Characterization of Peanut Protein Hydrolysate and Structural Identification of Umami-Enhancing Peptides
Abstract
:1. Introduction
2. Results and Discussion
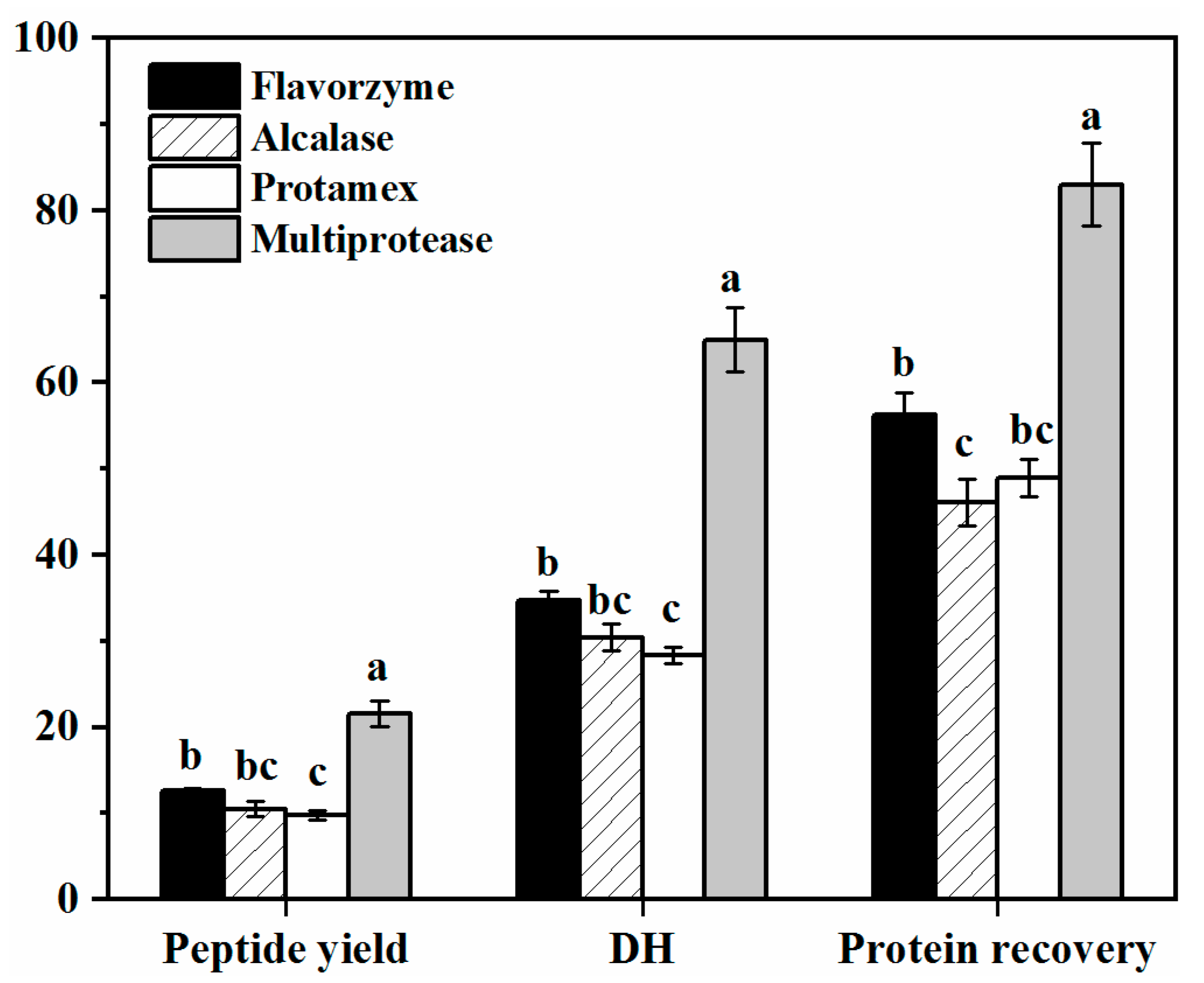
2.1. Enzymatic Hydrolysis Characteristics of DPF
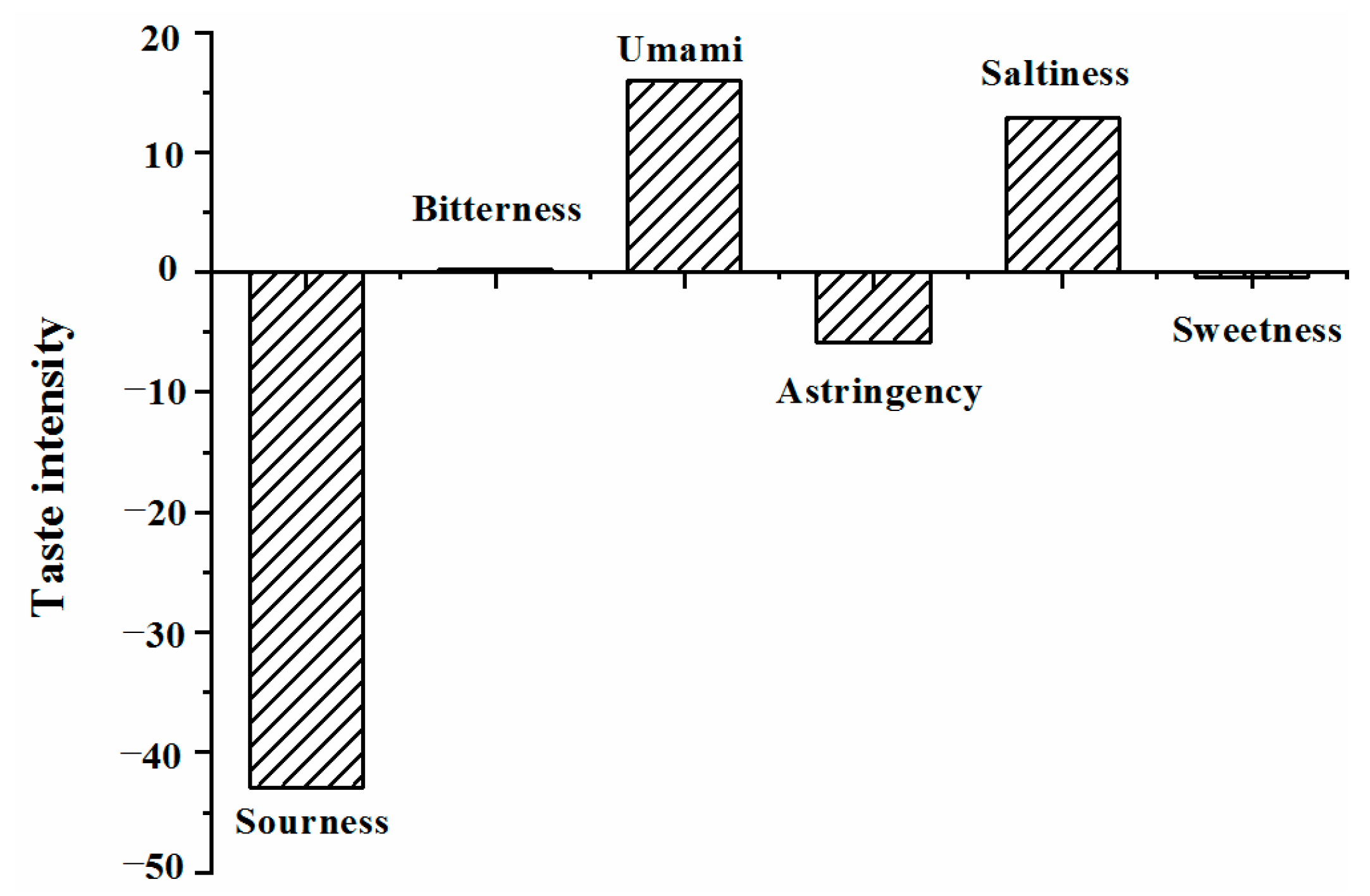
2.2. Taste Characteristics of DPH
2.3. Amino Acids Analysis
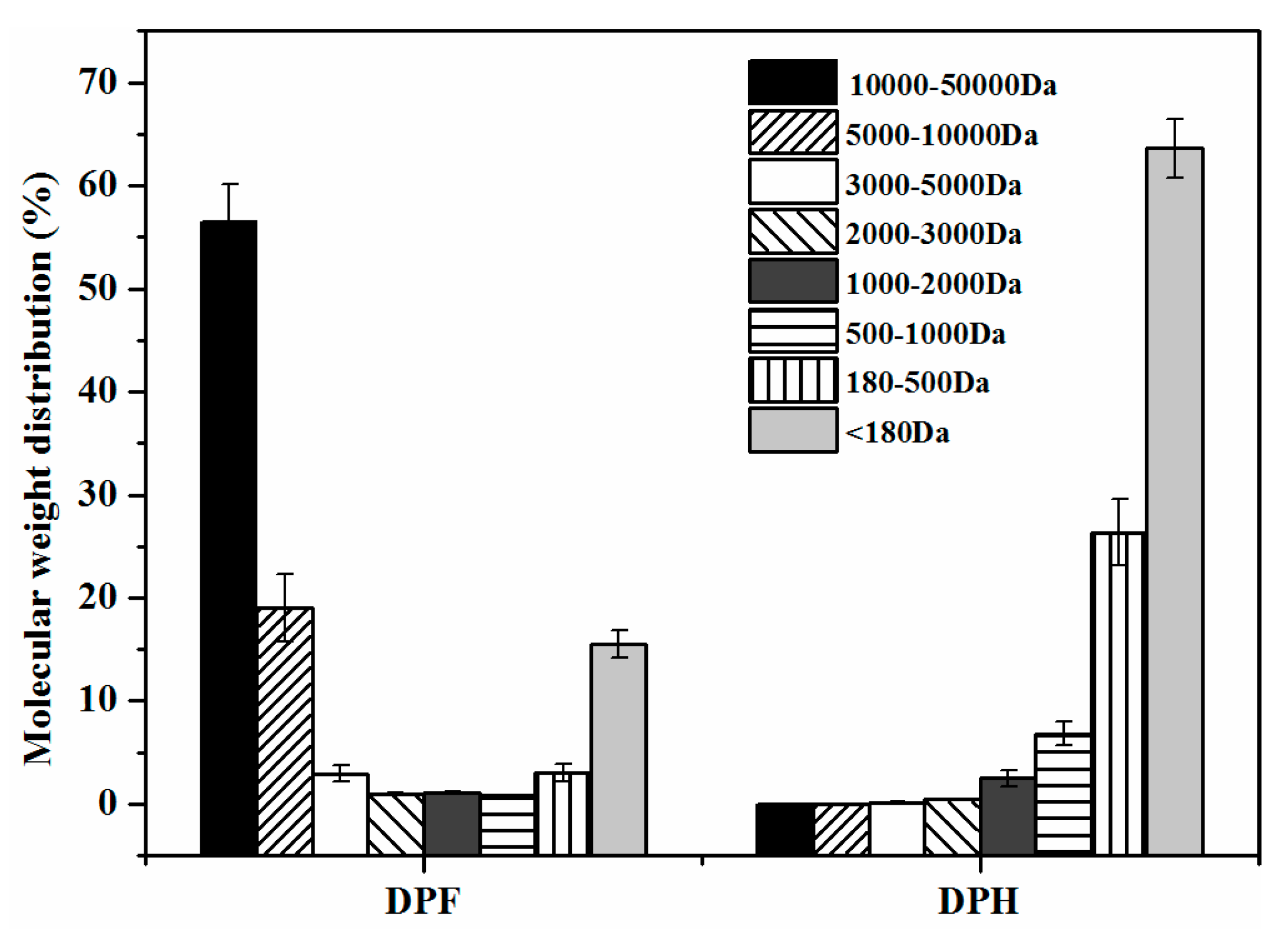
2.4. Molecular Weight Distribution of Peptides
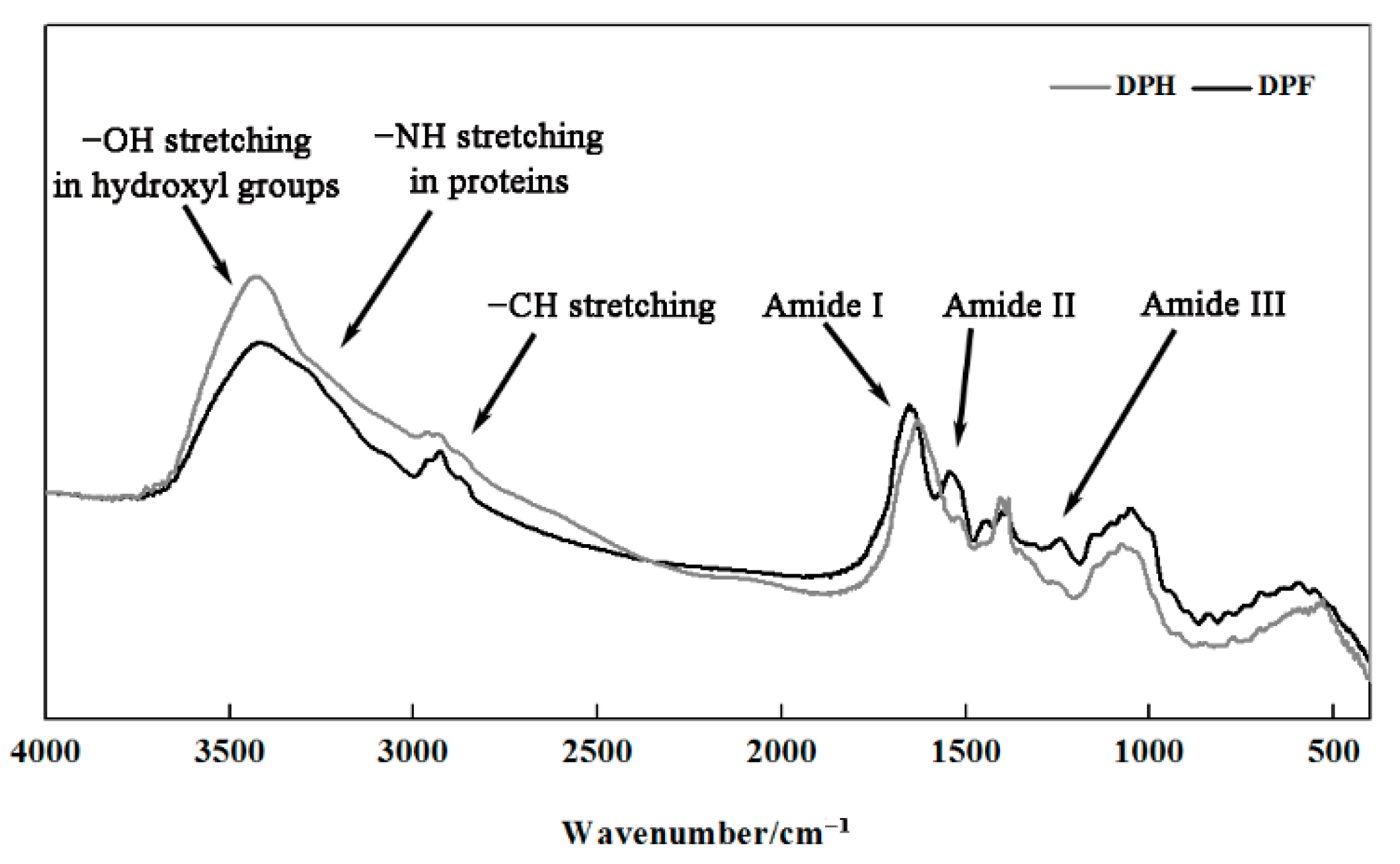
2.5. FT-IR Spectroscopy
2.6. Ultrafiltration (UF) of DPH
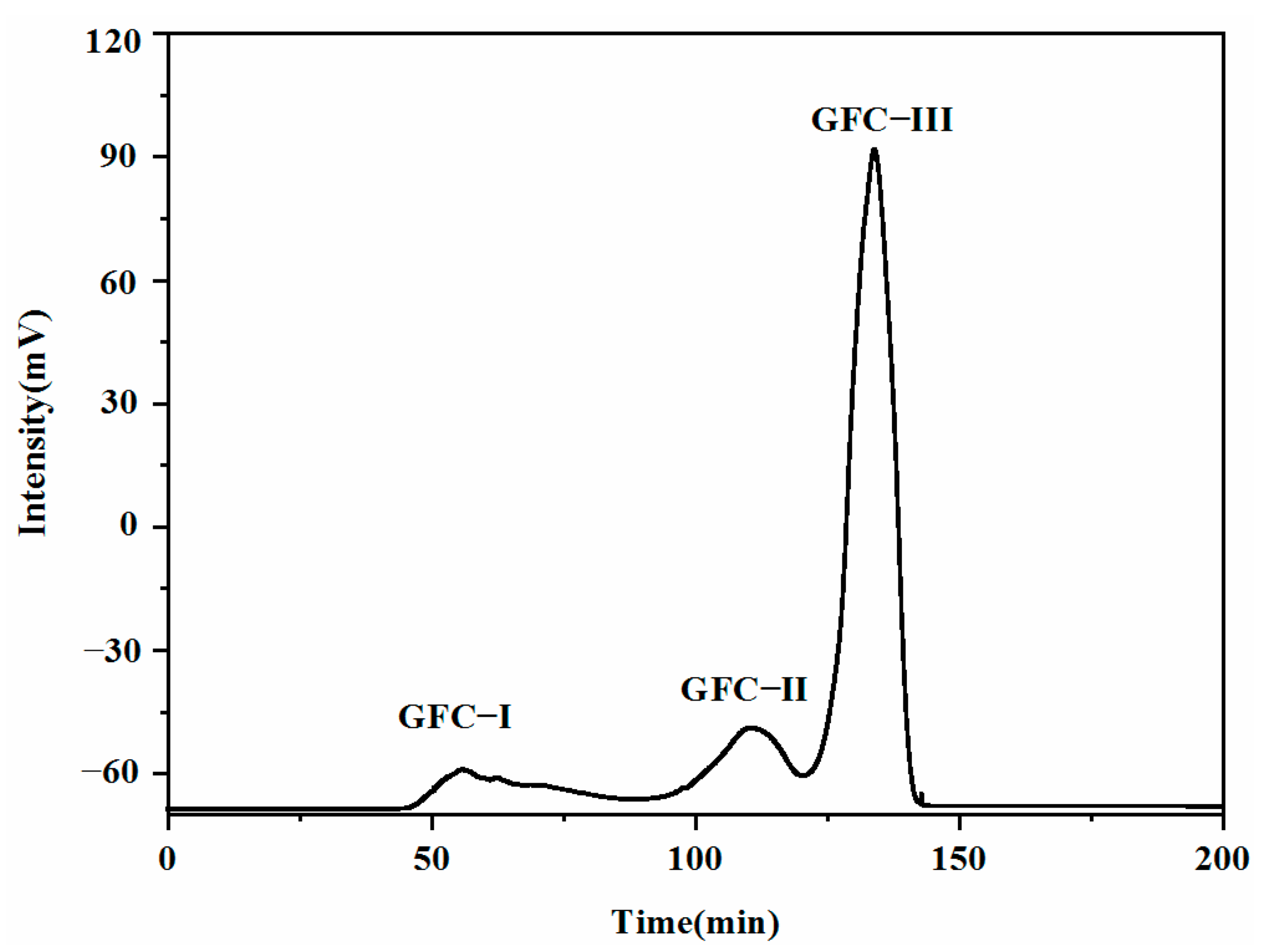
2.7. Gel Filtration Chromatography Purification of Taste Peptides
2.8. Taste Peptide Identification by UPLC-MS/MS
2.9. Sensory Evaluation of Synthetic Peptides
3. Materials and Methods
3.1. Materials
3.2. Preparation of DPF Hydrolysate (DPH)
3.3. Determination of Enzymatic Properties
3.3.1. Protein Recovery
3.3.2. Degree of Hydrolysis (DH)
3.3.3. The Yield of Peptide
3.4. Molecular Weight (MW) Distribution of Peptides
3.5. Amino Acids Analysis
3.6. FT-IR Spectroscopy
3.7. Isolation and Purification of Peptides
3.7.1. Ultrafiltration (UF)
3.7.2. Gel Filtration Chromatography
3.8. Identification of Peptides by UPLC-MS/MS
3.9. Peptide Synthesis
3.10. Taste Traits Analysis Using an Electronic Tongue
3.11. Sensory Profiling
3.12. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Sarmadi, B.H.; Ismail, A. Antioxidative peptides from food proteins: A review. Peptides 2010, 31, 1949–1956. [Google Scholar] [CrossRef] [PubMed]
- Kurihara, K. Umami the fifth basic taste: History of studies on receptor mechanisms and role as a food flavor. BioMed Res. Int. 2015, 2015, 189402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Venkitasamy, C.; Pan, Z.L.; Liu, W.L.; Zhao, L.M. Novel umami ingredients: Umami peptides and their taste. J. Food Sci. 2017, 82, 16–23. [Google Scholar] [CrossRef]
- Fujimakis, M.; Arai, S.; Yamashita, M. Taste peptide fractionation from a fish protein hydrolysate. Agric. Biol. Chem. 1973, 37, 2891–2898. [Google Scholar] [CrossRef]
- Xu, X.R.; You, M.C.; Song, H.L.; Gong, L.; Pan, W.Q. Investigation of umami and kokumi taste-active components in bovine bone marrow extract produced during enzymatic hydrolysis and Maillard reaction. Int. J. Food Sci. Technol. 2018, 53, 2465–2481. [Google Scholar] [CrossRef]
- Li, X.; Xie, X.; Wang, J.; Xu, Y.; Yi, S.; Zhu, W.; Mi, H.; Li, T.; Li, J. Identification, taste characteristics and molecular docking study of novel umami peptides derived from the aqueous extract of the clam meretrix Linnaeus. Food Chem. 2020, 312, 126053. [Google Scholar] [CrossRef]
- Oh, Y.C.; Shu, C.K.; Ho, C.T. Formation of novel 2(1H)-pyrazinones as peptide-specific Maillard reaction products. J. Agric. Food Chem. 1992, 40, 118–121. [Google Scholar] [CrossRef]
- Xu, X.; Xu, R.; Song, Z.; Jia, Q.; Feng, T.; Huang, M.; Song, S. Identification of umami-tasting peptides from Volvariellavolvacea using ultra performance liquid chromatography quadrupole time-of-flight mass spectrometry and sensory-guided separation techniques. J. Chromatogr. A 2019, 1596, 96–103. [Google Scholar] [CrossRef]
- Rhyu, M.R.; Kim, E.Y. Umami taste characteristics of water extract of Doenjang, a Korean soybean paste: Low-molecular acidic peptides may be a possible clue to the taste. Food Chem. 2011, 127, 1210–1215. [Google Scholar] [CrossRef]
- Govindaraju, K.; Srinivas, H. Studies on the effects of enzymatic hydrolysison functional and physico-chemical properties of arachin. LWT Food Sci. Technol. 2004, 11, 54–62. [Google Scholar]
- Su, G.; Ren, J.; Yang, B.; Cui, C.; Zhao, M. Comparison of hydrolysis characteristics on defatted peanut meal proteins between a protease extract from Aspergillus oryzae and commercial proteases. Food Chem. 2011, 126, 1306–1311. [Google Scholar] [CrossRef]
- Su, G.; Cui, C.; Zheng, L.; Yang, B.; Ren, J.; Zhao, M. Isolation and identification of two novel umami and umami-enhancing peptides from peanut hydrolysate by consecutive chromatography and MALDI-TOF/TOF MS. Food Chem. 2012, 135, 479–485. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Yang, H.S.; Li, J.; Li, Z.X.; Jiang, Y.M. Amino acid composition, molecular weight distribution and antioxidant activity of protein hydrolysates of soy sauce lees. Food Chem. 2011, 124, 551–555. [Google Scholar] [CrossRef]
- Quist, E.E.; Phillips, R.D.; Saalia, F.K. The effect of enzyme systems and processing on the hydrolysis of peanut (Arachis hypogaea L.) protein. LWT Food Sci. Technol. 2009, 42, 1717–1721. [Google Scholar] [CrossRef]
- Maehashi, K.; Matsuzaki, M.; Yamamoto, Y.; Udaka, S. Isolation of peptides from an enzymatic hydrolysate of food proteins and characterization of their taste properties. Biosci. Biotechnol. Biochem. 1999, 63, 555–559. [Google Scholar] [CrossRef]
- Zhou, X.; Cui, H.; Zhang, Q.; Hayat, K.; Yu, J.; Hussain, S.; Tahir, M.U.; Zhang, X.; Ho, C. Taste improvement of Maillard reaction intermediates derived from enzymatic hydrolysates of pea protein. Food Res. Int. 2021, 140, 109985. [Google Scholar] [CrossRef]
- Iwaniak, A.; Minkiewicz, P.; Darewicz, M.; Hrynkiewicz, M. Food protein originating peptides as tastants—Physiological, technological, sensory, and bioinformatic approaches. Food Res. Int. 2016, 89 Pt 1, 27–38. [Google Scholar] [CrossRef]
- Batal, A.; Dale, N.; Cafe, M. Nutrient composition of peanut meal. J. Appl. Poult. Res. 2005, 14, 254–257. [Google Scholar] [CrossRef]
- Zheng, L.; Zhao, Y.; Xiao, C.; Sun-Waterhouse, D.; Zhao, M.; Su, G. Mechanism of the discrepancy in the enzymatic hydrolysis efficiency between defatted peanut flour and peanut protein isolate by Flavorzyme. Food Chem. 2015, 168, 100–106. [Google Scholar] [CrossRef]
- Lioe, H.N.; Wade, K.; Aoki, T.; Yasuda, M. Chemical and sensory characteristics of low molecular weight fractions obtained from three types of Japanese soy sauces (shoyu)-Koikuchi, Tamari and Shiro shouyu. Food Chem. 2007, 100, 1669–1677. [Google Scholar] [CrossRef]
- Temussi, P.A. The good taste of peptides. J. Pept. Sci. 2012, 18, 73–82. [Google Scholar] [CrossRef] [PubMed]
- Schindler, A.; Dunkel, A.; Stahler, F.; Backes, M.; Ley, J.; Meyerhof, W.; Hofmann, T. Discovery of salt taste enhancing arginyl dipeptides in protein digests and fermented fish sauces by means of a sensomics approach. J. Agric. Food Chem. 2011, 59, 12578–12588. [Google Scholar] [CrossRef] [PubMed]
- Shim, J.; Son, H.J.; Kim, Y.; Kim, K.H.; Kim, J.T.; Moon, H.; Kim, M.J.; Misaka, T.; Rhyu, M.R. Modulation of sweet taste by umami compounds via sweet taste receptor subunit hT1R2. PLoS ONE 2015, 10, e124030. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.J.; Son, H.J.; Kim, Y.; Misaka, T.; Rhyu, M.R. Umami-bitter interactions: The suppression of bitterness by umami peptides via human bitter taste receptor. Biochem. Biophys. Res. Commun. 2015, 456, 586–590. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Kim, E.; Son, H.J.; Lee, J.; Choi, Y.; Rhyu, M. Identification of a key umami-active fraction in modernized Korean soy sauce and the impact thereof on bitter-masking. Food Chem. 2017, 233, 256–262. [Google Scholar] [CrossRef] [PubMed]
- Apriyantono, A.; Setyaningsih, D.; Hariyadi, D. Sensory and peptides characteristics of soy sauce fractions obtained by ultrafiltration. Qual. Fresh Process. Foods 2004, 542, 213–226. [Google Scholar]
- Jung, C. Insight into protein structure and protein-ligand recognition by Fourier transform infrared spectroscopy. J. Mol. Recognit. 2000, 13, 325–351. [Google Scholar] [CrossRef]
- Subramanian, A.; Ahn, J.; Balasubramaniam, V.M.; Rodriguez-Saona, L. Monitoring biochemical changes in bacterial spore during thermal and pressure-assisted thermal processing using FT-IR Spectroscopy. J. Agric. Food Chem. 2007, 55, 9311–9317. [Google Scholar] [CrossRef]
- Han, Y.; Wang, J.; Li, Y.; Hang, Y.; Yin, X.; Li, Q. Circular dichroism and infrared spectroscopic characterization of secondary structure components of protein Z during mashing and boiling processes. Food Chem. 2015, 188, 201–209. [Google Scholar] [CrossRef] [Green Version]
- Whitmore, L.; Wallace, B.A. Protein secondary structure analyses from circular dichroism spectroscopy: Methods and reference databases. Biopolymers 2008, 89, 392–400. [Google Scholar] [CrossRef]
- He, S.D.; Shi, J.; Walida, E.; Zhang, H.; Ma, Y.; Xue, S.J. Reverse micellar extraction of lectin from black turtle bean (Phaseolus vulgaris): Optimisation of extraction conditions by response surface methodology. Food Chem. 2015, 166, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.; Ma, C. Structural characterization of globulin from common buckwheat (Fagopyrum esculentum Moench) using circular dichroism and Raman spectroscopy. Food Chem. 2007, 102, 150–160. [Google Scholar] [CrossRef]
- Jang, A.; Lee, M. Purification and identification of angiotensin converting enzyme inhibitory peptides from beef hydrolysates. Meat Sci. 2005, 69, 653–661. [Google Scholar] [CrossRef] [PubMed]
- Amin, M.N.G.; Kusnadi, J.; Hsu, J.L.J.; Doerksen, R. Identification of a novel umami peptide in tempeh (Indonesian fermented soybean) and its binding mechanism to the umami receptor T1R. Food Chem. 2020, 333, 127411. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, M.; Lin, L.; Zhao, M.; Dong, Y.; Sunwaterhouse, D.; Chen, H.; Qiu, C.; Su, G. Sequence, taste and umami-enhancing effect of the peptides separated from soysauce. Food Chem. 2016, 206, 174–181. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.N.; Sun-Waterhouse, D.; Su, G.W.; Zhao, M.M. New insight into umami receptor, umami/umami-enhancing peptides and their derivatives: A review. Trends Food Sci. Technol. 2019, 88, 429–438. [Google Scholar] [CrossRef]
- Escriche, I.; Kadar, M.; Domenech, E.; Gil-Sánchez, L. A potentiometric electronic tongue for the discrimination of honey according to the botanical origin. Comparison with traditional methodologies: Physicochemical parameters and volatile profile. J. Food Eng. 2012, 109, 449–456. [Google Scholar]
- Yoshida, Y.; Kawabata, Y.; Kawabata, F.; Nishimura, S.T. Expressions of multiple umami taste receptors in oral and gastrointestinal tissues, and umami taste synergism in chickens. Biochem. Biophys. Res. Commun. 2015, 466, 346–349. [Google Scholar] [CrossRef] [Green Version]
- Nielsen, P.M.; Petersen, D.; Dambmann, C. Improved method for determining food protein degree of hydrolysis. J. Food Sci. 2001, 66, 642–646. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, K.; Lu, F.; Wang, Y.; Ouyang, N.; Ma, H. Increasing peptide yield of soybean meal solid-state fermentation of ultrasound-treated Bacillus amyloliquefaciens. Innov. Food Sci. Emerg. Technol. 2021, 72, 102704. [Google Scholar] [CrossRef]
- Wang, W.; Zhang, L.; Wang, Z.; Wang, X.; Liu, Y. Physicochemical and sensory variables of Maillard reaction products obtained from Takifugu obscurus muscle hydrolysates. Food Chem. 2019, 290, 40–46. [Google Scholar] [CrossRef]
- Dang, Y.; Hao, L.I.; Zhou, T.; Cao, J.; Sun, Y.; Pan, D. Establishment of new assessment method for the synergistic effect between umami peptides and monosodium glutamate using electronic tongue. Food Res. Int. 2019, 121, 20–27. [Google Scholar] [CrossRef] [PubMed]
- Frank, O.; Ottinger, H.; Hofmann, T. Characterization of an intense bitter-tasting 1H,4H-quino-lizinium-7-olate by application of the taste dilution analysis, a novel bioassay for the screening and identification of taste-active compounds in foods. J. Agric. Food Chem. 2001, 49, 231–238. [Google Scholar] [CrossRef] [PubMed]
- Ottinger, H.; Hofmann, T. Identification of the taste enhancer alapyridaine in beef broth and evaluation of its sensory impact by taste reconstitution experiments. J. Agric. Food Chem. 2003, 51, 6791–6796. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Gao, X.; Pan, D.; Zhang, Z.; Zhou, T.; Dang, Y. Isolation, characterization and molecular docking of novel umami and umami-enhancing peptides from Ruditapes philippinarum. Food Chem. 2021, 343, 128522. [Google Scholar] [CrossRef] [PubMed]
| Species | AAs (g/100 g) | FAAs (g/100 g) | ||
|---|---|---|---|---|
| DPH | DPF | DPH | DPF | |
| Asp | 5.92 ± 0.40 b | 7.03 ± 0.16 a | 0.84 ± 0.00 a | 0.03 ± 0.00 b |
| Glu | 10.42 ± 0.35 b | 12.51 ± 0.28 a | 1.81 ± 0.04 a | 0.20 ± 0.00 b |
| Ser | 2.26 ± 0.35 a | 2.70 ± 0.23 a | 2.27 ± 0.11 a | 0.03 ± 0.00 b |
| Arg | 5.97 ± 0.23 b | 7.28 ± 0.33 a | 3.77 ± 0.17 a | 0.07 ± 0.00 b |
| Gly | 2.73 ± 0.21 a | 3.32 ± 0.25 a | 0.70 ± 0.00 a | 0.01 ± 0.00 b |
| Thr | 1.34 ± 0.06 a | 1.57 ± 0.02 a | 1.00 ± 0.00 a | 0.01 ± 0.00 b |
| Pro | 2.05 ± 0.41 a | 2.49 ± 0.17 a | 0.32 ± 0.00 a | 0.17 ± 0.00 b |
| Ala | 1.97 ± 0.14 a | 2.37 ± 0.24 a | 1.14 ± 0.01 a | 0.03 ± 0.00 b |
| Val | 2.17 ± 0.10 a | 2.53 ± 0.23 a | 1.63 ± 0.04 a | 0.04 ± 0.00 b |
| Met | 0.44 ± 0.00 a | 0.49 ± 0.00 a | 0.32 ± 0.00 a | 0.01 ± 0.00 b |
| Ile | 1.72 ± 0.14 a | 2.03 ± 0.08 a | 1.34 ± 0.06 a | 0.01 ± 0.00 b |
| Leu | 3.17 ± 0.13 b | 3.92 ± 0.28 a | 2.47 ± 0.14 a | 0.01 ± 0.00 b |
| Phe | 2.81 ± 0.10 a | 3.47 ± 0.42 a | 1.89 ± 0.14 a | 0.03 ± 0.00 b |
| His | 1.13 ± 0.00 a | 1.39 ± 0.19 a | 0.69 ± 0.02 a | 0.01 ± 0.00 b |
| Lys | 1.84 ± 0.08 a | 2.27 ± 0.25 a | 1.08 ± 0.00 a | 0.02 ± 0.00 b |
| Tyr | 1.95 ± 0.07 a | 2.12 ± 0.30 a | 1.75 ± 0.00 a | 0.15 ± 0.00 b |
| Total | 47.89 ± 0.08 b | 57.51 ± 0.83 a | 23.30 ± 0.13 a | 0.85 ± 0.00 b |
| α-Helix | β-Sheet | β-Turn | Random Coil | |
|---|---|---|---|---|
| DPF | 20.93 ± 2.00 a | 43.54 ± 2.56 b | 24.40 ± 1.62 a | 11.14 ± 1.53 a |
| DPH | 16.24 ± 1.55 b | 52.02 ± 3.29 a | 23.61 ± 2.50 a | 8.13 ± 0.76 a |
| Fraction | Without MSG-Salt | With MSG-Salt | ||
|---|---|---|---|---|
| Taste Description | TD Value | Umami Enhancement | TD Value | |
| MSG-salt | -- | -- | Umami | 7.50 ± 0.36 |
| DPH | Strong umami, salty | 3.25 ± 0.29 | Strong umami | 9.00 ± 0.49 |
| UF-I | Slight umami | 0.90 ± 0.00 | Not detected | -- |
| UF-II | Slight umami | 1.23 ± 0.02 | Not detected | -- |
| UF-III | Umami | 1.36 ± 0.02 | Not detected | -- |
| UF-IV | Umami | 2.22 ± 0.17 | Strong Umami | 9.50 ± 0.66 |
| UF-V | Strong umami | 4.87 ± 0.20 | Strong umami | 12.81 ± 0.54 |
| GFC-I | Tasteless | -- | Not detected | -- |
| GFC-II | Tasteless | -- | Not detected | -- |
| GFC-III | Strong umami | 5.44 ± 0.34 | Strong Umami | 14.57 ± 0.82 |
| Peptide | Scan | Score | Length | m/z | z | Mass |
|---|---|---|---|---|---|---|
| ADSYRLP | 6995 | 99 | 7 | 411.2096 | 2 | 820.4079 |
| DAQRPF | 5396 | 97 | 6 | 367.1835 | 2 | 732.3555 |
| DFRAP | 5639 | 99 | 5 | 303.1544 | 2 | 604.2969 |
| DPLKY | 4887 | 99 | 5 | 318.1722 | 2 | 634.3326 |
| DQFPR | 4274 | 99 | 5 | 331.6651 | 2 | 661.3184 |
| DSRPF | 4986 | 98 | 5 | 311.1518 | 2 | 620.2918 |
| DWRQERP | 4321 | 91 | 7 | 329.4971 | 3 | 985.473 |
| EAFRVL | 8208 | 99 | 6 | 367.7119 | 2 | 733.4122 |
| EFHNR | 1617 | 99 | 5 | 351.6681 | 2 | 701.3245 |
| EWAGLTTN | 8189 | 90 | 8 | 446.2134 | 2 | 890.4134 |
| KDNNPF | 5065 | 94 | 6 | 367.675 | 2 | 733.3395 |
| LDAQRP | 2816 | 99 | 6 | 350.1914 | 2 | 698.3711 |
| LDQFPR | 5904 | 99 | 6 | 388.207 | 2 | 774.4024 |
| LDQFRP | 5497 | 97 | 6 | 388.2063 | 2 | 774.4024 |
| NDFGR | 2762 | 98 | 5 | 304.6415 | 2 | 607.2714 |
| NDNPFKF | 10,478 | 95 | 7 | 441.2111 | 2 | 880.4079 |
| SDLYVR | 4539 | 99 | 6 | 376.6993 | 2 | 751.3864 |
| VPPFDHQ | 6019 | 95 | 7 | 420.204 | 2 | 838.3973 |
| VPQDFR | 4362 | 99 | 6 | 381.1999 | 2 | 760.3868 |
| Synthetic Peptides | Without MSG-Salt | With MSG-Salt | ||
|---|---|---|---|---|
| Taste Description | Threshold Value (mmol/L) | Umami Enhancement | Threshold Value (mmol/L) | |
| ADSYRLP | Sour | -- | Strong umami | 0.107 ± 0.004 a |
| DAQRPF | Tasteless | -- | Not detected | -- |
| DFRAP | Tasteless | -- | Not detected | -- |
| DPLKY | Sour | -- | Slight umami | 0.164 ± 0.008 d |
| DQFPR | Tasteless | -- | Not detected | -- |
| DSRPF | Tasteless | -- | Not detected | -- |
| DWRQERP | Tasteless | -- | Not detected | -- |
| EAFRVL | Sour, astringent | -- | Umami | 0.134 ± 0.004 bc |
| EFHNR | Sour, astringent | -- | Umami | 0.148 ± 0.004 c |
| EWAGLTTN | Sour | -- | Not detected | -- |
| KDNNPF | Tasteless | -- | Not detected | -- |
| LDAQRP | Sour, astringent | -- | Not detected | -- |
| LDQFPR | Tasteless | -- | Not detected | -- |
| LDQFRP | Sour | -- | Not detected | -- |
| NDFGR | Tasteless | -- | Not detected | -- |
| NDNPFKF | Sour, astringent | -- | Not detected | -- |
| SDLYVR | Tasteless | -- | Strong umami | 0.132 ± 0.007 b |
| VPPFDHQ | Tasteless | -- | Not detected | -- |
| VPQDFR | Astringent | -- | Not detected | -- |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, L.; Sun, X.; Lu, X.; Wei, S.; Sun, Q.; Jin, L.; Song, G.; You, J.; Li, F. Characterization of Peanut Protein Hydrolysate and Structural Identification of Umami-Enhancing Peptides. Molecules 2022, 27, 2853. https://doi.org/10.3390/molecules27092853
Zhang L, Sun X, Lu X, Wei S, Sun Q, Jin L, Song G, You J, Li F. Characterization of Peanut Protein Hydrolysate and Structural Identification of Umami-Enhancing Peptides. Molecules. 2022; 27(9):2853. https://doi.org/10.3390/molecules27092853
Chicago/Turabian StyleZhang, Lixia, Xiaojing Sun, Xin Lu, Songli Wei, Qiang Sun, Lu Jin, Guohui Song, Jing You, and Fei Li. 2022. "Characterization of Peanut Protein Hydrolysate and Structural Identification of Umami-Enhancing Peptides" Molecules 27, no. 9: 2853. https://doi.org/10.3390/molecules27092853
APA StyleZhang, L., Sun, X., Lu, X., Wei, S., Sun, Q., Jin, L., Song, G., You, J., & Li, F. (2022). Characterization of Peanut Protein Hydrolysate and Structural Identification of Umami-Enhancing Peptides. Molecules, 27(9), 2853. https://doi.org/10.3390/molecules27092853





