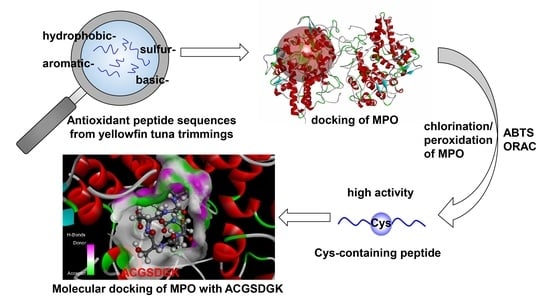
Purification and Identification of Novel Myeloperoxidase Inhibitory Antioxidant Peptides from Tuna (Thunnas albacares) Protein Hydrolysates
Abstract
:1. Introduction
2. Results and Discussion
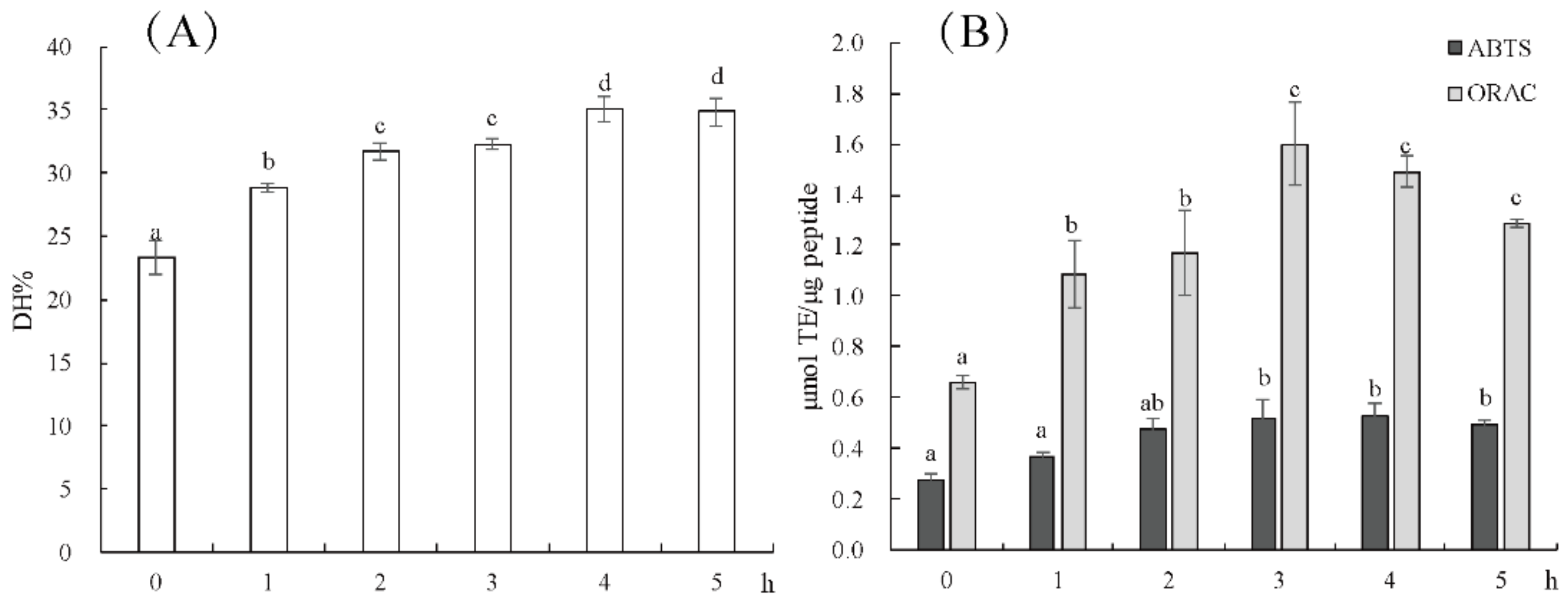
2.1. Effect of Enzymolysis Time on the Antioxidant Activity of Hydrolysate
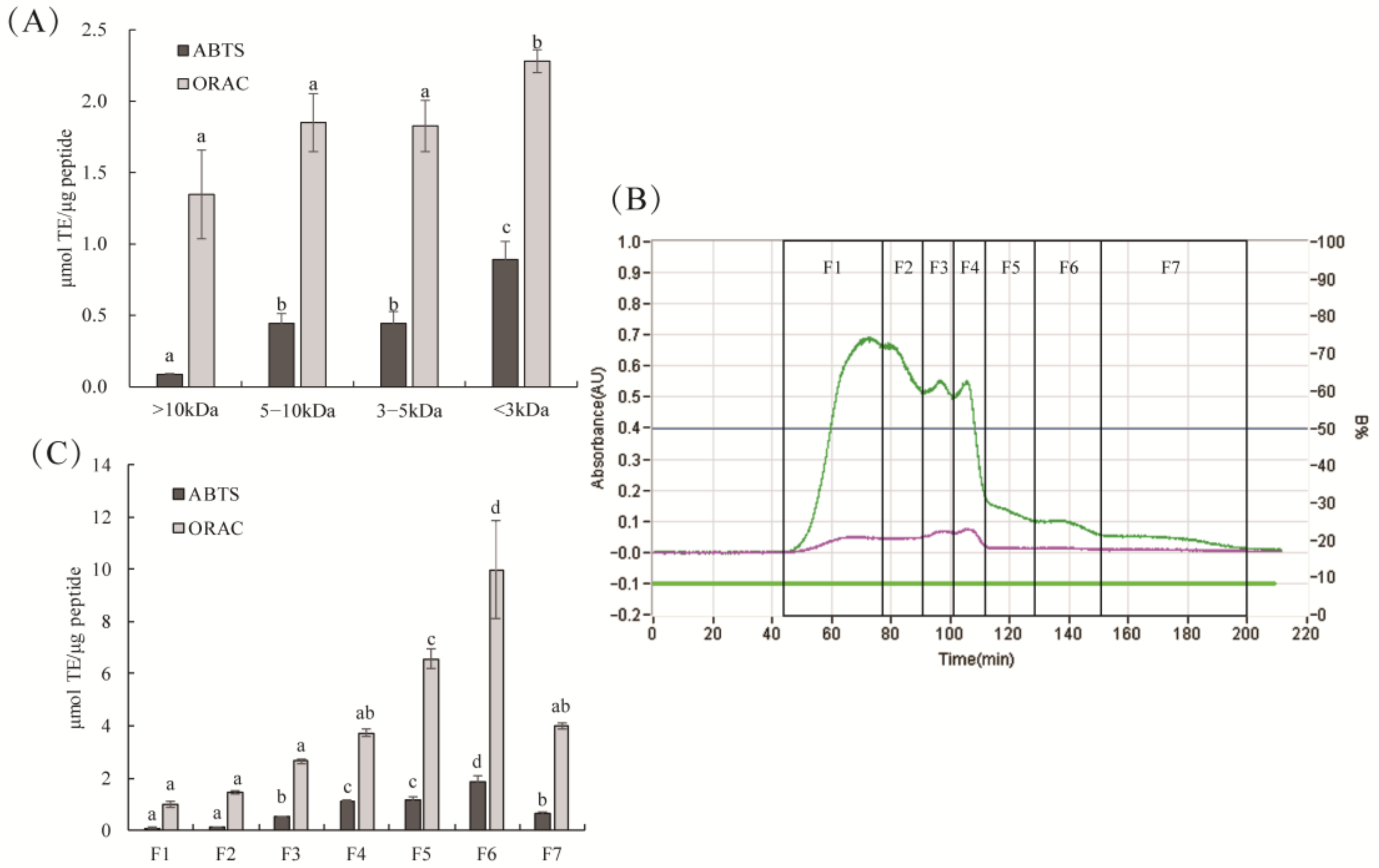
2.2. Antioxidant Activity of TPA3 Ultrafiltration and Gel Separation Fractions
2.3. Identification and Screening of Potential Antioxidant Peptides in Silico
2.4. The Activity of Synthesized Peptides
3. Materials and Methods
3.1. Materials
3.2. Preparation of TPA Protein Hydrolysates
3.3. Determination of Hydrolysis Degree Using the O-Phthaldialdehyde (OPA) Method
3.4. Purification of TPA
3.5. Antioxidant Assay
3.5.1. ABTS Radical Scavenging Ability Assay
3.5.2. Oxygen Radical Absorbance Capacity (ORAC) Assay
3.6. Identification of Peptide Sequence
3.7. Molecular Docking
3.8. Peptide Synthesis
3.9. Analysis of MPO Inhibitory Activity
3.10. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Food and Agriculture Organization. Information and Analysis on World Fish Trade. Available online: www.fao.org (accessed on 20 November 2019).
- Zamora-Sillero, J.; Gharsallaoui, A.; Prentice, C. Peptides from fish by-product protein hydrolysates and its functional properties: An overview. Mar. Biotechnol. 2018, 20, 118–130. [Google Scholar] [CrossRef] [PubMed]
- Abbey, L.; Glover-Amengor, M.; Atikpo, M.O.; Atter, A.; Toppe, J. Nutrient content of fish powder from low value fish and fish byproducts. Food Sci. Nutr. 2017, 5, 374–379. [Google Scholar] [CrossRef]
- Idowu, A.T.; Igiehon, O.O.; Idowu, S.; Olatunde, O.O.; Benjakul, S. Bioactivity potentials and general applications of fish protein hydrolysates. Int. J. Pept. Res. Ther. 2020, 27, 109–118. [Google Scholar] [CrossRef]
- Kemp, D.C.; Kwon, J.Y. Fish and shellfish-derived anti-inflammatory protein products: Properties and mechanisms. Molecules 2021, 26, 3225. [Google Scholar] [CrossRef] [PubMed]
- Nwachukwu, I.D.; Aluko, R.E. Structural and functional properties of food protein-derived antioxidant peptides. J. Food Biochem. 2019, 43, e12761. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zou, T.B.; He, T.P.; Li, H.B.; Tang, H.W.; Xia, E.Q. The structure-activity relationship of the antioxidant peptides from natural proteins. Molecules 2016, 21, 72. [Google Scholar] [CrossRef] [PubMed]
- Sridhar, K.; Inbaraj, B.S.; Chen, B.H.E. Recent developments on production, purification and biological activity of marine peptides. Food Res. Int. 2021, 147, 110468. [Google Scholar] [CrossRef] [PubMed]
- Karami, Z.; Akbari-adergani, B. Bioactive food derived peptides: A review on correlation between structure of bioactive peptides and their functional properties. J. Food Sci. Tech. Mys. 2019, 56, 535–547. [Google Scholar] [CrossRef]
- Bashir, K.M.I.; Sohn, J.H.; Kim, J.S.; Choi, J.S. Identification and characterization of novel antioxidant peptides from mackerel (Scomber japonicus) muscle protein hydrolysates. Food Chem. 2020, 323, 126809. [Google Scholar] [CrossRef]
- Ma, Y.K.; Wu, Y.Y.; Li, L.H. Relationship between primary structure or spatial conformation and functional activity of antioxidant peptides from Pinctada fucata. Food Chem. 2018, 264, 108–117. [Google Scholar] [CrossRef]
- Chen, G.L.; Seukep, A.J.; Guo, M.Q. Recent advances in molecular docking for the research and discovery of potential marine drugs. Mar. Drugs 2020, 18, 545. [Google Scholar] [CrossRef]
- Min, X.J.; Guo, Y.L.; Zhou, Y.S.; Chen, X.P. Protection against dextran sulfate sodium-induced ulcerative colitis in mice by neferine, a natural product from Nelumbo nucifera Gaertn. Cell J. 2021, 22, 523–531. [Google Scholar] [CrossRef]
- Van der Does, A.M.; Hensbergen, P.J.; Bogaards, S.J.; Cansoy, M.; Deelder, A.M.; van Leeuwen, H.C.; Drijfhout, J.W.; van Dissel, J.T.; Nibbering, P.H. The human lactoferrin-derived peptide hLF1-11 exerts immunomodulatory effects by specific inhibition of myeloperoxidase activity. J. Immunol. 2012, 188, 5012–5019. [Google Scholar] [CrossRef] [Green Version]
- He, S.D.; Zhang, Y.; Sun, H.J.; Du, M.; Qiu, J.L.; Tang, M.M.; Sun, X.B.; Zhu, B.W. Antioxidative peptides from proteolytic hydrolysates of false abalone (Volutharpa ampullacea perryi): Characterization, identification, and molecular docking. Mar. Drugs 2019, 17, 116. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; He, S.D.; Bonneil, E.; Simpson, B.K. Generation of antioxidative peptides from Atlantic sea cucumber using alcalase versus trypsin: In vitro activity, de novo sequencing, and in silico docking for in vivo function prediction. Food Chem. 2020, 306, 125581. [Google Scholar] [CrossRef]
- Hosseini, S.F.; Rezaei, M.; McClements, D.J. Bioactive functional ingredients from aquatic origin: A review of recent progress in marine-derived nutraceuticals. Crit. Rev. Food Sci. 2020, 62, 1839855. [Google Scholar] [CrossRef]
- Tacias-Pascacio, V.G.; Morellon-Sterling, R.; Siar, E.H.; Tavano, O.; Berenguer-Murcia, A.; Fernandez-Lafuente, R. Use of alcalase in the production of bioactive peptides: A review. Int. J. Biol. Macromol. 2020, 165, 2143–2196. [Google Scholar] [CrossRef]
- Oliveira, D.; Bernardi, D.; Drummond, F.; Dieterich, F.; Boscolo, W.; Leivas, C.; Kiatkoski, E.; Waszczynskyj, N. Potential use of tuna (Thunnus albacares) by product: Production of antioxidant peptides and recovery of unsaturated fatty acids from tuna head. Int. J. Food Eng. 2017, 13, 20150365. [Google Scholar] [CrossRef]
- Mongkonkamthorn, N.; Malila, Y.; Regenstein, J.M.; Wangtueai, S. Enzymatic hydrolysis optimization for preparation of tuna dark meat hydrolysate with antioxidant and angiotensin I-converting enzyme (ACE) inhibitory activities. J. Aquat. Food Prod. Technol. 2021, 30, 1090–1108. [Google Scholar] [CrossRef]
- Qiu, Y.T.; Wang, Y.M.; Yang, X.R.; Zhao, Y.Q.; Chi, C.F.; Wang, B. Gelatin and antioxidant peptides from gelatin hydrolysate of skipjack tuna (Katsuwonus pelamis) scales: Preparation, identification and activity evaluation. Mar. Drugs 2019, 17, 565. [Google Scholar] [CrossRef] [Green Version]
- Klomklao, S.; Benjakul, S. Protein hydrolysates prepared from the viscera of skipjack tuna (Katsuwonus pelmamis): Antioxidative activity and functional properties. Turk. J. Fish. Aquat. Sci. 2018, 18, 69–79. [Google Scholar] [CrossRef]
- Devita, L.; Lioe, H.N.; Nurilmala, M.; Suhartono, M.T. The bioactivity prediction of peptides from tuna skin collagen using integrated method combining in vitro and in silico. Foods 2021, 10, 2739. [Google Scholar] [CrossRef]
- Pezeshk, S.; Ojah, S.M.; Rezaei, M.; Shabanpour, B. Fractionation of protein hydrolysates of fish waste using membrane ultrafiltration: Investigation of antibacterial and antioxidant activities. Probiotics Antimicrob. Proteins 2019, 11, 1015–1022. [Google Scholar] [CrossRef]
- Huang, D.J.; Ou, B.X.; Prior, R.L. The chemistry behind antioxidant capacity assays. J. Agric. Food Chem. 2005, 53, 1841–1856. [Google Scholar] [CrossRef]
- He, Y.; Pan, X.; Chi, C.F.; Sun, K.L.; Wang, B. Ten new pentapeptides from protein hydrolysate of miiuy croaker (Miichthys miiuy) muscle: Preparation, identification, and antioxidant activity evaluation. LWT-Food Sci. Technol. 2019, 105, 1–8. [Google Scholar] [CrossRef]
- Zhang, X.Q.; Cao, D.Q.; Sun, X.; Sun, S.Q.; Xu, N.J. Preparation and identification of antioxidant peptides from protein hydrolysate of marine alga Gracilariopsis lemaneiformis. J. Appl. Phycol. 2019, 31, 2585–2596. [Google Scholar] [CrossRef]
- Wang, L.Y.; Ma, M.T.; Yu, Z.P.; Du, S.K. Preparation and identification of antioxidant peptides from cottonseed proteins. Food Chem. 2021, 352, 129399. [Google Scholar] [CrossRef]
- Zhu, Y.P.; Zhao, X.Y.; Zhang, X.W.; Liu, H.K.; Ao, Q. Amino acid, structure and antioxidant properties of Haematococcus pluvialis protein hydrolysates produced by different proteases. Int. J. Food Sci. Tech. 2021, 56, 185–195. [Google Scholar] [CrossRef]
- Shen, Y.; Hu, L.T.; Xia, B.; Ni, Z.J.; Elam, E.; Thakur, K.; Zhang, J.G.; Wei, Z.J. Effects of different sulfur-containing substances on the structural and flavor properties of defatted sesame seed meal derived maillard reaction products. Food Chem. 2021, 365, 130463. [Google Scholar] [CrossRef]
- Sonklin, C.; Alashi, A.M.; Laohakunjit, N.; Aluko, R.E. Functional characterization of mung bean meal protein-derived antioxidant peptides. Molecules 2021, 26, 1515. [Google Scholar] [CrossRef]
- Zeng, J.; Fenna, R.E. X-ray crystal structure of canine myeloperoxidase at 3 A resolution. J. Mol. Biol. 1992, 226, 185–207. [Google Scholar] [CrossRef]
- Sanjukta, S.; Padhi, S.; Sarkar, P.; Singh, S.P.; Sahoo, D.; Rai, A.K. Production, characterization and molecular docking of antioxidant peptides from peptidome of kinema fermented with proteolytic Bacillus spp. Food Res. Int. 2021, 141, 110161. [Google Scholar] [CrossRef] [PubMed]
- Chai, T.T.; Koh, J.A.; Wong, C.C.C.; Sabri, M.Z.; Wong, F.C. Computational screening for the anticancer potential of seed-derived antioxidant peptides: A cheminformatic approach. Molecules 2021, 26, 7396. [Google Scholar] [CrossRef] [PubMed]
- Arthur, D.E.; Uzairu, A. Molecular docking studies on the interaction of NCI anticancer analogues with human phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit. J. King Saud Univ. Sci. 2019, 31, 1151–1166. [Google Scholar] [CrossRef]
- Pattison, D.I.; Davies, M.J. Reactions of myeloperoxidase-derived oxidants with biological substrates: Gaining chemical insight into human inflammatory diseases. Curr. Med. Chem. 2006, 13, 3271–3290. [Google Scholar] [CrossRef]
- Nielsen, P.M.; Petersen, D.; Dambmann, C. Improved method for determining food protein degree of hydrolysis. J. Food Sci. 2001, 66, 642–646. [Google Scholar] [CrossRef]
- Zheng, L.; Zhao, M.M.; Xiao, C.Q.; Zhao, Q.Z.; Su, G.W. Practical problems when using ABTS assay to assess the radical-scavenging activity of peptides: Importance of controlling reaction pH and time. Food Chem. 2016, 192, 288–294. [Google Scholar] [CrossRef]
- Wu, R.B.; Huang, J.F.; Huan, R.; Chen, L.L.; Yi, C.P.; Liu, D.; Wang, M.; Liu, C.L.; He, H.L. New insights into the structure-activity relationships of antioxidative peptide PMRGGGGYHY. Food Chem. 2021, 337, 127678. [Google Scholar] [CrossRef]
| No. | Peptide Sequence | Length | Mass (Da) | Hydrophobic (%) | (-)CDOCKER Energy | Special Amino Acid (%) | |||
|---|---|---|---|---|---|---|---|---|---|
| Hydrophobic (Leu, Val, Ala, Pro, Phe) | Aromatic (Tyr, Trp, Phe) | Sulfur (Cys, Met) | Basic (Lys, His, Arg) | ||||||
| Ref | GRRRRSVQWCA | 11 | 1374.59 | 27.27 | 139.238 | 18.18 | 9.09 | 36.36 | |
| 1 | DVSDLDAD | 8 | 848.3399 | 37.50 | 148.631 | 37.50 | |||
| 2 | GDAFDKA | 7 | 722.3463 | 42.86 | 147.225 | 42.86 | 14.29 | 14.29 | |
| 3 | KFCSGHA | 7 | 748.3569 | 28.57 | 146.132 | 28.57 | 14.29 | 14.29 | 28.57 |
| 4 | DASHGHSG | 8 | 766.2994 | 12.50 | 145.510 | 12.50 | 25.00 | ||
| 5 | GELALKD | 7 | 744.4017 | 42.86 | 144.636 | 42.86 | 14.29 | ||
| 6 | ACGSDGK | 7 | 636.2664 | 14.29 | 143.951 | 14.29 | 14.29 | 14.29 | |
| 7 | GGGGGYE | 7 | 595.2754 | 0.00 | 139.034 | 14.29 | 14.29 | ||
| 8 | ASCGGRR | 7 | 706.37 | 14.29 | 138.127 | 14.29 | 28.57 | ||
| 9 | GGYGFGGGAG | 10 | 798.3297 | 20.00 | 137.873 | 20.00 | 14.29 | ||
| 10 | SGRSAVVS | 8 | 761.4031 | 37.50 | 133.948 | 37.50 | 20.00 | ||
| 11 | CTMGNGA | 7 | 652.3063 | 28.57 | 131.608 | 14.29 | |||
| 12 | DLSSNVTV | 8 | 833.4131 | 37.50 | 129.646 | 37.50 | 28.57 | ||
| 13 | GFAGGDGL | 8 | 692.3129 | 37.50 | 129.560 | 37.50 | |||
| 14 | SSQSSGY | 7 | 714.282 | 0.00 | 127.791 | 12.50 | |||
| 15 | ASAAADQ | 7 | 633.312 | 57.14 | 127.737 | 57.14 | 14.29 | ||
| 16 | DVDTRAYF | 8 | 985.4505 | 37.50 | 127.695 | 37.50 | 12.50 | ||
| 17 | CTIANGG | 7 | 634.3504 | 28.57 | 127.565 | 14.29 | 25.00 | ||
| 18 | FGVGGGN | 7 | 607.333 | 28.57 | 125.585 | 28.57 | 14.29 | ||
| 19 | SAINGYF | 7 | 770.3599 | 42.86 | 125.186 | 28.57 | |||
| 20 | ASGCCCK | 7 | 670.2618 | 14.29 | 124.649 | 14.29 | 28.57 | ||
| 21 | GYGGGVS | 7 | 595.2602 | 14.29 | 122.442 | 14.29 | 42.86 | 14.29 | |
| 22 | FGTGGAG | 7 | 565.2591 | 33.33 | 121.529 | 28.57 | 14.29 | ||
| 23 | AACGGLN | 7 | 604.279 | 42.86 | 121.129 | 42.86 | 14.29 | ||
| 24 | GFGGGAGSV | 9 | 707.3239 | 33.33 | 120.649 | 33.33 | 14.29 | ||
| 25 | GPTGHDGAPG | 10 | 864.3726 | 30.00 | 120.500 | 30.00 | 11.11 | ||
| 26 | AADILDA | 7 | 688.412 | 71.43 | 120.067 | 57.14 | 10.00 | 10.00 | |
| 27 | IATVGGG | 7 | 573.2186 | 42.86 | 119.743 | 28.57 | |||
| 28 | FVAAALW | 7 | 777.4286 | 100.00 | 119.420 | 85.71 | |||
| 29 | AGGPAGE | 7 | 558.312 | 42.86 | 119.045 | 42.86 | 28.57 | ||
| 30 | FGVGGNG | 7 | 606.3189 | 28.57 | 117.189 | 28.57 | |||
| 31 | AIGGAGG | 7 | 501.2955 | 42.86 | 116.584 | 28.57 | 14.29 | ||
| 32 | GIGGAGG | 7 | 487.2794 | 28.57 | 116.110 | 14.29 | |||
| 33 | ALSCGIW | 7 | 748.3914 | 57.14 | 115.762 | 28.57 | |||
| 34 | SSSGGHG | 7 | 587.2299 | 0.00 | 115.684 | 14.29 | 14.29 | ||
| 35 | AAAMALW | 7 | 733.445 | 100.00 | 112.952 | 71.43 | 14.29 | 14.29 | |
| 36 | GATGGAG | 7 | 489.2353 | 28.57 | 112.748 | 28.57 | 14.29 | 14.29 | |
| 37 | AAAIVVS | 7 | 630.37 | 85.71 | 111.837 | 71.43 | |||
| 38 | ASAAAMN | 7 | 634.3488 | 71.43 | 111.757 | 57.14 | |||
| 39 | GAGGGLS | 7 | 517.2932 | 28.57 | 110.742 | 28.57 | |||
| 40 | AGCSGPH | 7 | 627.3033 | 28.57 | 110.149 | 28.57 | |||
| 41 | AGGSGPAG | 8 | 572.2554 | 37.50 | 110.149 | 37.50 | 14.29 | ||
| 42 | AGPAGGSG | 8 | 572.2554 | 37.50 | 107.837 | 37.50 | 14.29 | 14.29 | 14.29 |
| 43 | PTGHLGE | 7 | 709.3279 | 28.57 | 107.319 | 28.57 | |||
| 44 | AGGPTGE | 7 | 588.322 | 28.57 | 106.705 | 28.57 | 14.29 | 14.29 | |
| 45 | YSSWMMDA | 8 | 1005.357 | 50.00 | 103.526 | 12.50 | |||
| 46 | GPGGSTA | 7 | 546.496 | 28.57 | 101.149 | 28.57 | 25.00 | 25.00 | |
| 47 | GETGPSGP | 8 | 700.3027 | 25.00 | 100.992 | 37.50 | |||
| 48 | APGGAGG | 7 | 485.2632 | 42.86 | 100.016 | 42.86 | |||
| 49 | GGGGAPG | 7 | 471.2485 | 28.57 | 97.938 | 28.57 | |||
| 50 | MYPGIAD | 7 | 765.3367 | 57.14 | 95.151 | 28.57 | |||
| 51 | PDPAGSG | 7 | 599.2851 | 42.86 | 94.751 | 42.86 | 14.29 | 14.29 | |
| 52 | PASGANG | 7 | 572.2554 | 42.86 | 93.382 | 42.86 | |||
| 53 | GAAGPPGP | 8 | 622.3074 | 62.50 | 74.040 | 62.50 | |||
| 54 | AAGGGCA | 7 | 505.2212 | 42.86 | - | 42.86 | |||
| 55 | GFAGSMFGSGAL | 12 | 1100.496 | 50.00 | - | 41.67 | |||
| Peptide Sequence | IC50 Value of the MPO Chlorination Assay (mM) | IC50 Value of the MPO Peroxidation Assay (mM) | ORAC (µmol TE/mg Peptide) | ABTS (µmol TE/mg Peptide) |
|---|---|---|---|---|
| GRRRRSVQWCA | 0.0272 | 0.0328 | 0.77 ± 0.04 a | 1.28 ± 0.19 a |
| DVSDLDAD | 0.2957 | - | 0.00 ± 0.00 b | 0.00 ± 0.00 b |
| KFCSGHA | 0.1079 | 0.0403 | 0.82 ± 0.05 a | 1.30 ± 0.11 a |
| GDAFDKA | - | - | 0.04 ± 0.02 b | 0.00 ± 0.00 b |
| DASHGHSG | - | - | 0.15 ± 0.01 c | 0.00 ± 0.00 b |
| GELALKD | 0.1221 | - | 0.15 ± 0.04 c | 0.00 ± 0.00 b |
| ACGSDGK | 0.0323 | 0.0179 | 0.96 ± 0.09 a | 1.78 ± 0.12 b |
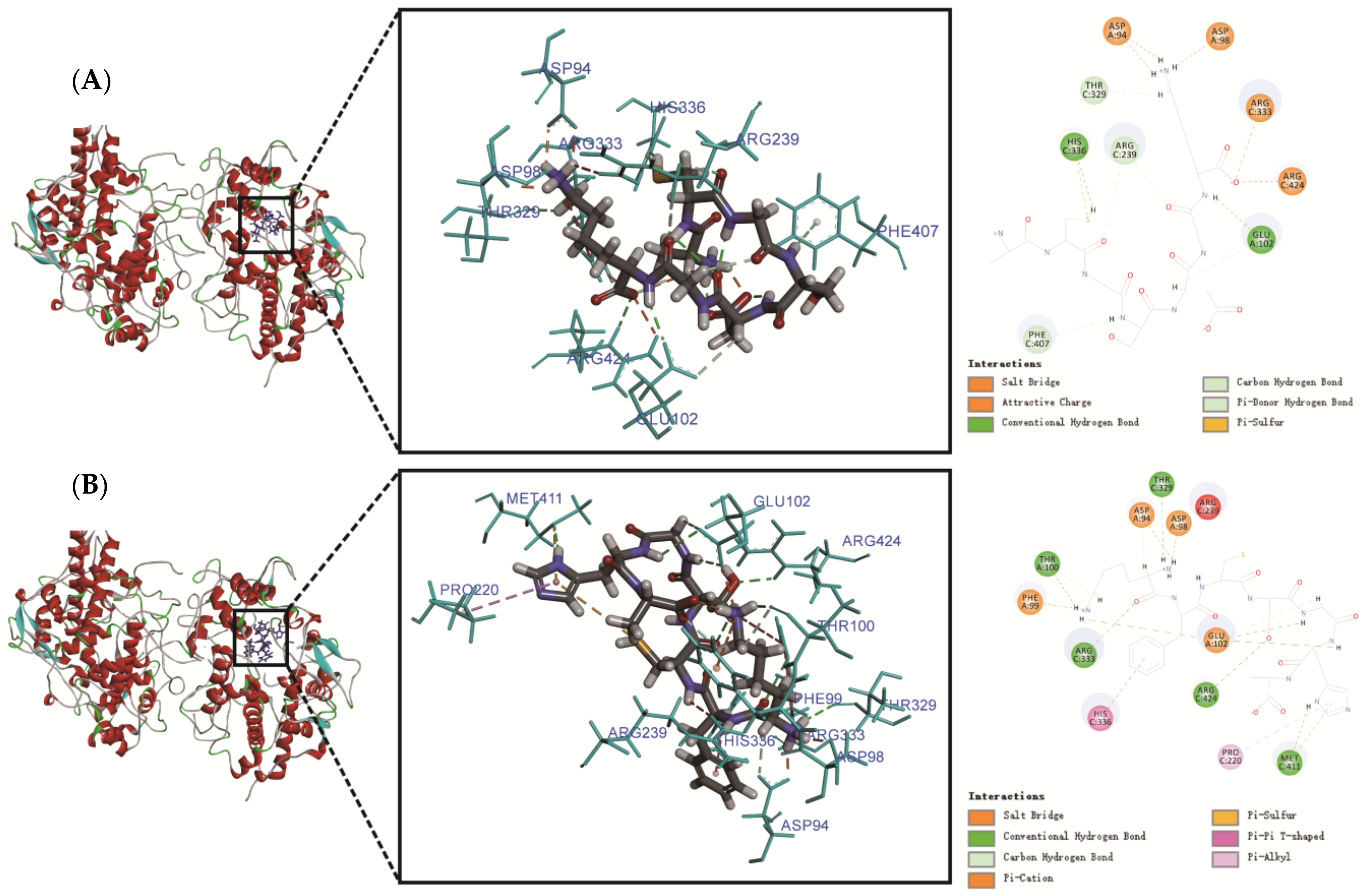
| Peptide Sequence | Interactions | Number | Interaction with A Chain of MPO | Interaction with C Chain of MPO |
|---|---|---|---|---|
| ACGSDGK | H-bond | 12 | Asp98, Glu102 (2), Asp94 (2) | Arg239 (2), Thr329, His336, Phe407, Arg424 (2) |
| Charge | 5 | Asp94 (2), Asp98 | Arg333, Arg424 | |
| Other | 1 | - | His336 | |
| KFCSGHA | H-bond | 14 | Asp94 (2), Asp98, Thr100 (2), Glu102 (4) | Thr329, Arg333 (2), Met411, Arg424 |
| Hydrophobic | 2 | - | Pro220, His336 | |
| Charge | 4 | Asp94, Asp98, Phe99, Glu102 | - | |
| Other | 1 | - | Met411 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cai, B.; Wan, P.; Chen, H.; Huang, J.; Ye, Z.; Chen, D.; Pan, J. Purification and Identification of Novel Myeloperoxidase Inhibitory Antioxidant Peptides from Tuna (Thunnas albacares) Protein Hydrolysates. Molecules 2022, 27, 2681. https://doi.org/10.3390/molecules27092681
Cai B, Wan P, Chen H, Huang J, Ye Z, Chen D, Pan J. Purification and Identification of Novel Myeloperoxidase Inhibitory Antioxidant Peptides from Tuna (Thunnas albacares) Protein Hydrolysates. Molecules. 2022; 27(9):2681. https://doi.org/10.3390/molecules27092681
Chicago/Turabian StyleCai, Bingna, Peng Wan, Hua Chen, Jingtong Huang, Ziqing Ye, Deke Chen, and Jianyu Pan. 2022. "Purification and Identification of Novel Myeloperoxidase Inhibitory Antioxidant Peptides from Tuna (Thunnas albacares) Protein Hydrolysates" Molecules 27, no. 9: 2681. https://doi.org/10.3390/molecules27092681
APA StyleCai, B., Wan, P., Chen, H., Huang, J., Ye, Z., Chen, D., & Pan, J. (2022). Purification and Identification of Novel Myeloperoxidase Inhibitory Antioxidant Peptides from Tuna (Thunnas albacares) Protein Hydrolysates. Molecules, 27(9), 2681. https://doi.org/10.3390/molecules27092681