Structure Elucidation of Triterpenoid Saponins Found in an Immunoadjuvant Preparation of Quillaja brasiliensis Using Mass Spectrometry and 1H and 13C NMR Spectroscopy
Abstract
:1. Introduction
2. Results and Discussion
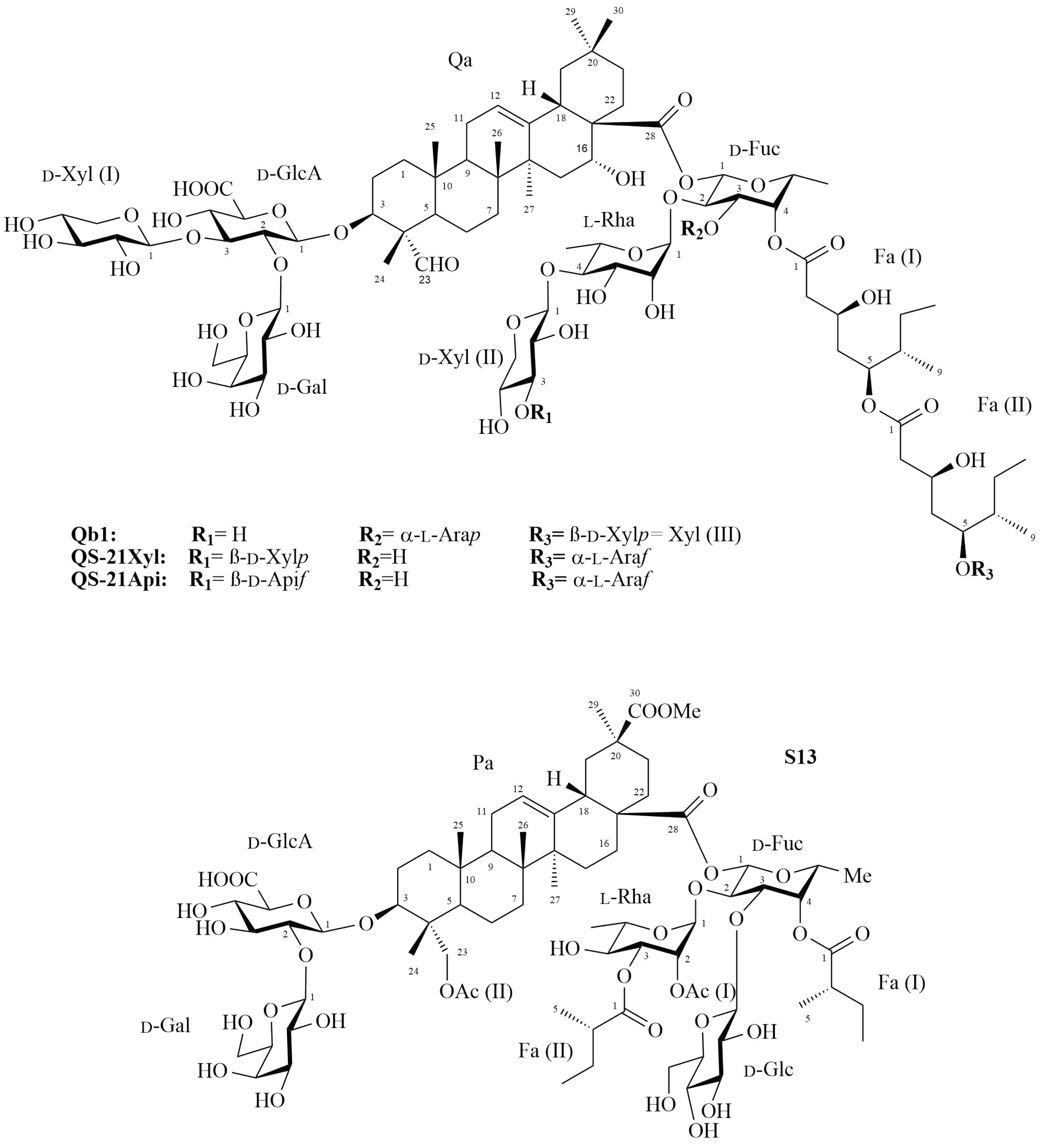
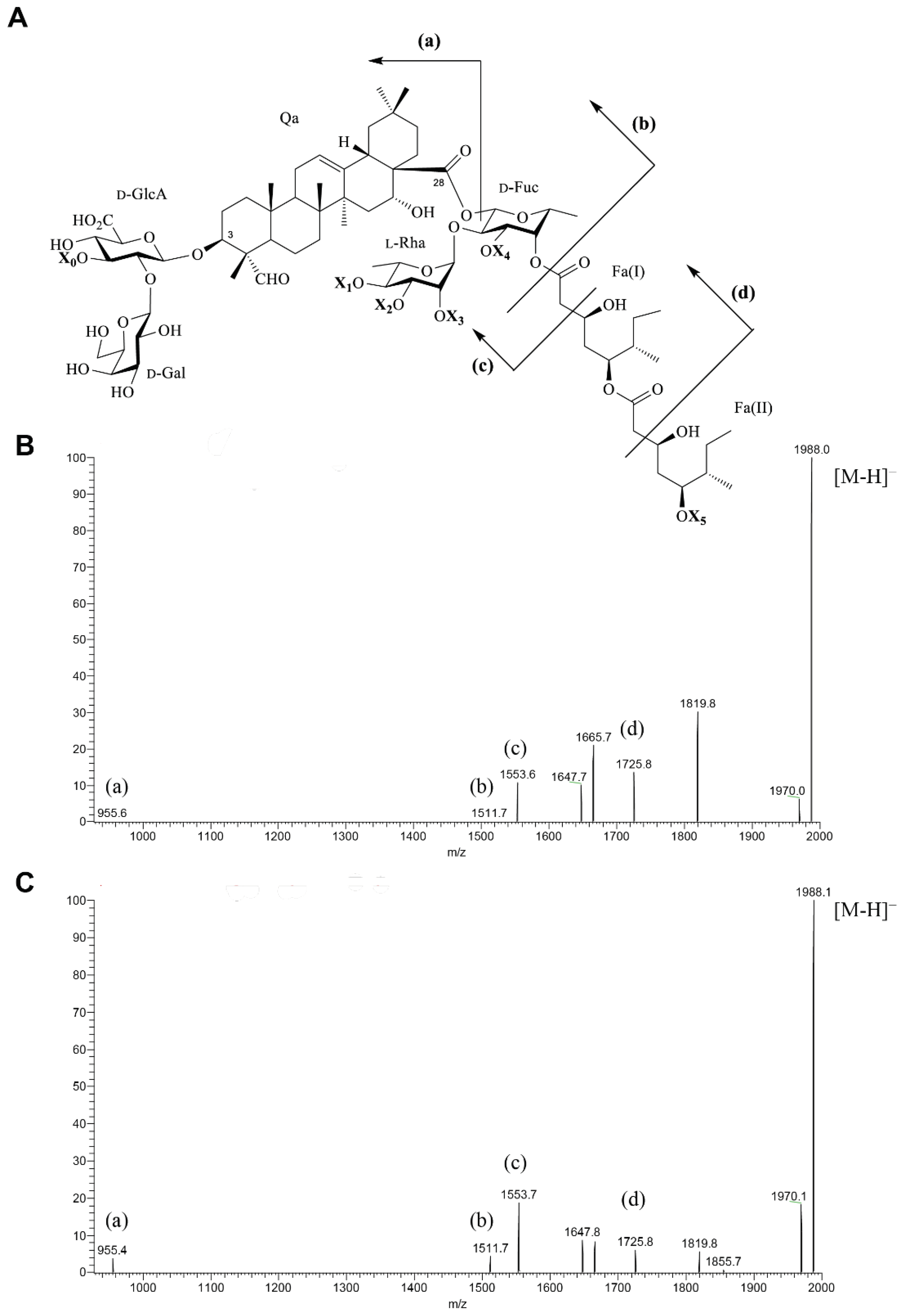
2.1. Saponin Qb1
2.2. Saponin S13
3. Materials and Methods
3.1. Materials and Chemical Reagents
3.2. Isolation of Saponins
3.2.1. Medium-Pressure Liquid Chromatography (MPLC)
3.2.2. Semi-Preparative High Performance Liquid Chromatography (HPLC)
3.3. Analysis of Fractions and Purified Saponins
3.3.1. Thin Layer Chromatography (TLC)
3.3.2. Liquid Chromatography Mass Spectrometry (LC-MS)
3.3.3. NMR Spectroscopy
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Luebert, F. Taxonomy and distribution of the genus Quillaja Molina (Quillajaceae). Feddes Repert. 2013, 124, 157–162. [Google Scholar] [CrossRef]
- Cibulski, S.P.; Mourglia-Ettlin, G.; Teixeira, T.F.; Quirici, L.; Roehe, P.M.; Ferreira, F.; Silveira, F. Novel ISCOMs from Quillaja brasiliensis saponins induce mucosal and systemic antibody production, T-cell responses and improved antigen uptake. Vaccine 2016, 34, 1162–1171. [Google Scholar] [CrossRef]
- Cibulski, S.; Rivera-Patron, M.; Suárez, N.; Pirez, M.; Rossi, S.; Yendo, A.C.; de Costa, F.; Gosmann, G.; Fett-Neto, A.; Roehe, P.M.; et al. Leaf saponins of Quillaja brasiliensis enhance long-term specific immune responses and promote dose-sparing effect in BVDV experimental vaccines. Vaccine 2018, 36, 55–65. [Google Scholar] [CrossRef]
- Garçon, N.; Chomez, P.; Van Mechelen, M. GlaxoSmithKline Adjuvant Systems in vaccines: Concepts, achievements and perspectives. Expert Rev. Vaccines 2007, 6, 723–739. [Google Scholar] [CrossRef]
- Lacaille-Dubois, M.A.; Wagner, H. A review of the biological and pharmacological activities of saponins. Phytomedicine 1996, 2, 363–386. [Google Scholar] [CrossRef]
- Marty-Roix, R.; Vladimer, G.I.; Pouliot, K.; Weng, D.; Buglione-Corbett, R.; West, K.; MacMicking, J.D.; Chee, J.D.; Wang, S.; Lu, S.; et al. Identification of QS-21 as an Inflammasome-activating Molecular Component of Saponin Adjuvants. J. Biol. Chem. 2016, 291, 1123–1136. [Google Scholar] [CrossRef] [Green Version]
- Barbosa, A.D.P. Saponins as immunoadjuvant agent: A review. Afr. J. Pharm. Pharmacol. 2014, 8, 1049–1057. [Google Scholar] [CrossRef]
- Sun, H.-X.; Xie, Y.; Ye, Y.-P. Advances in saponin-based adjuvants. Vaccine 2009, 27, 1787–1796. [Google Scholar] [CrossRef]
- Fleck, J.D.; Betti, A.H.; da Silva, F.P.; Troian, E.A.; Olivaro, C.; Ferreira, F.; Verza, S.G. Saponins from Quillaja saponaria and Quillaja brasiliensis: Particular chemical characteristics and biological activities. Molecules 2019, 24, 171. [Google Scholar] [CrossRef] [Green Version]
- van Setten, D.C.; van de Werken, G.; Zomer, G.; Kersten, G.F.A. Glycosyl compositions and structural characteristics of the potential immuno-adjuvant active saponins in the Quillaja saponaria Molina extract Quil A. Rapid Commun. Mass Spectrom. 1995, 9, 660–666. [Google Scholar] [CrossRef]
- San Martín, R.; Briones, R. Quality control of commercial quillaja (Quillaja saponaria Molina) extracts by reverse phase HPLC. J. Sci. Food Agric. 2000, 80, 2063–2068. [Google Scholar] [CrossRef]
- ClinicalTrials.gov. Available online: https://clinicaltrials.gov/ct2/results?cond=&term=QS-21&cntry=&state=&city=&dist= (accessed on 19 November 2021).
- Tait, D.R.; Hatherill, M.; Van Der Meeren, O.; Ginsberg, A.M.; Van Brakel, E.; Salaun, B.; Scriba, T.J.; Akite, E.J.; Ayles, H.M.; Bollaerts, A.; et al. Final Analysis of a Trial of M72/AS01 E Vaccine to Prevent Tuberculosis. N. Engl. J. Med. 2019, 381, 2429–2439. [Google Scholar] [CrossRef]
- Keech, C.; Albert, G.; Cho, I.; Robertson, A.; Reed, P.; Neal, S.; Plested, J.S.; Zhu, M.; Cloney-Clark, S.; Zhou, H.; et al. Phase 1–2 Trial of a SARS-CoV-2 Recombinant Spike Protein Nanoparticle Vaccine. N. Engl. J. Med. 2020, 383, 2320–2332. [Google Scholar] [CrossRef]
- Krammer, F. SARS-CoV-2 vaccines in development. Nature 2020, 586, 516–527. [Google Scholar] [CrossRef]
- European Medicines Agency (EMA). Available online: https://www.ema.europa.eu/en/news/first-malaria-vaccine-receives-positive-scientific-opinion-ema (accessed on 19 November 2021).
- European Medicines Agency (EMA). Available online: https://www.ema.europa.eu/en/medicines/human/EPAR/shingrix (accessed on 19 November 2021).
- Marciani, D.J. Elucidating the Mechanisms of Action of Saponin-Derived Adjuvants. Trends Pharmacol. Sci. 2018, 39, 573–585. [Google Scholar] [CrossRef]
- Silveira, F.; Cibulski, S.P.; Varela, A.P.; Marqués, J.M.; Chabalgoity, A.; de Costa, F.; Yendo, A.C.A.; Gosmann, G.; Roehe, P.M.; Fernández, C.; et al. Quillaja brasiliensis saponins are less toxic than Quil A and have similar properties when used as an adjuvant for a viral antigen preparation. Vaccine 2011, 29, 9177–9182. [Google Scholar] [CrossRef] [Green Version]
- de Costa, F.; Yendo, A.C.A.; Cibulski, S.P.; Fleck, J.D.; Roehe, P.M.; Spilki, F.R.; Gosmann, G.; Fett-Neto, A.G. Alternative Inactivated Poliovirus Vaccines Adjuvanted with Quillaja brasiliensis or Quil-A Saponins Are Equally Effective in Inducing Specific Immune Responses. PLoS ONE 2014, 9, e105374. [Google Scholar] [CrossRef] [Green Version]
- Fleck, J.D.; Kauffmann, C.; Spilki, F.; Lencina, C.L.; Roehe, P.M.; Gosmann, G. Adjuvant activity of Quillaja brasiliensis saponins on the immune responses to bovine herpesvirus type 1 in mice. Vaccine 2006, 24, 7129–7134. [Google Scholar] [CrossRef]
- Yendo, A.C.A.; de Costa, F.; Cibulski, S.P.; Teixeira, T.F.; Colling, L.C.; Mastrogiovanni, M.; Soulé, S.; Roehe, P.M.; Gosmann, G.; Ferreira, F.A.; et al. A rabies vaccine adjuvanted with saponins from leaves of the soap tree (Quillaja brasiliensis) induces specific immune responses and protects against lethal challenge. Vaccine 2016, 34, 2305–2311. [Google Scholar] [CrossRef]
- Cibulski, S.; Varela, A.P.M.; Teixeira, T.F.; Cancela, M.P.; Sesterheim, P.; Souza, D.O.; Roehe, P.M.; Silveira, F. Zika Virus Envelope Domain III Recombinant Protein Delivered With Saponin-Based Nanoadjuvant From Quillaja brasiliensis Enhances Anti-Zika Immune Responses, Including Neutralizing Antibodies and Splenocyte Proliferation. Front. Immunol. 2021, 12, 515. [Google Scholar] [CrossRef]
- Cibulski, S.; Teixeira, T.F.; Varela, A.P.M.; de Lima, M.F.; Casanova, G.; Nascimento, Y.M.; Tavares, J.F.; da Silva, M.S.; Sesterheim, P.; Souza, D.O.; et al. IMXQB-80: A Quillaja brasiliensis saponin-based nanoadjuvant enhances Zika virus specific immune responses in mice. Vaccine 2021, 39, 571–579. [Google Scholar] [CrossRef] [PubMed]
- Wallace, F.; Bennadji, Z.; Ferreira, F.; Olivaro, C. Analysis of an immunoadjuvant saponin fraction from Quillaja brasiliensis leaves by electrospray ionization ion trap multiple-stage mass spectrometry. Phytochem. Lett. 2017, 20, 228–233. [Google Scholar] [CrossRef]
- Wallace, F.; Bennadji, Z.; Ferreira, F.; Olivaro, C. Structural characterisation of new immunoadjuvant saponins from leaves and the first study of saponins from the bark of Quillaja brasiliensis by liquid chromatography electrospray ionisation ion trap mass spectrometry. Phytochem. Anal. 2019, 30, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Nord, L.I.; Kenne, L. Novel acetylated triterpenoid saponins in a chromatographic fraction from Quillaja saponaria Molina. Carbohydr. Res. 2000, 329, 817–829. [Google Scholar] [CrossRef]
- Jacobsen, N.E.; Fairbrother, W.J.; Kensil, C.R.; Lim, A.; Wheeler, D.A.; Powell, M.F. Structure of the saponin adjuvant QS-21 and its base-catalyzed isomerization product by 1 H and natural abundance 13 C NMR spectroscopy. Carbohydr. Res. 1996, 280, 1–14. [Google Scholar] [CrossRef]
- Jansson, P.-E.; Kenne, L.; Widmalm, G. Computer-assisted structural analysis of regular polysaccharides. Pure Appl. Chem. 1989, 61, 1181–1192. [Google Scholar] [CrossRef] [Green Version]
- Agrawal, P.K. NMR Spectroscopy in the Structural Elucidation of Oligosaccharides and Glycosides. Phytochemistry 1992, 31, 3307–3330. [Google Scholar] [CrossRef]
- Nord, L.I.; Kenne, L. Separation and Structural Analysis of Saponins in a Bark Extract from Quillaja saponaria Molina. Carbohydr. Res. 1999, 320, 70–81. [Google Scholar] [CrossRef]
- Bock, K.; Pedersen, C. A Study of 13CH Coupling Constants in Hexopyranoses. J. Chem. Soc. Perkin Trans. 2 1974, 3, 293–297. [Google Scholar] [CrossRef]
- Rao, V.S.R.; Qasba, P.K.; Balaji, P.V.; Chandrasekaran, R. Conformation of Carbohydrates, 1st ed.; CRC Press: London, UK, 1998; pp. 49–67. [Google Scholar] [CrossRef]
- Schleucher, J.; Schwendinger, M.; Sattler, M.; Schmidt, P.; Schedletzky, O.; Glaser, S.J.; Sørensen, O.W.; Griesinger, C. A general enhancement scheme in heteronuclear multidimensional NMR employing pulsed field gradients. J. Biomol. NMR 1994, 4, 301–306. [Google Scholar] [CrossRef]
- Bax, A.D.; Donald, G.D. MLEV-17-based two-dimensional homonuclear magnetization transfer spectroscopy. J. Magn. Reson. 1969, 65, 355–360. [Google Scholar] [CrossRef]
- Nyberg, N.T.; Duus, J.Ø.; Sørensen, O.W. Heteronuclear Two-Bond Correlation: Suppressing Heteronuclear Three-Bond or Higher NMR Correlations while Enhancing Two-Bond Correlations Even for Vanishing 2JCH. J. Am. Chem. Soc. 2005, 127, 6154–6155. [Google Scholar] [CrossRef] [PubMed]
- Claridge, T.D.W.; Pérez-Victoria, I. Enhanced 13C resolution in semi-selective HMBC: A band-selective, constant-time HMBC for complex organic structure elucidation by NMR. Org. Biomol. Chem. 2003, 1, 3632–3634. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Ernst, R.R.; Wüthrich, K. A two-dimensional nuclear Overhauser enhancement (2D NOE) experiment for the elucidation of complete proton-proton cross-relaxation networks in biological macromolecules. Biochem. Biophys. Res. Commun. 1980, 64, 2229–2246. [Google Scholar] [CrossRef]
- Wang, P. Natural and Synthetic Saponins as Vaccine Adjuvants. Vaccines 2021, 9, 222. [Google Scholar] [CrossRef]
- Kamstrup, S.; San Martin, R.; Doberti, A.; Grande, H.; Dalsgaard, K. Preparation and Characterisation of Quillaja Saponin with Less Heterogeneity than Quil-A. Vaccine 2000, 18, 2244–2249. [Google Scholar] [CrossRef]
- Harandi, A.M.; Medaglini, D.; Shattock, R.J. Vaccine Adjuvants: A Priority for Vaccine Research. Vaccine 2010, 28, 2363–2366. [Google Scholar] [CrossRef]
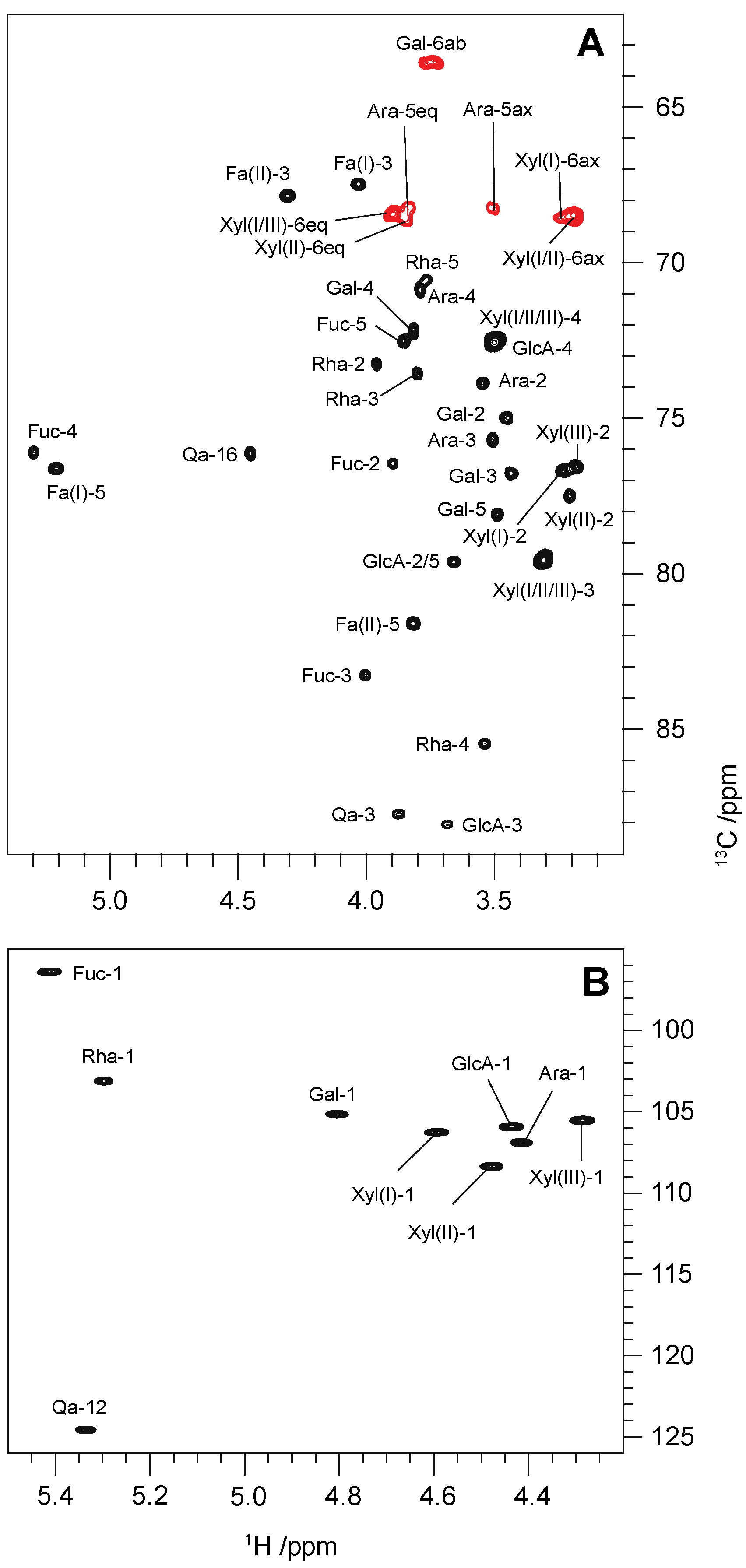
| Residue | Abbreviation | 1H/13C | Correlation to Atom (from Anomeric Atom) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 1H,13C-HMBC | 1H,1H-NOESY | ||
| Qa C3-O-glycan | |||||||||
| →2,3)-β-d-GlcpA-(1→ | GlcA | 4.43 [8.6] | 3.65 | 3.67 | 3.49 | 3.65 | C3, Qa (a) | H3, Qa | |
| 104.6 | ~78.3 | 86.7 | ~71.2 | ~78.3 | n.d. | H3, Qa (a) | |||
| β-d-Galp-(1→ | Gal | 4.80 [7.1] | 3.44 | 3.43 | 3.81 | 3.48 | 3.73, 3.76 | C2, GlcA | H2, GlcA |
| 103.8 | 73.6 | 75.4 | 70.9 | 76.7 | 62.2 | H2, GlcA | |||
| β-d-Xylp-(1→ | Xyl(I) | 4.59 [7.7] | 3.23 | 3.30 | 3.49 | 3.18, 3.89 | C3, GlcA | H3, GlcA | |
| 104.9 | 75.3 | ~78.2 | ~71.2 | ~67.1 | H3, GlcA | ||||
| Qa C28-O-glycan | |||||||||
| →2,3,4)-β-d-Fucp-(1→ | Fuc | 5.41 [8.1] | 3.89 | 4.00 | 5.29 | 3.85 | 1.08 | C28, Qa (a) | |
| 95.1 {164} | 75.1 | 81.9 | 74.7 | 71.2 | 16.9 | ||||
| →4)-α-l-Rhap-(1→ | Rha | 5.29 [1.9] | 3.96 | 3.80 | 3.56 | 3.77 | 1.30 | C2, Fuc | H2, Fuc |
| 101.8 {172} | 71.9 | 72.2 | 84.1 | 69.2 | 18.5 | H2, Fuc | |||
| β-d-Xylp-(1→ | Xyl(II) | 4.47 [7.7] | 3.20 | 3.30 | 3.49 | 3.18, 3.89 | C4, Rha | H4, Rha | |
| 107.0 | 76.1 | ~78.2 | ~71.2 | ~67.3 | H4, Rha | ||||
| β-d-Xylp-(1→ | Xyl(III) | 4.27 [7.7] | 3.18 | 3.30 | 3.49 | 3.18, 3.89 | C5, Fa(II) (a) | ||
| 104.2 | 75.2 | ~78.2 | ~71.2 | ~67.1 | H5, Fa(II) (a) | ||||
| α-l-Arap-(1→ | Ara | 4.41 [6.7] | 3.54 | 3.51 | 3.78 | 3.49, 3.84 | C3, Fucp | H3, Fuc | |
| 105.6 | 72.5 | 74.3 | 69.5 | 66.9 | H3, Fucp | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wallace, F.; Fontana, C.; Ferreira, F.; Olivaro, C. Structure Elucidation of Triterpenoid Saponins Found in an Immunoadjuvant Preparation of Quillaja brasiliensis Using Mass Spectrometry and 1H and 13C NMR Spectroscopy. Molecules 2022, 27, 2402. https://doi.org/10.3390/molecules27082402
Wallace F, Fontana C, Ferreira F, Olivaro C. Structure Elucidation of Triterpenoid Saponins Found in an Immunoadjuvant Preparation of Quillaja brasiliensis Using Mass Spectrometry and 1H and 13C NMR Spectroscopy. Molecules. 2022; 27(8):2402. https://doi.org/10.3390/molecules27082402
Chicago/Turabian StyleWallace, Federico, Carolina Fontana, Fernando Ferreira, and Cristina Olivaro. 2022. "Structure Elucidation of Triterpenoid Saponins Found in an Immunoadjuvant Preparation of Quillaja brasiliensis Using Mass Spectrometry and 1H and 13C NMR Spectroscopy" Molecules 27, no. 8: 2402. https://doi.org/10.3390/molecules27082402
APA StyleWallace, F., Fontana, C., Ferreira, F., & Olivaro, C. (2022). Structure Elucidation of Triterpenoid Saponins Found in an Immunoadjuvant Preparation of Quillaja brasiliensis Using Mass Spectrometry and 1H and 13C NMR Spectroscopy. Molecules, 27(8), 2402. https://doi.org/10.3390/molecules27082402






