Current Knowledge on Mammalian Phospholipase A1, Brief History, Structures, Biochemical and Pathophysiological Roles
Abstract
1. Introduction
2. History of PLA1 Research
3. Structural Evaluation of PLA1 Molecules
4. Extracellular PLA1/Lipase Family Members
4.1. Phosphatidylserine-Specific Phospholipase A1 (PS-PLA1)
4.1.1. Historical Aspects
4.1.2. Biochemical Characterization and Tissue Distribution
4.1.3. Structural Characteristics
4.1.4. Possible Functions
4.2. Membrane-Associated Phosphatidic Acid-Selective Phospholipase A1s (mPA-PLA1α/LIPH and mPA-PLA1β)
4.2.1. Historical Aspects
4.2.2. Biochemical Characterization and Distribution
4.2.3. Structural Characteristics
4.2.4. Function
4.3. Extracellular Lipases
4.3.1. Historical Aspects
4.3.2. Structural Characteristics
4.3.3. Functional Characteristics
5. Intracellular PLA1 Families
5.1. iPLA1 Family
5.1.1. Historical Aspects
5.1.2. Phosphatidic Acid-Preferring Phospholipase A1 (PA-PLA1)/DDHD1/iPLA1α
Characterization and Distribution
Substrate Specificity
Function
5.1.3. KIAA0725p/DDHD2, a Second Member of Intracellular PLA1
Historical Aspects
Characterization and Distribution
Function
5.2. Other Recently Identified Intracellular PLA1s
5.2.1. iPLA2/PNPLA Family
PNPLA6/iPLA2δ/NTE
- Historical Aspects
- Biochemical Characterization and Tissue Distribution
- Structural Characteristics
- Possible Functions
PNPLA7/iPLA2θ/NRE
- Historical Aspects
- Biochemical Characterization and Tissue Distribution
- Structural Characteristics
- Possible Functions
PNPLA8/iPLA2γ/group VIB PLA2
- Historical Aspects
- Biochemical Characterization and Tissue Distribution
- Structural Characteristics
- Possible Functions
5.2.2. cPLA2 Family
PLA2G16/group XVI PLA2/PLAAT3/HRASLS3/H-Rev107
- Historical Aspects
- Biochemical Characterization and Tissue Distribution
- Structural Characteristics
- Possible Functions
6. Conclusions and Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Dennis, E.A. The growing phospholipase A2 superfamily of signal transduction enzymes. Trends Biochem. Sci. 1997, 22, 1–2. [Google Scholar] [CrossRef]
- Murakami, M.; Nakatani, Y.; Atsumi, G.; Inoue, K.; Kudo, I. Regulatory functions of phospholipase A2. Crit. Rev. Immunol. 1997, 17, 225–283. [Google Scholar] [CrossRef] [PubMed]
- Lykidis, A.; Jackowski, S. Regulation of mammalian cell membrane biosynthesis. Prog. Nucleic Acid Res. Mol. Biol. 2001, 65, 361–393. [Google Scholar] [PubMed]
- Sonoda, H.; Aoki, J.; Hiramatsu, T.; Ishida, M.; Bandoh, K.; Nagai, Y.; Taguchi, R.; Inoue, K.; Arai, H. A novel phosphatidic acid-selective phospholipase A1 that produces lysophosphatidic acid. J. Biol. Chem. 2002, 277, 34254–34263. [Google Scholar] [CrossRef]
- Bandoh, K.; Aoki, J.; Tsujimoto, M.; Arai, H.; Inoue, K. Lysophosphatidic acid (LPA) receptors of the EDG family are differentially activated by LPA species. Structure-activity relationship of cloned LPA receptors. FEBS Lett. 2000, 478, 159–165. [Google Scholar] [CrossRef]
- Bandoh, K.; Aoki, J.; Hosono, H.; Kobayashi, S.; Kobayashi, T.; Murakami, M.K.; Tsujimoto, M.; Arai, H.; Inoue, K. Molecular cloning and characterization of a novel human G-protein-coupled receptor, EDG7, for lysophosphatidic acid. J. Biol. Chem. 1999, 274, 27776–27785. [Google Scholar] [CrossRef]
- Robinson, M.; Waite, M. Physical-chemical requirements for the catalysis of substrates by lysosomal phospholipase A1. J. Biol. Chem. 1983, 258, 14371–14378. [Google Scholar] [CrossRef]
- Kucera, G.L.; Miller, C.; Sisson, P.J.; Wilcox, R.W.; Wiemer, Z.; Waite, M. Hydrolysis of thioester analogs by rat liver phospholipase A1. J. Biol. Chem. 1988, 263, 12964–12969. [Google Scholar] [CrossRef]
- Ueda, H.; Kobayashi, T.; Kishimoto, M.; Tsutsumi, T.; Okuyama, H. A possible pathway of phosphoinositide metabolism through EDTA-insensitive phospholipase A1 followed by lysophosphoinositide-specific phospholipase C in rat brain. J. Neurochem. 1993, 61, 1874–1881. [Google Scholar] [CrossRef]
- Ueda, H.; Kobayashi, T.; Kishimoto, M.; Tsutsumi, T.; Watanabe, S.; Okuyama, H. The presence of Ca(2+)-independent phospholipase A1 highly specific for phosphatidylinositol in bovine brain. Biochem. Biophys. Res. Commun. 1993, 195, 1272–1279. [Google Scholar] [CrossRef]
- Ueda, H.; Kobayashi, T.; Kishimoto, M.; Tsutsumi, T.; Okuyama, H. EDTA-insensitive deacylation of phosphatidylinositol in porcine platelet membranes. Life Sci. 1993, 53, 629–634. [Google Scholar] [CrossRef]
- Hostetler, K.Y.; Gardner, M.F.; Giordano, J.R. Purification of lysosomal phospholipase A and demonstration of proteins that inhibit phospholipase A in a lysosomal fraction from rat kidney cortex. Biochemistry 1986, 25, 6456–6461. [Google Scholar] [CrossRef]
- Waite, M. The Phospholipases: Handbook of Lipid Research; Plenum Publishing Corp: New York, NY, USA, 1987; pp. 79–110. [Google Scholar]
- Van den Bosch, H. Phosphaolipases; Elsevier: Amsterdam, The Netherlands, 1982; pp. 313–351. [Google Scholar]
- Carriere, F.; Withers, M.C.T.H.; van Roussel, A.; Cambillau, C.; Verger, R. Structural basis for the substrate selectivity of pancreatic lipases and some related proteins. Biochim. Biophys. Acta 1998, 1376, 417–432. [Google Scholar] [CrossRef]
- Sato, T.; Aoki, J.; Nagai, Y.; Dohmae, N.; Takio, K.; Doi, T.; Arai, H.; Inoue, K. Serine phospholipid-specific phospholipase A that is secreted from activated platelets. A new member of the lipase family. J. Biol. Chem. 1997, 272, 2192–2198. [Google Scholar] [CrossRef]
- Nagai, Y.; Aoki, J.; Sato, T.; Amano, K.; Matsuda, Y.; Arai, H.; Inoue, K. An alternative splicing form of phosphatidylserine-specific phospholipase A1 that exhibits lysophosphatidylserine-specific lysophospholipase activity in humans. J. Biol. Chem. 1999, 274, 11053–11059. [Google Scholar] [CrossRef]
- Hiramatsu, T.; Sonoda, H.; Takanezawa, Y.; Morikawa, R.; Ishida, M.; Kasahara, K.; Sanai, Y.R.; Taguchi, J.; Aoki, H.A. Biochemical and molecular characterization of two phosphatidic acid-selective phospholipase A1s, mPA-PLA1alpha and mPA-PLA1beta. J. Biol. Chem. 2003, 278, 49438–49447. [Google Scholar] [CrossRef]
- Hide, W.A.; Chan, L.; Li, W.H. Structure and evolution of the lipase superfamily. J. Lipid. Res. 1992, 33, 167–178. [Google Scholar] [CrossRef]
- Winkler, F.K.; D’Arcy, A.; Hunziker, W. Structure of human pancreatic lipase. Nature 1990, 343, 771–774. [Google Scholar] [CrossRef]
- Carriere, F.; Thirstrup, K.; Hjorth, S.; Ferrato, F.; Nielsen, P.F.; Withers-Martinez, C.; Cambillau, C.; Boel, E.; Thim, L.; Vergerc, R. Pancreatic lipase structure-function relationships by domain exchange. Biochemistry 1997, 36, 239–248. [Google Scholar] [CrossRef]
- Higgs, H.N.; Glomset, J.A. Identification of a phosphatidic acid-preferring phospholipase A1 from bovine brain and testis. Proc. Natl. Acad. Sci. USA 1994, 91, 9574–9578. [Google Scholar] [CrossRef]
- Higgs, H.N.; Han, M.H.; Johnson, G.E.; Glomset, J.A. Cloning of a phosphatidic acid-preferring phospholipase A1 from bovine testis. J. Biol. Chem. 1998, 273, 5468–5477. [Google Scholar] [CrossRef]
- Nakajima, K.; Sonoda, H.; Mizoguchi, T.; Aoki, J.; Arai, H.; Nagahama, M.; Tagaya, M.; Tani, K. A novel phospholipase A1 with sequence homology to a mammalian Sec23p-interacting protein, p125. J. Biol. Chem. 2002, 277, 11329–11335. [Google Scholar] [CrossRef]
- Araki, M.; Ohshima, N.; Aso, C.; Konishi, A.; Obinata, H.; Tatei, K.; Izumi, T. Enzymatic characterization of recombinant rat DDHD2: A soluble diacylglycerol lipase. J. Biochem. 2016, 160, 269–279. [Google Scholar] [CrossRef]
- Inloes, J.M.; Hsu, K.L.; Dix, M.M.; Viader, A.; Masuda, K.; Takei, T.; Wood, M.R.; Cravatt, B.F. The hereditary spastic paraplegia-related enzyme DDHD2 is a principal brain triglyceride lipase. Proc. Natl. Acad. Sci. USA 2014, 111, 14924–14929. [Google Scholar] [CrossRef]
- Tani, K.; Mizoguchi, T.; Iwamatsu, A.; Hatsuzawa, K.; Tagaya, M. p125 is a novel mammalian Sec23p-interacting protein with structural similarity to phospholipid-modifying proteins. J. Biol. Chem. 1999, 274, 20505–20512. [Google Scholar] [CrossRef]
- Quistad, G.B.; Barlow, C.; Winrow, C.J.; Sparks, S.E.; Casida, J.E. Evidence that mouse brain neuropathy target esterase is a lysophospholipase. Proc. Natl. Acad. Sci. USA 2003, 100, 7983–7987. [Google Scholar] [CrossRef]
- Zaccheo, O.; Dinsdale, D.; Meacock, P.A.; Glynn, P. Neuropathy target esterase and its yeast homologue degrade phosphatidylcholine to glycerophosphocholine in living cells. J. Biol. Chem. 2004, 279, 24024–24033. [Google Scholar] [CrossRef]
- Muhlig-Versen, M.; da Cruz, A.B.; Tschape, J.A.; Moser, M.; Buttner, R.; Athenstaedt, K.; Glynn, P.; Kretzschmar, D. Loss of Swiss cheese/neuropathy target esterase activity causes disruption of phosphatidylcholine homeostasis and neuronal and glial death in adult Drosophila. J. Neurosci. 2005, 25, 2865–2873. [Google Scholar] [CrossRef]
- Kienesberger, P.C.; Lass, A.; Preiss-Landl, K.; Wolinski, H.; Kohlwein, S.D.; Zimmermann, R.; Zechner, R. Identification of an insulin-regulated lysophospholipase with homology to neuropathy target esterase. J. Biol. Chem. 2008, 283, 5908–5917. [Google Scholar] [CrossRef]
- Liu, X.; Moon, S.H.; Jenkins, C.M.; Sims, H.F.; Gross, R.W. Cyclooxygenase-2 Mediated Oxidation of 2-Arachidonoyl-Lysophospholipids Identifies Unknown Lipid Signaling Pathways. Cell Chem. Biol. 2016, 23, 1217–1227. [Google Scholar] [CrossRef]
- Mancuso, D.J.; Sims, H.F.; Han, X.; Jenkins, C.M.; Guan, S.P.; Yang, K.; Moon, S.H.; Pietka, T.; Abumrad, N.A.; Schlesinger, P.H.; et al. Genetic ablation of calcium-independent phospholipase A2gamma leads to alterations in mitochondrial lipid metabolism and function resulting in a deficient mitochondrial bioenergetic phenotype. J. Biol. Chem. 2007, 282, 34611–34622. [Google Scholar] [CrossRef] [PubMed]
- Pickard, R.T.; Strifler, B.A.; Kramer, R.M.; Sharp, J.D. Molecular cloning of two new human paralogs of 85-kDa cytosolic phospholipase A2. J. Biol. Chem. 1999, 274, 8823–8831. [Google Scholar] [CrossRef] [PubMed]
- Song, C.; Chang, X.J.; Bean, K.M.; Proia, M.S.; Knopf, J.L.; Kriz, R.W. Molecular characterization of cytosolic phospholipase A2-beta. J. Biol. Chem. 1999, 274, 17063–17067. [Google Scholar] [CrossRef] [PubMed]
- Uyama, T.; Morishita, J.; Jin, X.H.; Okamoto, Y.; Tsuboi, K.; Ueda, N. The tumor suppressor gene H-Rev107 functions as a novel Ca2+-independent cytosolic phospholipase A1/2 of the thiol hydrolase type. J. Lipid. Res. 2009, 50, 685–693. [Google Scholar] [CrossRef]
- Pete, M.J.; Ross, A.H.; Exton, J.H. Purification and properties of phospholipase A1 from bovine brain. J. Biol. Chem. 1994, 269, 19494–19500. [Google Scholar] [CrossRef]
- Uchiyama, S.; Miyazaki, Y.; Amakasu, Y.; Kuwata, H.; Nakatani, Y.; Atsumi, G.; Murakami, M.; Kudo, I. Characterization of heparin low-affinity phospholipase A1 present in brain and testicular tissue. J. Biochem. 1999, 125, 1001–1010. [Google Scholar] [CrossRef]
- Horigome, K.; Hayakawa, M.; Inoue, K.; Nojima, S. Selective release of phospholipase A2 and lysophosphatidylserine-specific lysophospholipase from rat platelets. J. Biochem. 1987, 101, 53–61. [Google Scholar] [CrossRef]
- Horigome, K.; Hayakawa, M.; Inoue, K.; Nojima, S. Purification and characterization of phospholipase A2 released from rat platelets. J. Biochem. 1987, 101, 625–631. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Higashi, S.; Kobayashi, T.; Kudo, I.; Inoue, K. Purification and characterization of lysophospholipase released from rat platelets. J. Biochem. 1988, 103, 442–447. [Google Scholar] [CrossRef]
- Aoki, J.; Nagai, Y.; Hosono, H.; Inoue, K.; Arai, H. Structure and function of phosphatidylserine-specific phospholipase A1. Biochim. Biophys. Acta 2002, 1582, 26–32. [Google Scholar] [CrossRef]
- Nakamura, K.; Igarashi, K.; Ohkawa, R.; Saiki, N.; Nagasaki, M.; Uno, K.; Hayashi, N.; Sawada, T.; Syukuya, K.; Yokota, H.; et al. A novel enzyme immunoassay for the determination of phosphatidylserine-specific phospholipase A(1) in human serum samples. Clin. Chim. Acta 2010, 411, 1090–1094. [Google Scholar] [CrossRef]
- Sawada, T.; Kurano, M.; Shirai, H.; Iwasaki, Y.; Tahara, K.; Hayashi, H.; Igarashi, K.; Fujio, K.; Aoki, J.; Yatomi, Y. Serum phosphatidylserine-specific phospholipase A 1 as a novel biomarker for monitoring systemic lupus erythematosus disease activity. Int. J. Rheum. Dis. 2019, 22, 2059–2066. [Google Scholar] [CrossRef]
- Yokoyama, K.; Kudo, I.; Inoue, K. Phospholipid degradation in rat calcium ionophore-activated platelets is catalyzed mainly by two discrete secretory phospholipase As. J. Biochem. 1995, 117, 1280–1287. [Google Scholar] [CrossRef]
- Martin, T.W.; Lagunoff, D. Interactions of lysophospholipids and mast cells. Nature 1979, 279, 250–252. [Google Scholar] [CrossRef]
- Tamori, N.Y.; Horigome, K.; Inoue, K.; Nojima, S. Metabolism of lysophosphatidylserine, a potentiator of histamine release in rat mast cells. J. Biochem. 1986, 100, 581–590. [Google Scholar] [CrossRef]
- Horigome, K.; Tamori, N.Y.; Inoue, K.; Nojima, S. Effect of serine phospholipid structure on the enhancement of concanavalin A-induced degranulation in rat mast cells. J. Biochem. 1986, 100, 571–579. [Google Scholar] [CrossRef]
- Bruni, A.; Monastra, G.; Bellini, F.; Toffano, G. Autacoid properties of lysophosphatidylserine. Prog. Clin. Biol. Res. 1988, 282, 165–179. [Google Scholar]
- Hosono, H.; Aoki, J.; Nagai, Y.; Bandoh, K.; Ishida, M.; Taguchi, R.; Arai, H.; Inoue, K. Phosphatidylserine-specific phospholipase A1 stimulates histamine release from rat peritoneal mast cells through production of 2-acyl-1-lysophosphatidylserine. J. Biol. Chem. 2001, 276, 29664–29670. [Google Scholar] [CrossRef]
- Hecht, J.H.; Weiner, J.A.; Post, S.R.; Chun, J. Ventricular zone gene-1 (vzg-1) encodes a lysophosphatidic acid receptor expressed in neurogenic regions of the developing cerebral cortex. J. Cell Biol. 1996, 135, 1071–1083. [Google Scholar] [CrossRef]
- An, S.; Bleu, T.; Hallmark, O.G.; Goetzl, E.J. Characterization of a novel subtype of human G protein-coupled receptor for lysophosphatidic acid. J. Biol. Chem. 1998, 273, 7906–7910. [Google Scholar] [CrossRef]
- Hayashi, K.; Takahashi, M.; Nishida, W.; Yoshida, K.; Ohkawa, Y.; Kitabatake, A.; Aoki, J.; Arai, H.; Sobue, K. Phenotypic modulation of vascular smooth muscle cells induced by unsaturated lysophosphatidic acids. Circ. Res. 2001, 89, 251–258. [Google Scholar] [CrossRef]
- Tokumura, A.; Iimori, M.; Nishioka, Y.; Kitahara, M.; Sakashita, M.; Tanaka, S. Lysophosphatidic acids induce proliferation of cultured vascular smooth muscle cells from rat aorta. Am. J. Physiol. 1994, 267, C204–C210. [Google Scholar] [CrossRef]
- Kurano, M.; Suzuki, A.; Inoue, A.; Tokuhara, Y.; Kano, K.; Matsumoto, H.; Igarashi, K.; Ohkawa, R.; Nakamura, K.; Dohi, T.; et al. Possible involvement of minor lysophospholipids in the increase in plasma lysophosphatidic acid in acute coronary syndrome. Arterioscler. Thromb. Vasc. Biol. 2015, 35, 463–470. [Google Scholar] [CrossRef]
- Aoki, J.; Inoue, A.; Okudaira, S. Two pathways for lysophosphatidic acid production. Biochim. Biophys Acta 2008, 1781, 513–518. [Google Scholar] [CrossRef]
- Inoue, A.; Arima, N.; Ishiguro, J.; Prestwich, G.D.; Arai, H.; Aoki, J. LPA-producing enzyme PA-PLA1α regulates hair follicle development by modulating EGFR signalling. EMBO J. 2011, 30, 4248–4260. [Google Scholar] [CrossRef]
- Foell, J.L.; Hesse, M.; Volkmer, I.; Schmiedel, B.J.; Neumann, I.; Staege, M.S. Membrane-associated phospholipase A1 beta (LIPI) Is an Ewing tumour-associated cancer/testis antigen. Pediatr. Blood Cancer 2008, 51, 228–234. [Google Scholar] [CrossRef]
- Kazantseva, A.; Goltsov, A.; Zinchenko, R.; Grigorenko, A.P.; Abrukova, A.V.; Moliaka, Y.K.; Kirillov, A.G.; Guo, Z.; Lyle, S.; Ginter, E.K.; et al. Human hair growth deficiency is linked to a genetic defect in the phospholipase gene LIPH. Science 2006, 314, 982–985. [Google Scholar] [CrossRef]
- Shimomura, Y.; Wajid, M.; Ishii, Y.; Shapiro, L.; Petukhova, L.; Gordon, D.; Christiano, A.M. Disruption of P2RY5, an orphan G protein-coupled receptor, underlies autosomal recessive woolly hair. Nat. Genet. 2008, 40, 335–339. [Google Scholar] [CrossRef]
- Pasternack, S.M.; von Kügelgen, I.; al Aboud, K.; Lee, T.A.; Rüschendorf, F.; Voss, K.; Hillmer, A.M.; Molderings, G.J.; Franz, T.; Ramirez, A.; et al. G protein-coupled receptor P2Y5 and its ligand LPA are involved in maintenance of human hair growth. Nat. Genet. 2008, 40, 329–334. [Google Scholar] [CrossRef]
- Yanagida, K.; Masago, K.; Nakanishi, H.; Kihara, Y.; Hamano, F.; Tajima, Y.; Taguchi, R.; Shimizu, T.; Ishii, S. Identification and characterization of a novel lysophosphatidic acid receptor, p2y5/LPA6. J. Biol. Chem. 2009, 284, 17731–17741. [Google Scholar] [CrossRef] [PubMed]
- Soldatova, L.; Kochoumian, L.; King, T.P. Sequence similarity of a hornet (D. maculata) venom allergen phospholipase A1 with mammalian lipases. FEBS Lett. 1993, 320, 145–149. [Google Scholar] [CrossRef]
- Shamburek, R.D.; Zech, L.A.; Cooper, P.S.; Vandenbroek, J.M.; Schwartz, C.C. Disappearance of two major phosphatidylcholines from plasma is predominantly via LCAT and hepatic lipase. Am. J. Physiol. 1996, 271, E1073–E1082. [Google Scholar] [CrossRef] [PubMed]
- MMcCoy, G.; Sun, G.S.; Marchadier, D.; Maugeais, C.; Glick, J.M.; Rader, D.J. Characterization of the lipolytic activity of endothelial lipase. J. Lipid. Res. 2002, 43, 921–929. [Google Scholar] [CrossRef]
- Higgs, H.N.; Glomset, J.A. Purification and properties of a phosphatidic acid-preferring phospholipase A1 from bovine testis. Examination of the molecular basis of its activation. J. Biol. Chem. 1996, 271, 10874–10883. [Google Scholar] [CrossRef]
- Barlowe, C.; Schekman, R. SEC12 encodes a guanine-nucleotide-exchange factor essential for transport vesicle budding from the ER. Nature 1993, 365, 347–349. [Google Scholar] [CrossRef]
- Han, M.H.; Han, D.K.; Aebersold, R.H.; Glomset, J.A. Effects of protein kinase CK2, extracellular signal-regulated kinase 2, and protein phosphatase 2A on a phosphatidic acid-preferring phospholipase A1. J. Biol. Chem. 2001, 276, 27698–27708. [Google Scholar] [CrossRef]
- Matsumoto, N.; Nemoto-Sasaki, Y.; Oka, S.; Arai, S.; Wada, I.; Yamashita, A. Phosphorylation of human phospholipase A1 DDHD1 at newly identified phosphosites affects its subcellular localization. J. Biol. Chem. 2021, 297, 100851. [Google Scholar] [CrossRef]
- Miyazawa, D.; Ikemoto, A.; Fujii, Y.; Okuyama, H. Partial purification and characterization of phosphatidic acid-specific phospholipase A(1) in porcine platelet membranes. Biochim. Biophys. Acta 2003, 1631, 17–25. [Google Scholar] [CrossRef]
- Pete, M.J.; Exton, J.H. Phospholipid interactions affect substrate hydrolysis by bovine brain phospholipase A1. Biochim. Biophys. Acta 1995, 1256, 367–373. [Google Scholar] [CrossRef]
- Giudice, T.L.; Lombardi, F.; Santorelli, F.M.; Kawarai, T.; Orlacchio, A. Hereditary spastic paraplegia: Clinical-genetic characteristics and evolving molecular mechanisms. Exp. Neurol. 2014, 261, 518–539. [Google Scholar] [CrossRef]
- Tesson, C.; Nawara, M.; Salih, M.A.; Rossignol, R.; Zaki, M.S.; al Balwi, M.; Schule, R.; Mignot, C.; Obre, E.; Bouhouche, A.; et al. Alteration of fatty-acid-metabolizing enzymes affects mitochondrial form and function in hereditary spastic paraplegia. Am. J. Hum. Genet. 2012, 91, 1051–1064. [Google Scholar] [CrossRef]
- Yamashita, A.; Kumazawa, T.; Koga, H.; Suzuki, N.; Oka, S.; Sugiura, T. Generation of lysophosphatidylinositol by DDHD domain containing 1 (DDHD1): Possible involvement of phospholipase D/phosphatidic acid in the activation of DDHD1. Biochim. Biophys. Acta 2010, 1801, 711–720. [Google Scholar] [CrossRef]
- Inloes, J.M.; Jing, H.; Cravatt, B.F. The Spastic Paraplegia-Associated Phospholipase DDHD1 Is a Primary Brain Phosphatidylinositol Lipase. Biochemistry 2018, 57, 5759–5767. [Google Scholar] [CrossRef]
- Baba, T.; Kashiwagi, Y.; Arimitsu, N.; Kogure, T.; Edo, A.; Maruyama, T.; Nakao, K.; Nakanishi, H.; Kinoshita, M.; Frohman, M.A.; et al. Phosphatidic acid (PA)-preferring phospholipase A1 regulates mitochondrial dynamics. J. Biol. Chem. 2014, 289, 11497–11511. [Google Scholar] [CrossRef]
- Morikawa, R.K.; Aoki, J.; Kano, F.; Murata, M.; Yamamoto, A.; Tsujimoto, M.; Arai, H. Intracellular phospholipase A1gamma (iPLA1gamma) is a novel factor involved in coat protein complex I- and Rab6-independent retrograde transport between the endoplasmic reticulum and the Golgi complex. J. Biol. Chem. 2009, 284, 26620–26630. [Google Scholar] [CrossRef]
- Gonzalez, M.; Nampoothiri, S.; Kornblum, C.; Oteyza, A.C.; Walter, J.; Konidari, I.; Hulme, W.; Speziani, F.; Schöls, L.; Züchner, S.; et al. Mutations in phospholipase DDHD2 cause autosomal recessive hereditary spastic paraplegia (SPG54). Eur. J. Hum. Genet. 2013, 21, 1214–1218. [Google Scholar] [CrossRef]
- Maruyama, T.; Baba, T.; Maemoto, Y.; Hara-Miyauchi, C.; Hasegawa-Ogawa, M.; Enda, Y.O.; Matsumoto, K.; Arimitsu, N.; Nakao, K.; Hamamoto, H.; et al. Loss of DDHD2, whose mutation causes spastic paraplegia, promotes reactive oxygen species generation and apoptosis. Cell Death Dis. 2018, 9, 797. [Google Scholar] [CrossRef]
- Kienesberger, P.C.; Oberer, M.; Lass, A.; Zechner, R. Mammalian patatin domain containing proteins: A family with diverse lipolytic activities involved in multiple biological functions. J. Lipid. Res. 2009, 50, S63–S68. [Google Scholar] [CrossRef]
- Winrow, C.J.; Hemming, M.L.; Allen, D.M.; Quistad, G.B.; Casida, J.E.; Barlow, C. Loss of neuropathy target esterase in mice links organophosphate exposure to hyperactivity. Nat. Genet. 2003, 33, 477–485. [Google Scholar] [CrossRef]
- Moser, M.; Li, Y.; Vaupel, K.; Kretzschmar, D.; Kluge, R.; Glynn, P.; Buettner, R. Placental failure and impaired vasculogenesis result in embryonic lethality for neuropathy target esterase-deficient mice. Mol. Cell. Biol. 2004, 24, 1667–1679. [Google Scholar] [CrossRef]
- Akassoglou, K.; Malester, B.; Xu, J.; Tessarollo, L.; Rosenbluth, J.; Chao, M.V. Brain-specific deletion of neuropathy target esterase/swisscheese results in neurodegeneration. Proc. Natl. Acad. Sci. USA 2004, 101, 5075–5080. [Google Scholar] [CrossRef]
- Rainier, S.; Bui, M.; Mark, E.; Thomas, D.; Tokarz, D.; Ming, L.; Delaney, C.E.; Richardson, R.J.; Albers, J.W.; Matsunami, N.; et al. Neuropathy target esterase gene mutations cause motor neuron disease. Am. J. Hum. Genet. 2008, 82, 780–785. [Google Scholar] [CrossRef]
- Synofzik, M.; Gonzalez, M.A.; Lourenco, C.M.; Coutelier, M.; Haack, T.B.; Rebelo, A.; Hannequin, D.; Strom, T.M.; Prokisch, H.; Kernstock, C.; et al. PNPLA6 mutations cause Boucher-Neuhauser and Gordon Holmes syndromes as part of a broad neurodegenerative spectrum. Brain 2014, 137, 69–77. [Google Scholar] [CrossRef]
- Kmoch, S.; Majewski, J.; Ramamurthy, V.; Cao, S.; Fahiminiya, S.; Ren, H.; MacDonald, I.M.; Lopez, I.; Sun, V.; Keser, V.; et al. Mutations in PNPLA6 are linked to photoreceptor degeneration and various forms of childhood blindness. Nat. Commun. 2015, 6, 5614. [Google Scholar] [CrossRef]
- Gallazzini, M.; Ferraris, J.D.; Kunin, M.; Morris, R.G.; Burg, M.B. Neuropathy target esterase catalyzes osmoprotective renal synthesis of glycerophosphocholine in response to high NaCl. Proc. Natl. Acad. Sci. USA 2006, 103, 15260–15265. [Google Scholar] [CrossRef]
- Richardson, R.J.; Hein, N.D.; Wijeyesakere, S.J.; Fink, J.K.; Makhaeva, G.F. Neuropathy target esterase (NTE): Overview and future. Chem. Biol. Interact. 2013, 203, 238–244. [Google Scholar] [CrossRef]
- Heier, C.; Huang, B.K.F.; Eichmann, T.O.; Xie, H.; Zechner, R.; Chang, P. The phospholipase PNPLA7 functions as a lysophosphatidylcholine hydrolase and interacts with lipid droplets through its catalytic domain. J. Biol. Chem. 2017, 292, 19087–19098. [Google Scholar] [CrossRef] [PubMed]
- Moon, S.H.; Jenkins, C.M.; Liu, X.; Guan, S.; Mancuso, D.J.; Gross, R.W. Activation of mitochondrial calcium-independent phospholipase A2γ (iPLA2γ) by divalent cations mediating arachidonate release and production of downstream eicosanoids. J. Biol. Chem. 2012, 287, 14880–14895. [Google Scholar] [CrossRef] [PubMed]
- Hazen, S.L.; Stuppy, R.J.; Gross, R.W. Purification and characterization of canine myocardial cytosolic phospholipase A2. A calcium-independent phospholipase with absolute f1-2 regiospecificity for diradyl glycerophospholipids. J. Biol. Chem. 1990, 265, 10622–10630. [Google Scholar] [CrossRef]
- Jabůrek, M.; Průchová, P.; Holendová, B.; Galkin, A.; Ježek, P. Antioxidant Synergy of Mitochondrial Phospholipase PNPLA8/iPLA2γ with Fatty Acid-Conducting SLC25 Gene Family Transporters. Antioxidants 2021, 10, 678. [Google Scholar] [CrossRef]
- Mancuso, D.J.; Jenkins, C.M.; Sims, H.F.; Cohen, J.M.; Yang, J.; Gross, R.W. Complex transcriptional and translational regulation of iPLAgamma resulting in multiple gene products containing dual competing sites for mitochondrial or peroxisomal localization. Eur. J. Biochem. 2004, 271, 4709–4724. [Google Scholar] [CrossRef]
- Murakami, M.; Masuda, S.; Ueda-Semmyo, K.; Yoda, E.; Kuwata, H.; Takanezawa, Y.; Aoki, J.; Arai, H.; Sumimoto, H.; Ishikawa, Y.; et al. Group VIB Ca2+-independent phospholipase A2gamma promotes cellular membrane hydrolysis and prostaglandin production in a manner distinct from other intracellular phospholipases A2. J. Biol. Chem. 2005, 280, 14028–14041. [Google Scholar] [CrossRef]
- Liu, G.Y.; Moon, S.H.; Jenkins, C.M.; Li, M.; Sims, H.F.; Guan, S.; Gross, R.W. The phospholipase iPLA 2 γ is a major mediator releasing oxidized aliphatic chains from cardiolipin, integrating mitochondrial bioenergetics and signaling. J. Biol. Chem. 2017, 292, 10672–10684. [Google Scholar] [CrossRef]
- Saunders, C.J.; Moon, S.H.; Liu, X.; Thiffault, I.; Coffman, K.; LePichon, J.B.; Taboada, E.; Smith, L.D.; Farrow, E.G.; Miller, N.; et al. Loss of function variants in human PNPLA8 encoding calcium-independent phospholipase A2 γ recapitulate the mitochondriopathy of the homologous null mouse. Hum. Mutat. 2015, 36, 301–306. [Google Scholar] [CrossRef]
- Moon, S.H.; Mancuso, D.J.; Sims, H.F.; Liu, X.; Nguyen, A.L.; Yang, K.; Guan, S.; Dilthey, B.G.; Jenkins, C.M.; Weinheimer, C.J.; et al. Cardiac Myocyte-specific Knock-out of Calcium-independent Phospholipase A2γ (iPLA2γ) Decreases Oxidized Fatty Acids during Ischemia/Reperfusion and Reduces Infarct Size. J. Biol. Chem. 2016, 291, 19687–19700. [Google Scholar] [CrossRef]
- Dessen, A.; Tang, J.; Schmidt, H.; Stahl, M.; Clark, J.D.; Seehra, J.; Somers, W.S. Crystal structure of human cytosolic phospholipase A2 reveals a novel topology and catalytic mechanism. Cell 1999, 97, 349–360. [Google Scholar] [CrossRef]
- Hajnal, A.; Klemenz, R.; Schäfer, R. Subtraction cloning of H-rev107, a gene specifically expressed in H-ras resistant fibroblasts. Oncogene 1994, 9, 479–490. [Google Scholar]
- Sers, C.; Emmenegger, U.; Husmann, K.; Bucher, K.; Andres, A.C.; Schäfer, R. Growth-inhibitory activity and downregulation of the class II tumor-suppressor gene H-rev107 in tumor cell lines and experimental tumors. J. Cell. Biol. 1997, 136, 935–944. [Google Scholar] [CrossRef]
- Shinohara, N.; Uyama, T.; Jin, X.H.; Tsuboi, K.; Tonai, T.; Houchi, H.; Ueda, N. Enzymological analysis of the tumor suppressor A-C1 reveals a novel group of phospholipid-metabolizing enzymes. J. Lipid. Res. 2011, 52, 1927–1935. [Google Scholar] [CrossRef]
- Duncan, E.; Sarkadi-Nagy, E.; Jaworski, K.; Ahmadian, M.; Sul, H.S. Identification and functional characterization of adipose-specific phospholipase A2 (AdPLA). J. Biol. Chem. 2008, 283, 25428–25436. [Google Scholar] [CrossRef] [PubMed]
- Golczak, M.; Kiser, P.D.; Sears, A.E.; Lodowski, D.T.; Blaner, W.S.; Palczewski, K. Structural basis for the acyltransferase activity of lecithin:retinol acyltransferase-like proteins. J. Biol. Chem. 2012, 287, 23790–23807. [Google Scholar] [CrossRef] [PubMed]
- Jaworski, K.; Ahmadian, M.; Duncan, R.E.; Sarkadi-Nagy, E.; Varady, K.A.; Hellerstein, M.K.; Lee, H.Y.; Samuel, V.T.; Shulman, G.I.; Kim, K.H.; et al. AdPLA ablation increases lipolysis and prevents obesity induced by high-fat feeding or leptin deficiency. Nat. Med. 2009, 15, 159–168. [Google Scholar] [CrossRef] [PubMed]
- Xiong, S.; Tu, H.; Kollareddy, M.; Pant, V.; Li, Q.; Zhang, Y.; Jackson, J.G.; Suh, Y.A.; Elizondo-Fraire, A.C.; Yang, P.; et al. Pla2g16 phospholipase mediates gain-of-function activities of mutant p53. Proc. Natl. Acad. Sci. USA 2014, 111, 11145–11150. [Google Scholar] [CrossRef] [PubMed]
- Golczak, M.; Sears, A.E.; Kiser, P.D.; Palczewski, K. LRAT-specific domain facilitates vitamin A metabolism by domain swapping in HRASLS3. Nat. Chem. Biol. 2015, 11, 26–32. [Google Scholar] [CrossRef]
- Uyama, T.; Kawai, K.; Kono, N.; Watanabe, M.; Tsuboi, K.; Inoue, T.; Araki, N.; Arai, H.; Ueda, N. Interaction of Phospholipase A/Acyltransferase-3 with Pex19p: A POSSIBLE INVOLVEMENT IN THE DOWN-REGULATION OF PEROXISOMES. J. Biol. Chem. 2015, 290, 17520–17534. [Google Scholar] [CrossRef]
- Morishita, H.; Eguchi, T.; Tsukamoto, S.; Sakamaki, Y.; Takahashi, S.; Saito, C.; Koyama-Honda, I.; Mizushima, N. Organelle degradation in the lens by PLAAT phospholipases. Nature 2021, 592, 634–638. [Google Scholar] [CrossRef]
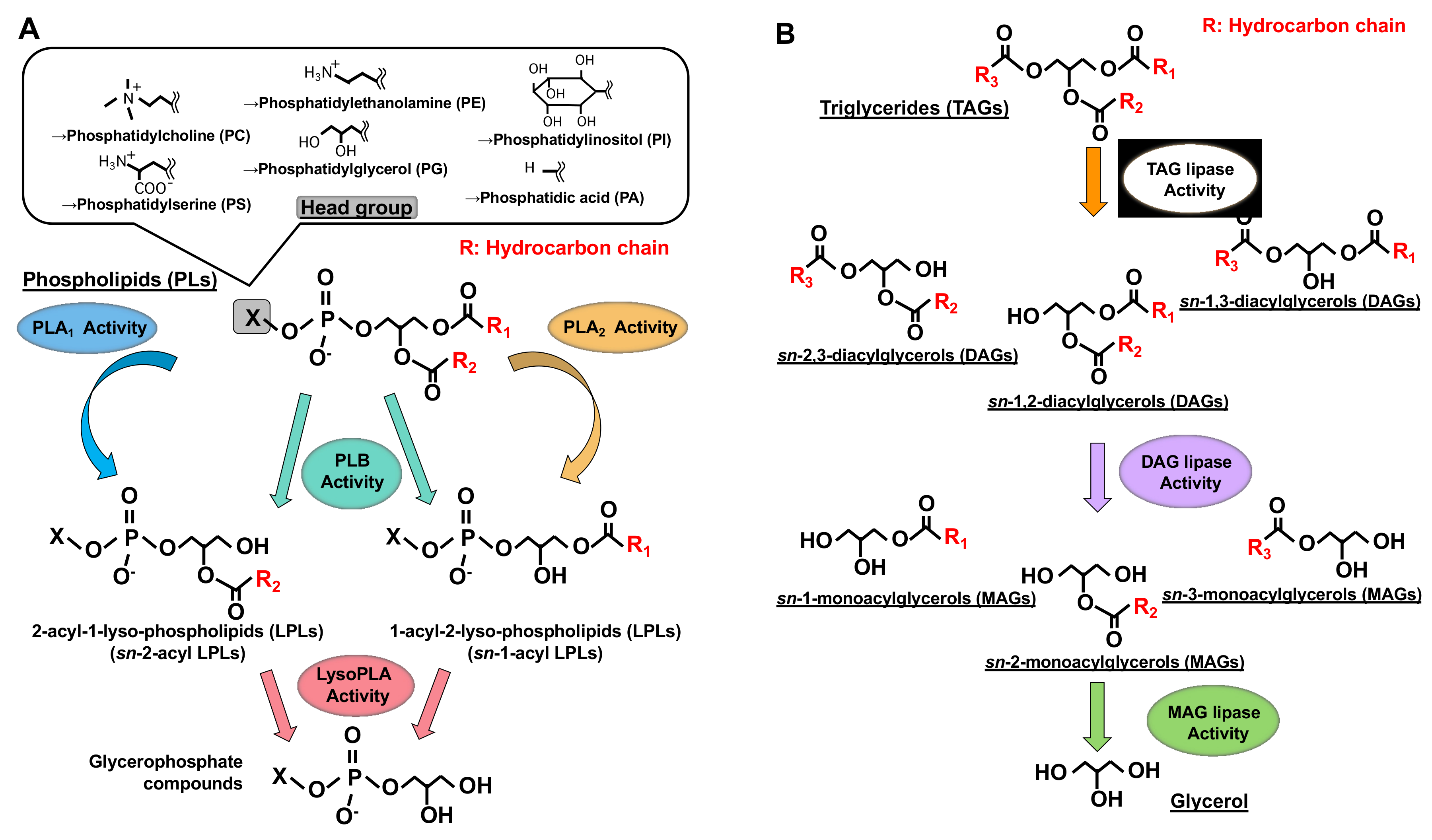

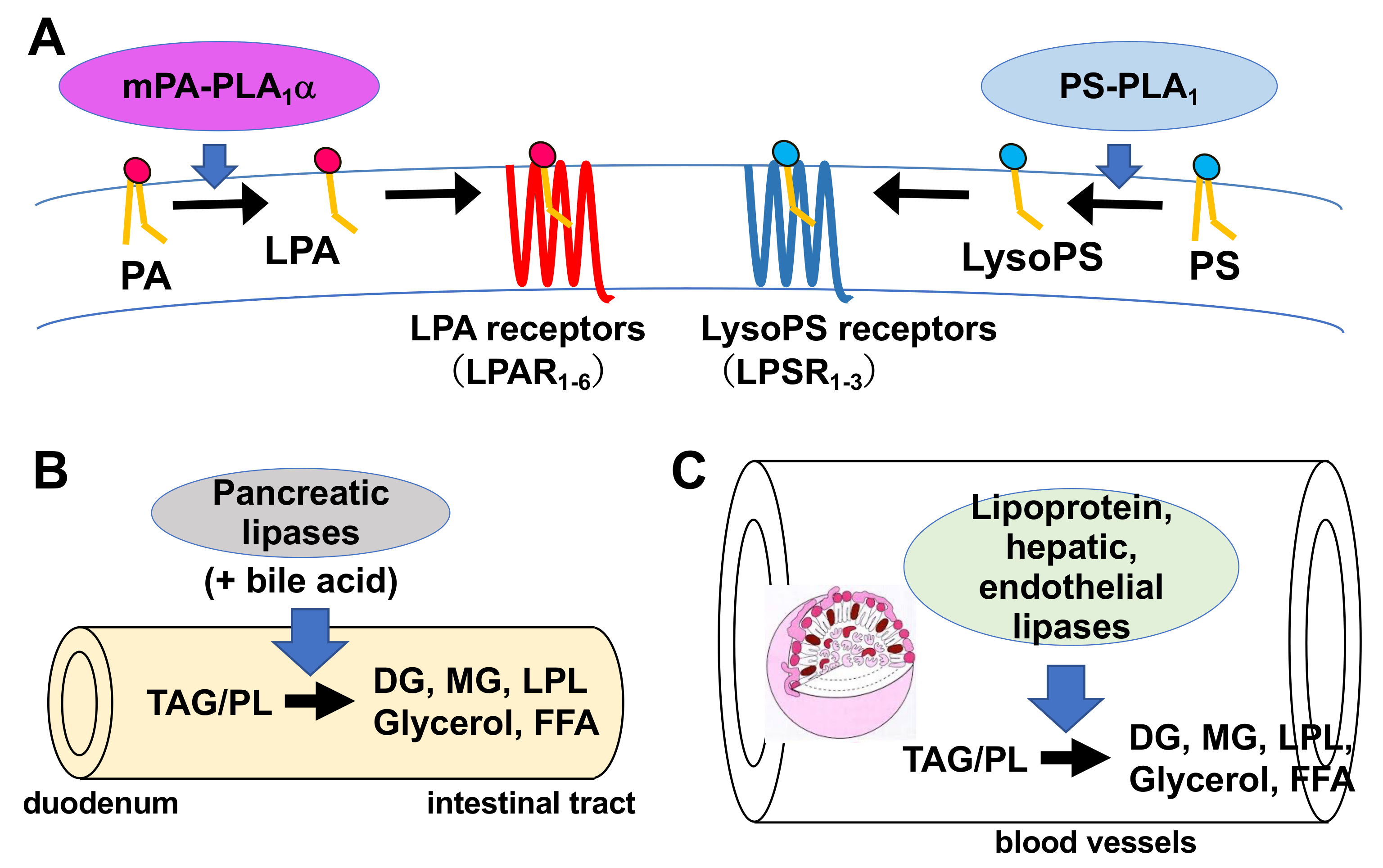
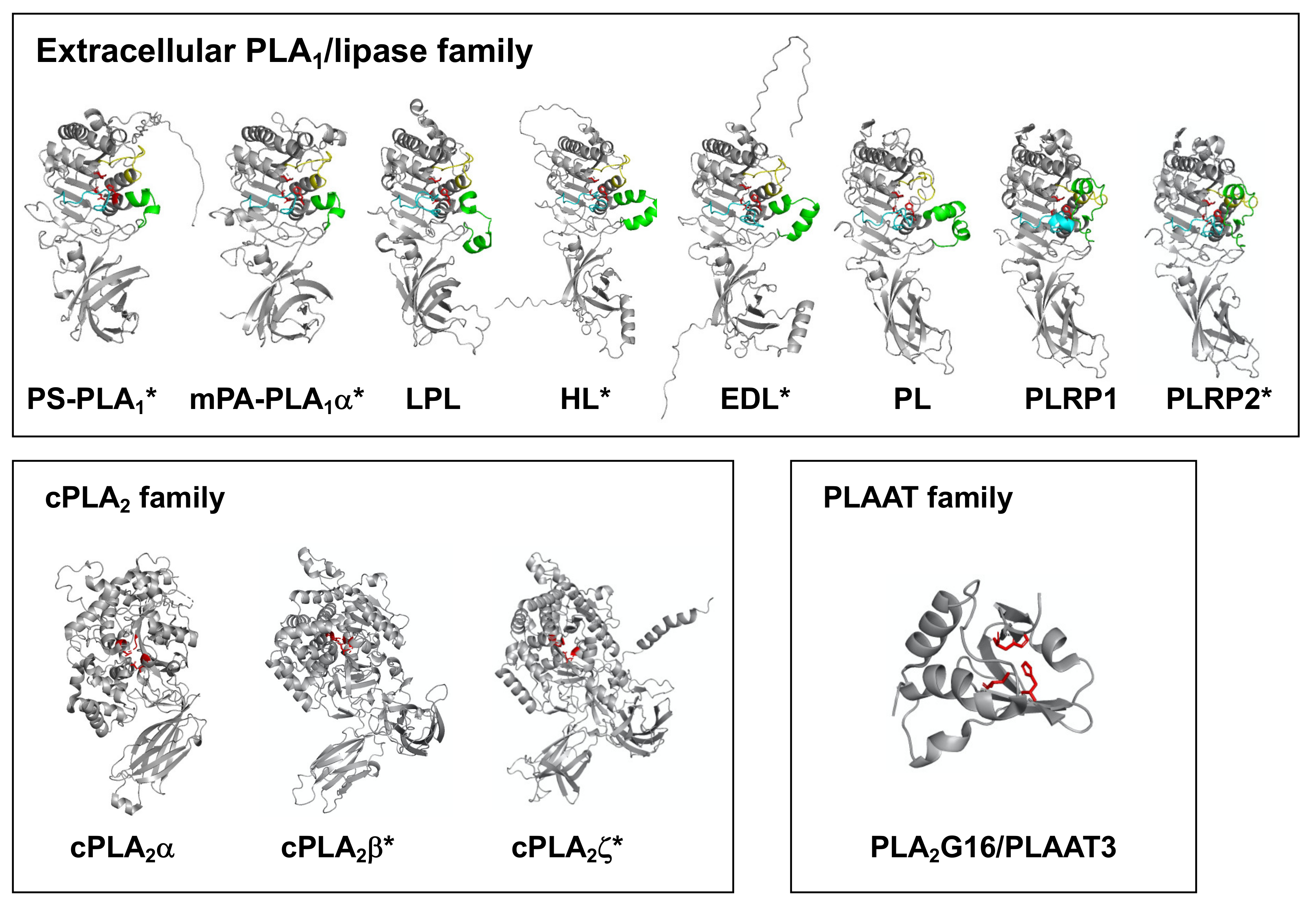
| Primary Name | Other Name | Human Gene | Substrate | Reaction Mediated | PDB ID (H: Human, R: Rat) | Ref. | |
|---|---|---|---|---|---|---|---|
| extracellular PLA1s | PS-PLA1 | PLA1A | PLA1A | PS | Producing enzyme for bioactive lysophospholipid, LysoPS | - | [16,17] |
| PA-PLA1α | LIPH, mPA-PLA1α | LIPH | PA | Producing enzyme for bioactive lysophospholipid, LPA | - | [4,18] | |
| lipoprotein lipase | LPL, LIPD | LIPD | TAG, PL | TAG lipase and PLA1 activity | H: 6E7K, 6OAU, 6OAZ, 6OB0, 6WN4 | [19] | |
| hepatic lipase | HL, LIPC | LIPC | TAG, PL | TAG lipase and PLA1 activity | - | [19] | |
| endothelial cell-derived lipase | EDL, EL, LIPG | LIPG | PL | Predominant PLA1 activity | - | ||
| pacreatic lipase | PL, PNLIP | PNLIP | TAG, PL | TAG lipase and PLA1 activity | H: 1GPL, 1LPA, 1LPB, 1N8S | [15,20,21] | |
| pancreatic lipase-related protein 1 | PLRP1 | PLRP1 | TAG, PL | TAG lipase and PLA1 activity | H: 2PPL | ||
| pancreatic lipase-related protein 2 | PLRP2 | PLRP2 | TAG, PL | TAG lipase and PLA1 activity | H: 2OXE, 2PVS; R: 1BU8 | [15,21] | |
| intracellular PLA1s | PA-PLA1 | DDHD1, iPLA1α | DDHD1 | PL | PLA1 activity | - | [22,23] |
| KIAA0725p | DDHD2, iPLA1γ | DDHD2 | PL | PE, DAG, CL | - | [24,25,26] | |
| p125 | iPLA1β | P125 | n.d. | Enzymatic activity has not been detected | - | [27] | |
| PNPLA6 | iPLA2δ, NTE | PNPLA6 | PC, LPC | PLB, LysoPLA activity cleaving FAs at both sn-1 and sn-2 positions | - | [28,29,30] | |
| PNPLA7 | iPLA2θ, NRE | PNPLA7 | PC, LPC | PLB, LysoPLA activity cleaving FAs at both sn-1 and sn-2 positions | - | [31] | |
| PNPLA8 | iPLA2γ, Group VIB PLA2 | PNPLA8 | PC | PLB activity cleaving FAs at both sn-1 and sn-2 positions | - | [32,33] | |
| cPLA2α | PLA2G4A, Group IVA PLA2 | PLA2G4A | PL | PLB activity cleaving FAs at both sn-1 and sn-2 positions | H: 1BCI, 1CJY, 1RLW | [34] | |
| cPLA2β | PLA2G4B, Group IVB PLA2 | PLA2G4B | PL | PLB activity cleaving FAs at both sn-1 and sn-2 positions | - | [34,35] | |
| cPLA2ζ | PLA2G4F, Group IVF PLA2 | PLA2G4F | PL | PLB activity cleaving FAs at both sn-1 and sn-2 positions | - | ||
| PLA2G16 | Group XVI PLA2, PLAAT3, HRASLS3, H-Rev107 | PLA2G16 | PL | PLB activity cleaving FAs at both sn-1 and sn-2 positions | H: 2KYT, 4DOT, 4FA0, 4Q95, 7C3Z, 7C41 | [36] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yaginuma, S.; Kawana, H.; Aoki, J. Current Knowledge on Mammalian Phospholipase A1, Brief History, Structures, Biochemical and Pathophysiological Roles. Molecules 2022, 27, 2487. https://doi.org/10.3390/molecules27082487
Yaginuma S, Kawana H, Aoki J. Current Knowledge on Mammalian Phospholipase A1, Brief History, Structures, Biochemical and Pathophysiological Roles. Molecules. 2022; 27(8):2487. https://doi.org/10.3390/molecules27082487
Chicago/Turabian StyleYaginuma, Shun, Hiroki Kawana, and Junken Aoki. 2022. "Current Knowledge on Mammalian Phospholipase A1, Brief History, Structures, Biochemical and Pathophysiological Roles" Molecules 27, no. 8: 2487. https://doi.org/10.3390/molecules27082487
APA StyleYaginuma, S., Kawana, H., & Aoki, J. (2022). Current Knowledge on Mammalian Phospholipase A1, Brief History, Structures, Biochemical and Pathophysiological Roles. Molecules, 27(8), 2487. https://doi.org/10.3390/molecules27082487





