
| m (Mo, Ka) (mm−1) | 5.349 | 5.628 | 2.585 |
| Flack Parameter | 0.457(7) | 0.488(6) | |
| R1 a (%) (all data) | 2.16 (2.45) | 1.90 (2.73) | 5.50 (5.73) |
| wR2 b (%) (all data) | 4.63 (4.81) | 3.41 (3.56) | 18.67 (18.84) |
| Distance (Å) | La | Ce | Pr | Nd | Sm | Eu | Gd |
|---|---|---|---|---|---|---|---|
| Ln-Cl | 2.796 | 2.758 | 2.737 | 2.724 | 2.702 | 2.689 | 2.676 |
| Cl---Cl | 3.954 | 3.900 | 3.871 | 3.852 | 3.821 | 3.803 | 3.784 |
| (py)N-H-N(py) | 2.678 | 2.678 | 2.672 | 2.674 | 2.664 | 2.678 | 2.676 |
| (py)N-H---Cl | 3.165 | 3.171 | 3.210 | 3.179 | 3.164 | 3.171 | 3.181 |
| Angles (deg) | La | Ce | Pr | Nd | Sm | Eu | Gd |
| trans Cl-Ln-Cl | 173.11 | 172.18 | 174.43 | 175.00 | 174.48 | 173.45 | 174.43 |
| (py)N-H-N(py) | 171.80 | 170.96 | 174.11 | 174.20 | 173.58 | 173.97 | 171.91 |
| (py)N-H---Cl | 169.63 | 168.77 | 164.87 | 165.67 | 165.67 | 164.88 | 164.47 |
| Distance (Å) | Tb | Dy | Ho | Er | Tm | Yb | Lu |
|---|---|---|---|---|---|---|---|
| Ln-Cl | 2.643 | 2.638 | 2.633 | 2.602 | 2.597 | 2.591 | 2.576 |
| Cl---Cl | 3.738 | 3.73 | 3.726 | 3.680 | 3.673 | 3.664 | 3.643 |
| Ln-N(py) | 2.524 | 2.517 | 2.507 | 2.507 | 2.475 | 2.475 | 2.454 |
| (py)N-H---Cl | 3.230 | 3.232 | 3.229 | 3.276 | 3.235 | 3.274 | 3.265 |
| Angles (deg) | Tb | Dy | Ho | Er | Tm | Yb | Lu |
| trans Cl-Ln-Cl | 176.105 | 176.37 | 176.37 | 176.615 | 176.71 | 176.805 | 176.775 |
| tans Cl-Ln-N(py) | 177.19 | 177.28 | 177.29 | 177.11 | 177.47 | 178.06 | 177.36 |
| (py)N-H---Cl | 177.19 | 177.28 | 177.29 | 177.11 | 177.47 | 178.06 | 177.36 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cramer, R.E.; Baca, E.M.; Boyle, T.J. Pyridinium Salts of Dehydrated Lanthanide Polychlorides. Molecules 2023, 28, 283. https://doi.org/10.3390/molecules28010283
Cramer RE, Baca EM, Boyle TJ. Pyridinium Salts of Dehydrated Lanthanide Polychlorides. Molecules. 2023; 28(1):283. https://doi.org/10.3390/molecules28010283
Chicago/Turabian StyleCramer, Roger E., Esteban M. Baca, and Timothy J. Boyle. 2023. "Pyridinium Salts of Dehydrated Lanthanide Polychlorides" Molecules 28, no. 1: 283. https://doi.org/10.3390/molecules28010283
APA StyleCramer, R. E., Baca, E. M., & Boyle, T. J. (2023). Pyridinium Salts of Dehydrated Lanthanide Polychlorides. Molecules, 28(1), 283. https://doi.org/10.3390/molecules28010283






