Abstract
The A2A adenosine receptor (A2AAR) is one of the four subtypes activated by nucleoside adenosine, and the molecules able to selectively counteract its action are attractive tools for neurodegenerative disorders. In order to find novel A2AAR ligands, two series of compounds based on purine and triazolotriazine scaffolds were synthesized and tested at ARs. Compound 13 was also tested in an in vitro model of neuroinflammation. Some compounds were found to possess high affinity for A2AAR, and it was observed that compound 13 exerted anti-inflammatory properties in microglial cells. Molecular modeling studies results were in good agreement with the binding affinity data and underlined that triazolotriazine and purine scaffolds are interchangeable only when 5- and 2-positions of the triazolotriazine moiety (corresponding to the purine 2- and 8-positions) are substituted.
1. Introduction
The A2A adenosine receptor (A2AAR) subtype is one of the four G protein-coupled receptors (GPCRs,) named A1, A2A, A2B, and A3, which are ARs activated by the natural nucleoside adenosine (Ado) [1,2]. In contrast to the A1 and A3 receptor subtypes, which are coupled to G inhibitory proteins, the A2AAR and A2BAR are coupled to G stimulatory proteins and their activation produces an increase in the second messenger cAMP [3]. It is well-known that Ado is among the key modulators in the manifestation of the periphery and central nervous system (CNS) inflammation. This activity of Ado is mainly due to the activation of the A2AAR among the AR subtypes. In the periphery, Ado reduces inflammation through the decreased recruitment of leucocyte [4,5,6]. Regarding the CNS, several studies have demonstrated that A2AAR antagonists play a key role in counteracting neuroinflammation [7,8,9], a common event in neurodegenerative diseases, such as PD, characterized by the activation of microglial cells associated with the release of pro-inflammatory mediators [10,11]. In this context, molecules able to selectively block the A2AARs are attractive tools useful in the treatment of neurodegenerative disorders, such as Parkinson’s disease (PD) [12,13]. In fact, numerous studies demonstrated that A2AAR antagonists increase the therapeutic efficacy of levodopa (L-Dopa), which represents the election drug for PD therapy [14].
These findings led to the commercialization of the first and unique A2AAR antagonist Istradefylline (1, also called KW6002, Figure 1) as a coadjutant of L-Dopa for the treatment of PD [15]. In 2013, in Japan, Istradefylline was approved under the name Nouriast® [16], and in 2019, the Food and Drug Administration (FDA) approved its commercialization (Nourianz®) [17,18]. This molecule belongs to the class of xanthine derivatives such as caffeine and theophylline, which represent the first A2AAR antagonists to be discovered [19,20,21]. In recent decades, a great number of AR antagonists were synthesized using different scaffolds. CGS 15943 (2, Figure 1) is an example of a triazoloquinazoline derivative, initially proposed as a selective A2AAR antagonist, which demonstrated only slight selectivity in subsequent studies [22]. Modification of the CGS 15943 triazoloquinazoline scaffold, obtained by the replacement of the phenyl ring with a pyrazole moiety, considerably improved the selectivity for A2AAR, leading to a number of potent and selective A2AAR antagonists such as Preladenant (3, also called SCH 420814, Figure 1) [23]. This compound has been studied for the treatment of Parkinson’s disease by Schering-Plough (https://www.fiercebiotech.com/biotech/schering-plough-reports-preladenant-meets-primary-endpoint-phase-ii-dose-finding-trial-for, accessed on 8 March 2022) and, later on, it was studied by the Merck Company in two phase III trials as an adjunct to levodopa in PD patients. However, in these studies, its efficacy was not significantly higher than the placebo [24,25].
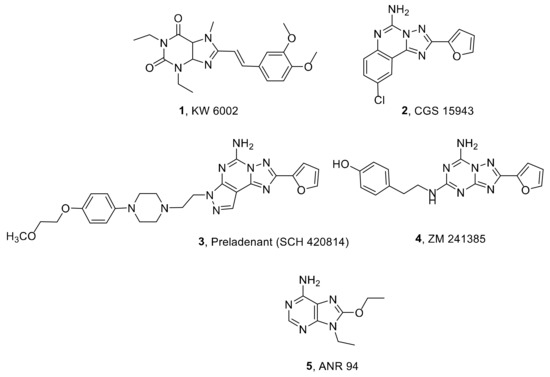
Figure 1.
Structures of A2AAR antagonists.
A simplification of the triazolochinazoline scaffold led to the triazolotriazine ZM241385, which binds the A2AAR with high affinity (4, Ki = 1.6 nM, Figure 1) and shows good selectivity versus the A1 and A3 ARs but not versus the A2BAR. This compound was used as a radioligand for the study of the A2AAR both in tritiated and iodinated form and as a ligand for crystallization studies of the A2AAR [26,27,28].
Examples of other non-xanthine A2AAR antagonists are adenine derivatives, which have been the subject of our research for a long time. In fact, we synthesized a number of such molecules substituted at the 2-, 9-, and/or 8-positions, which were found to behave as A2AAR antagonists endowed with various degrees of affinity and selectivity for ARs [29]. Among them, the 9-ethyl-8-ethoxyadenine (ANR 94, 5, Figure 1) showed good affinity and selectivity for the human A2AAR subtype (Ki A1 = 2400 nM, Ki A2A = 46 nM, Ki A2B > 30,000 nM, and Ki A3 = 21,000 nM) and demonstrated very high efficacy in in vivo models of Parkinson’s disease. In fact, it reversed the catalepsy induced by haloperidol and induced contralateral turning behavior in rats sensitized to L-DOPA. Moreover, this compound showed good efficacy in lowering the intensity of the tremor evoked by Parkinson’s induced by tacrine and in reducing the motor deficit in a neuronal injury model induced by 6-hydroxydopamine [30]. Despite these promising results, ANR 94 does not show a remarkable affinity for A2AARs (Ki = 46 nM) with respect to other A2AAR antagonists like as ZM241385, which is considered the reference compound for studying the A2AAR.
Starting from these observations, and in order to verify whether the triazolotriazine scaffold of the ZM241385 can be replaced by the purine ring to give a high affinity A2AAR ligand, we synthesized an adenine derivative bearing the same substituents present in the 2- and 5-positions of the ZM241385 structure. Hence, an adenine analogue substituted at the 2-position with a (4-hydroxy)-2-phenylethylamino group and bearing a furyl ring at the 8-position, was designed and synthesized together with its 4-methoxy analogue. At the 9-position of these molecules, we introduced an ethyl group that favors the interaction of adenine derivatives with A2AAR [31] (Figure 2a). We also replaced the furyl ring present in the 8-position of ZM241385 with an ethoxy group (Figure 2b).
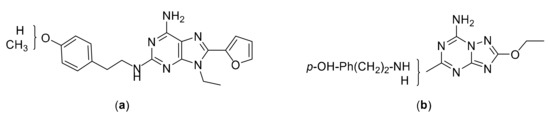
Figure 2.
Structures of the designed compounds: (a) purine derivatives; (b) triazolotriazine derivatives.
Furthermore, aimed at improving the A2AAR binding affinity of ANR 94, we designed and synthesized its triazolotriazine analogue, which lacks, for structural requirement reasons, the ethyl group present at the 9-position of the ANR 94 purine ring (Figure 2b).
All the newly synthesized compounds were evaluated in binding and functional studies in order to assess their affinity/activity at ARs and the compound endowed with the most promising profile of A2AAR affinity and selectivity was also evaluated in a microglial model of neuroinflammation.
2. Results and Discussion
2.1. Chemical Synthesis
The 9-ethyl-8-furyl-2-(4-hydroxyphenyl)-2-ethylaminoadenine (13) and its 4-methoxyphenyl analogue 12 were synthesized starting from the 2-chloro-9-ethyladenine (6), obtained from the commercially available 2,6-dichloropurine, in two steps [32]. Compound 6 was reacted with the p-methoxyphenethylamine, at 130 °C for 24 h, to give the disubstituted adenine derivative 7, which was brominated using N-bromosuccinimide (NBS) in dimethylformamide (DMF) for 30 min at r. t. to obtain the 8-bromo derivative 8 (Scheme 1). The latter was treated with hydrobromic acid (48% solution in water), at r. t. for 2 h, in order to convert the phenylmethoxy group to a phenol. In this reaction, compounds 10 and 11, deriving from the substitution of the 8-bromine atom and partial hydrolysis of the methyl ether, were obtained together with the desired 8-bromo-2-(4-hydroxy-phenyl)-2-ethylaminoadenine (9). Compounds 8 and 9 were, in turn, reacted with (2-tributylstannyl)furane, in tetrahydrofurane (THF) at r. t. for 16 h, to obtain the desired trisubstituted adenine derivatives 12 and 13.
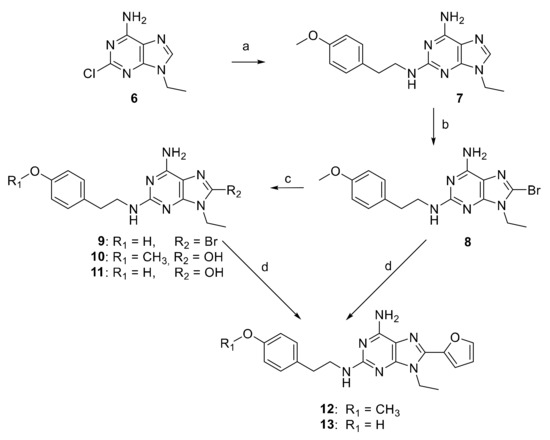
Scheme 1.
Reagents and conditions: (a) 4-methoxyphenethylamine at 130 °C for 24 h, 86% yield; (b) NBS, dry DMF, r. t. for 30 min, 44% yield; (c) HBr 48% at 100 °C for 2 h, 23, 10, and 32% yields for compounds 9, 10, 11, respectively; (d) 2-tributylstannylfuran, [(Ph)3]2PdCl2, dry THF at reflux for 4 h (12, 35% yield) and 6 h (13, 67% yield).
For the synthesis of the new [1,2,4]-triazolo-[1,5-a]-[1,3,5]-triazine derivatives 20 and 21, a targeted approach, aimed at forming first the triazole nucleus and then the triazine ring, was used (Scheme 2). A similar approach was also utilized for the synthesis of ZM241385 [33], but here we chose the commercially available dimethyl N-cyanodithioiminocarbonate (14) as the starting material which presents two very important advantages: the low price and the fact that it can be used both for the formation of the triazole and the triazine ring. In fact, 14 was reacted with aqueous hydrazine in toluene at 0 °C to afford the 5-(methylthio)-1H-1,2,4-triazol-3-amine (15).
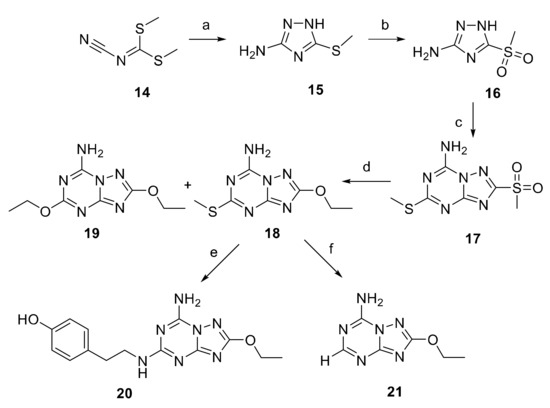
Scheme 2.
Reagents and conditions: (a) hydrazine hydrate, toluene, 0 °C for 1 h, r. t. for 16 h, 83% yield; (b) m-CPBA, DCM, from 0 °C to r. t., 80% yield; (c) 14, 170 °C, 6.5 h, 22% yield; (d) EtOH, NaH, 24 h, r. t., 32 and 5% yields for compounds 18 and 19, respectively; (e) m-CPBA, THF of 2 h, tyramine, Et3N, 40 °C, 12 h, 10% yield; (f) Et3SiH, Pd/C 10%, THF, 8 h, r. t., 44% yield.
The formation of the triazole ring was followed by the oxidation of the 15-thiol group, using meta-chloroperbenzoic acid (m-CPBA) in dichloromethane at r. t., to give the sulfone derivatives 16. As already mentioned, the triazine ring was obtained by treating 16 with the starting reagent 14 in a fusion reaction performed in a steel bomb at 170 °C for 4 h. The reaction led to the formation of a main product which was isolated after chromatography on a silica gel column. Since this reaction could lead to the formation of two or more isomers of 17 [34], the structure of the obtained compound was unequivocally attributed through a further crystallographic X-ray analysis (see next section).
The reaction of 17 with sodium ethoxyde, obtained by treating ethanol with sodium hydride for 24 h, furnished the 5-ethoxy derivative 18 as the main product together with a small percentage of the disubstituted derivative 19.
The ZM241385 analogue 20 was obtained in a two-step reaction. First, the oxidation of the thiomethyl group of 18 with m-CPBA to the sulfone, which is a better leaving group, followed after 2 h when the starting material was totally consumed by the addition of tyramine. Then, the reaction mixture was left at 40 °C for 12 h.
Finally, hydrogenolisis of 18, using triethylsilane as a proton donor and Pd/C 10% as the catalyst, allowed the removal of the thiomethyl goup to obtain the desired triazolotriazine 21.
2.2. Crystallographic X-ray Analysis
The ORTEP style view of 17 is reported in Figure 3 together with the atomic labeling scheme. The most important bond distances and angles are reported in the caption. The triazolotriazine system is planar, atoms S1 and S2 are at a distance of 0.147(2) and 0.018(2) Å from the mean plane, respectively. The C-N bond distances are in the range expected for parent systems found in the literature. The crystal packing of the molecule is built up through Van der Waals and hydrogen bond interactions. In particular, N3 and N4 atoms of two adjacent molecules are involved in strong hydrogen bonds. The N3⋯N4′ distance is 3.034(2) Å, and the N3-H3A⋯N4′ angle is 174.14(9)°. Moreover, N3 is at a distance of 3.039(2) Å from O1 of another molecule in the packing. Hence, X-ray analysis unequivocally confirmed the structure of the desired triazolotriazine derivative 17.
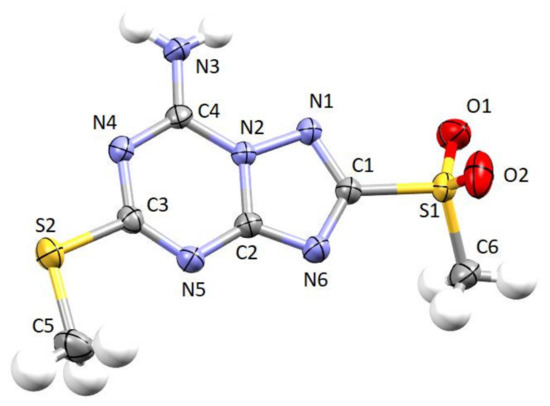
Figure 3.
Ortep style view of compound 17. Ellipsoids are shown at 50% probability level. Selected bond distances (Å) and angles (°): C1-N1 1.323(2), C1-N6 1.353(2), C1-S1 1.7794(16), C2-N6 1.332(2), C2-N5 1.344(2), C2-N2 1.3728(19), C3-N5 1.317(2), C3-N4 1.365(2), C3-S2 1.7480(16), C4-N3 1.311(2), C4-N4 1.325(2), C4-N2 1.3721(19), C5-S2 1.797(2), N1-N2 1.3748(17); N1-C1-N6 118.47(14), N1-C1-S1 118.59(12), N6-C1-S1 122.64(12), N6-C2-N5 129.47(14), N6-C2-N2 109.12(13), N5-C2-N2 121.41(14), N5-C3-N4 128.40(14), N5-C3-S2 120.45(12), N4-C3-S2 111.14(11), N3-C4-N4 123.52(15), N3-C4-N2 119.13(14), N4-C4-N2 117.35(14), C1-N1-N2 99.58(12), C4-N2-C2 121.84(13), C4-N2-N1 127.06(13), C2-N2-N1 110.91(12), C4-N4-C3 117.44(13), C3-N5-C2 113.48(13), C2-N6-C1 101.90(13).
2.3. Biological Evaluation
2.3.1. Binding Evaluation at ARs
The affinities of the newly synthesized compounds 7–13 and 17–21 were determined in binding studies at human recombinant A1, A2A, and A3 ARs, stably transfected in Chinese hamster ovary (CHO) cells, using [3H]CCPA (2-chloro-N6-cyclopentylAdo), [3H]NECA (5′-N-ethylcarboxamidoAdo), and [3H]HEMADO (2-hexynyl-N6-methylAdo) as respective radioligands [35,36]. Most of the compounds were also evaluated at A2BARs in a functional study using the GloSensor cAMP assay [37]. The results are reported in Table 1 as Ki values in nM (± standard errors). In the case of A2BARs, Ki-values were calculated from IC50-values determined by the inhibition of NECA-stimulated adenylyl cyclase activity. ZM241385 (4) and the 9-ethyl-8-ethoxyadenine (ANR 94, 5) are reported as reference compounds.

Table 1.
Binding data at A1, A2A, and A3 ARs or adenylate cyclase activity at A2BAR of compounds 4, 5, 7–13, 17–21 at human ARs stably transfected in CHO cells (Ki, nM).
As already mentioned in the introduction, ZM241385 (4) possesses a high affinity for the A2AAR (Ki = 1.6 nM), good selectivity versus the A1 and A3 AR subtypes by 484- and 464-fold, respectively, and lower selectivity towards the A2BAR by just 47 times.
Its purine analogue maintained high affinity at the A2AAR but showed an inverse profile of selectivity that decreased towards the A1 and A3 subtypes and increased versus the A2BAR (13; Ki A2A = 1.8 nM; selectivity A1/A2A = 28, A3/A2A = 7, and A2B/A2A = 796). Supposing that the triazoletriazine scaffold of ZM241385 and the purine core of 13 are positioned with the same orientation in the binding pocket of the A2AAR, the presence of the ethyl group in 9-position of 13 seems to favor the interaction of this molecule with the A1 and A3 subtypes and disadvantage the one with the A2BAR, while it does not seem to influence the interaction with the A2AAR. These findings indicate that, at least with regards to A2AAR affinity, the triazoletriazine and purine scaffolds are interchangeable. When the phenolic hydroxyl group of 13 is replaced by a methoxy substituent, a further increase in affinity at the A1AR is observed, leading to an A1/A2A unselective ligand (12; Ki A2A = 1.0 nM and Ki A1 = 2.6 nM). As expected, the presence of an 8-bromine atom, instead of a furyl ring, led to a decrease in affinity at A1, A2A, and A3 ARs; in fact, 9 and 8 showed higher Ki values with respect to 12 and 13 while remaining A2A selective ligands (9; Ki A2A = 44 nM and 8; Ki A2A = 34 nM). The 8-unsubstituted derivative 7 lost affinity at all ARs.
These findings are in agreement with our previous results which demonstrated that the presence of an 8-bromine atom and, even more, a 8-furyl ring, favor the interaction of 2-9-disubstituted adenine derivatives at ARs [29]. Furthermore, the presence of a hydroxyl group at the 8-position of these molecules is detrimental for the affinity, especially at the A2A and A3 subtypes (10; Ki A2A > 30,000 nM and 11; Ki A2A and Ki A3 > 30,000 nM).
The 8,9-disubstituted adenine derivative ANR 94 exhibits good A2AAR affinity and selectivity (5; Ki A2A = 46 nM; selectivity A1/A2A = 52, A3/A2A = 457, and A2B/A2A > 652). Unfortunately, its triazolotriazine analogue 20 showed a decrease in A2AAR affinity by 44-fold (Ki = 2029 nM) and a decrease at the A1 subtype but not at the A3AR (Ki = 1019 nM). In this case, the replacement of the purine ring with the triazolotriazine scaffold had a negative impact on the A2AAR affinity. Among the 2,5-disubstituted triazoletriazines reported here, only compounds 18 and 21, the latter being the 8-ethoxy analogue of ZM241385, showed a sub-microM affinity at the A2AAR with Ki values of 583 nM and 178 nM, respectively.
The obtained results did not allow to give a univocal answer to the question of the interchangeability between the two scaffolds that we have tried to explain with molecular modeling studies.
2.3.2. Study of the Anti-Inflammatory Activity of 13
In order to investigate whether the promising affinity profile of 13 could be associated with potential protective activity against PD, we studied its anti-inflammatory properties in a microglial model of neuroinflammation. Increasing evidence indicates that neuroinflammation mediated by microglia plays a major role in PD [40,41,42]. Although it is still unclear whether the neuroimmune response may or not represent a primary cause for neuronal loss, there is wide consensus that it participates in disease progression [43]. ARs expressed in microglial cells modulate inflammatory response in PD and preliminary data suggest that A2AAR antagonists may have therapeutic efficacy [8,44].
Activated microglia can assume two different polarization states: a pro-inflammatory “M1” phenotype and an anti-inflammatory “M2” phenotype. Interestingly, in PD patients, an increase in M1-polarized microglia is observed [45]. M1-phenotype microglia is characterized by the increased production of inflammatory cytokines, such as interleukin-1β (IL-1β) and IL-6, that leads to tissue damage. On the other hand, the M2-phenotype is characterized by an upregulation of anti-inflammatory mediators, such as IL-10. In activated microglia, these two phenotypes can coexist, revealing the complexity of microglia function and the dynamic changes of the environment in vivo [46]. Different studies associate the release of pro-inflammatory mediators with the activation of the NOD-like receptor family, the pyrin domain containing-3 protein (NLRP3) inflammasome in BV-2 cells [47]. NLRP3, a multi-protein complex, modulates the maturation and secretion of pro-inflammatory cytokines, including IL-1β. Of note, the inhibition of the NLRP3 inflammasome activation could protect dopaminergic neurons [48,49]. On these bases, the identification of new compounds able to inhibit the activation of NLRP3 Inflammasome are a promising therapeutic intervention against neurodegeneration in PD.
The microglial BV-2 cell line exposed to bacterial endotoxin lipopolysaccharide (LPS) was chosen as the in vitro model of neuroinflammation. LPS, a pro-inflammatory mediator, is widely used in in vitro studies to activate microglia cells and induce proinflammatory transduction pathways [50,51,52]. In addition, BV-2 cells are a valid alternative to primary microglia as 90% of the genes regulated by LPS in BV-2 cells are also modulated in primary microglia and both display similar reaction patterns [53].
First of all, we verified the potential cytotoxicity of 13 in our cell model system. BV-2 cells were treated with increasing concentrations of the compound for 24 h and cell viability was evaluated by MTT assay (Figure 4A). Of note, the tested A2AAR antagonist did not show any cytotoxicity up to 5 μM, demonstrating its high safety profile.
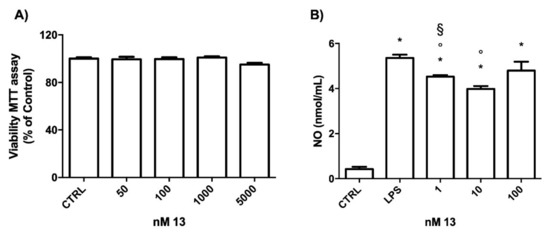
Figure 4.
Effect of 13 on cell viability and NO production in BV-2 cells. (A) Cells were treated with 50–5000 nM of 13 for 24 h, and MTT assay was used to measure cell viability. (B) Cells were treated with 10 nM of 13 before activation with 100 ng/mL LPS for 24 h and NO release was measured by the Griess assay. Each bar represents means ± SEM of at least three independent experiments. Data were analyzed by one-way ANOVA followed by Bonferroni’s test. * p < 0.05 vs. CTRL; ° p < 0.05 vs. LPS; § p < 0.05 vs. 10 nM.
To determine the effects of 13 on LPS-induced inflammatory mediators, we initially evaluated the production of NO. BV-2 cells were treated with different concentrations of 13, then, were exposed to 100 ng/mL LPS for 24 h, and the release of NO in the culture medium was measured by Griess assay (Figure 4B). LPS strongly and significantly increased the release of NO in the culture medium with respect to the control cells (CTRL).
The treatments with 13 at the concentrations 1 and 10 nM significantly reduced the NO level with respect to LPS treated cells. Interestingly, 10 nM treatment was the most effective in counteracting NO release as 1 nM led to significantly higher NO production than 10 nM. On theses bases, the concentration 10 nM was chosen for the following experiments.
To better characterize the anti-inflammatory activity of 13, we evaluated the expression of the main pro-inflammatory cytokines and enzymes up-regulated in activated microglial cells, such as IL-1β, IL-6, inducible NO synthase (iNOS), and cyclooxygenase 2 (COX2). ANR 94 was chosen as the reference compound as we already demonstrated its ability to reduce microglia activation in a murine model of PD [8]. BV-2 cells were treated with 13 or ANR 94 (10 nM) and activated with 100 ng/mL LPS for 24 h. Total RNA was isolated, and proinflammatory cytokine and enzyme expressions were measured using RT-PCR (Figure 5). As expected, LPS significantly increased the expression of IL-1β, IL-6, COX-2, and iNOS with respect to the control cells. 13 significantly reduced the expression of all the pro-inflammatory mediators tested with respect to cells exposed to LPS. On the other hand, ANR 94 was only able to significantly reduce the expression of IL-1β. In particular, the treatment with 13 led to a significant down-regulation of IL-6, iNOS, and COX-2 with respect to ANR 94, demonstrating it possesses higher efficacy than ANR 94 in reducing the expression level of these pro-inflammatory mediators.
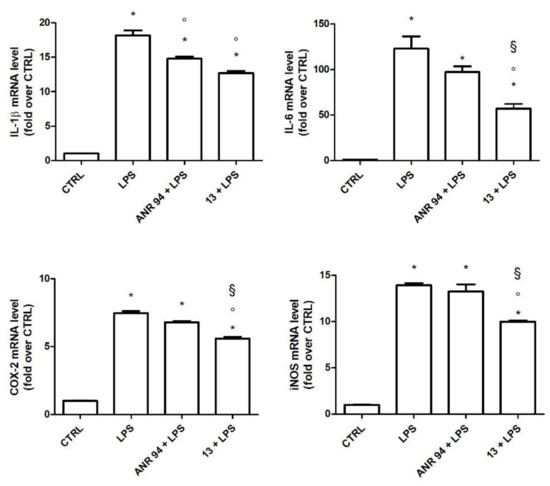
Figure 5.
Expression of IL-1β, IL-6, COX-2, and iNOS in BV-2 cells treated with 13 and ANR 94. Cells were treated with 10 nM 13 or 10 nM ANR 94 for 24 h, exposed to 100 ng/mL LPS for 24 h and cytokines and enzymes mRNA levels were measured by RT-PCR. Data are expressed as relative abundance compared to untreated cells. Each bar represents the mean ± SEM of three independent experiments. Data were analyzed with one-way ANOVA followed by Bonferroni’s test. * p < 0.05 vs. CTRL; ° p < 0.05 vs. LPS; § p < 0.05 vs. ANR 94 + LPS.
To verify if the ability of 13 to reduce the previous inflammatory mediators is also associated with a reduction in the inflammasome, we measured the expression of NLRP3 inflammasome (Figure 6). The data demonstrated that, after LPS stimulation, the expression of NLRP3 mRNA, which is the rate-limiting step for inflammasome activation [54], was significantly increased. Moreover, the increased mRNA of NLRP3 was significantly reduced by 13 pre-treatment.
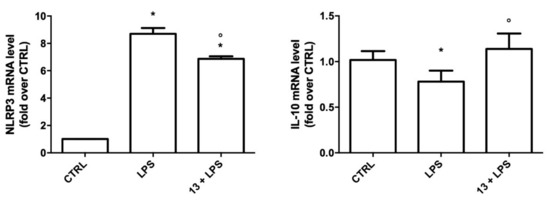
Figure 6.
Expression of NLRP3 and IL-10 in BV-2 cells treated with 13. Cells were treated with 10 nM 13, exposed to 100 ng/mL LPS for 24 h and NLRP3 and IL-10 mRNA levels were measured by RT-PCR. Data are expressed as relative abundance compared to untreated cells. Each bar represents the mean ± SEM of three independent experiments. Data were analyzed with one-way ANOVA followed by Bonferroni’s test. * p < 0.05 vs. CTRL; ° p < 0.05 vs. LPS.
As IL-10 was reported to promote microglial M2 polarization [55], we then evaluated the expression of IL-10 in BV-2 cells treated with 13 before LPS activation (Figure 6). As expected, LPS significantly reduced the expression of this anti-inflammatory cytokine with respect to the control cells, while treatment with 13, in agreement with the previous results, maintained IL-10 expression to a level comparable to the control cells, suggesting that 13 promotes the protective M2 phenotype.
In conclusion, 13 could represent a promising A2AAR antagonist in the fight against PD due to its ability to reduce pro-inflammatory mediators and to increase anti-inflammatory cytokines in microglia cells. Future studies are needed to increase the knowledge of its anti-inflammatory molecular mechanisms and to verify its efficacy in animal models of PD before suggesting this interesting compound for clinical applications.
2.4. Molecular Modeling Studies
Molecular modeling studies, consisting of docking experiments at the A2AAR, A1AR, and A3AR 3D structures, were performed to analyze the binding data of the developed compounds. We employed the crystal structures of the human A2AAR and A1AR in combination with ZM241385 and the antagonist PSB36, respectively, downloaded from the Protein Data Bank webpage (http://www.rcsb.org accessed on 7 January 2022; pdb code: 5NM4; 1.7-Å resolution [26], and pdb code: 5N2S; 3.6-Å resolution [56], respectively). A homology model of the human A3AR was built using the above cited X-ray structure of the antagonist-bound A1AR as a template (pdb code: 5N2S). Docking analyses were performed by using CCDC Gold [57] and then analyzed within the Molecular Operating Environment (MOE, version 2020.09) suite [58].
The docking analysis with the compound ZM241385 at the A2AAR was useful for checking the reliability of the docking protocols, by comparing its obtained top score docking conformation with the co-crystallized arrangement of the same molecule within the receptor structure. Results were in good agreement with the experimental data (RMSD = 1.31 Å).
The docking conformations generally observed for the new adenine derivatives at the A2AAR are similar to those obtained in our previous studies and are showed in Figure 7A, where the purine derivative 13 is perfectly overlapped to the co-crystallized ligand ZM241385. The bicyclic purine core is positioned between the side chains of Phe168 (EL2) and Leu2496.51 and gives non-polar interactions with these residues. The N6-amine group makes H-bond interactions with Asn2536.55 and Glu169 (EL2), while the 8-substituent is located in the depth of the binding cavity. The 2-substituent is positioned at the entrance of the binding site and points toward the extracellular environment.
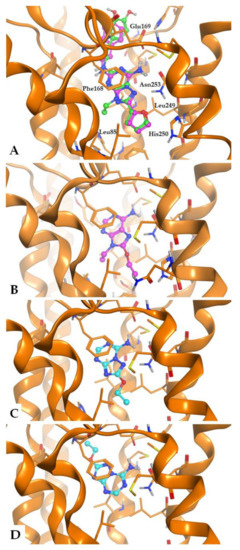
Figure 7.
General binding mode of the synthesized compounds at the A2A AR (pdb: 5NM4) binding cavity, with indication of some key receptor residues. (A) Compound 13 is shown in green, with the co-crystallized ligand ZM241385 (purple) for comparison; (B) Docking conformations of compound ANR 94 (5, purple); (C,D) Docking conformations of compound 21 (cyan) with representation of the two observed potential binding modes.
Purine derivatives lacking the 2-substituent (i.e., ANR 94 (5), Figure 7B) or presenting a small moiety in the same position, are observed to give an analogue binding mode (respect to 13 and ZM241385), with interactions given by the purine core and the exocyclic amine group in the 6-position, and further interactions made by the other substituents. Hence, the lack of the 2-substituent or its substitution with a small group does not seems critical for the binding mode of the 9-ethyladenines. In fact, both 13 and 2-unsubstituted ANR 94 possess nM affinity at the A2AAR. In the case of compound 21, based on a triazoletriazine scaffold and lacking the exocyclic ethyl chain (compared to ANR 94) and the (4-hydroxy)-2-phenylethylamino group (compared to ZM241385), we observed two potential binding modes (Figure 7C,D).
One of these binding modes presents the triazoletriazine scaffold of 21 as almost superimposable to the one of ZM241385, while the other docking conformation makes the triazoletriazine moiety oppositely oriented (see the docking conformation colored in cyan, Figure 7C,D, respectively). We may assume that the presence of a 9-ethyl group in the adenine derivatives makes their binding mode stably anchored within the binding cavity. On the contrary, the triazolotriazine derivatives lacking the long (4-hydroxy)-2-phenylethylamino group are less stably inserted within the binding cavity, giving more potential binding modes. This feature appears to be slightly beneficial in the case of the A3AR, for which the affinity of 21 is higher than ANR 94.
Compounds ZM241385 and 13 make the same interactions with the A2AAR, apart from the ligand region corresponding to the 9-position of the purine core. Considering the purine derivative, its 9-ethyl substituent is located in the depth of the cavity and makes hydrophobic interactions with the residues in proximity, such as Ala632.61, Ile662.64, Val843.32, Leu853.33, Thr883.36, Trp2466.48, Ile2747.39, Ser2777.42, and His2787.43 (see Figure 8). Binding affinity data of ZM241385 and 13 show that at the A2AAR they are endowed with comparable affinity, while 13 presents higher affinity (with respect to ZM241385) at the A1AR and A3AR. Given that the unique structural difference between these two molecules is the presence of a 9-ethyl group in the purine derivative and a nitrogen atom in the corresponding position of ZM241385, this feature appears responsible for the different affinity of the above cited molecules at these two receptors. We analyzed and compared the ability of these two compounds to interact with the A2AAR, A1AR, and A3AR with the aid of the IF-E 6.0 tool (see Experimental section for details), which calculates the atomic and residue interaction forces and the per-residue interaction energies (expressed as kcal × mol−1). This tool was previously used for analogue analyses at hARs obtaining useful interpretation of the compound activity [59,60,61]. We focused on the above residues located in proximity of the 9-ethyl group of 13. The results are displayed in Figure 8.
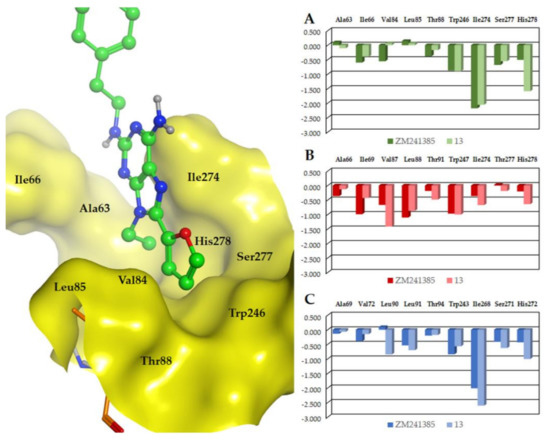
Figure 8.
Interaction energies (represented as kcal mol−1) calculated with the aid of the IF-E 6.0 tool at the A2AAR (A), A1AR (B), and A3AR (C) binding sites. Compounds ZM241385 and 13 were submitted to analysis. Receptor residues in proximity to the compound region corresponding to the 9-ethyl substituent of 13 were considered. In the panel at left, there is a representation of the analyzed residues and their position with respect to the 9-ethyl group (surface representation); the labels indicate the A2AAR residues. See text for details.
The comparison of the interaction energies calculated for ZM241385 and 13 at the A2AAR shows that the two compounds make analogue interactions with a sum of −5.53 and −5.63 kcal mol−1, respectively. This is in agreement with the similar binding affinity data of the two molecules at this AR subtype. At the A1AR and the A3AR, the two molecules show a different pharmacological behavior, with significantly higher affinity for both receptors shown by 13 compared to ZM241385. The interaction energies calculated at these receptors are again in agreement with the biological data, since at the A1AR and the A3AR the sum of the interaction energies for 13 is −5.84 and −6.76 kcal mol−1, respectively, while at the same receptors the sum of the interaction energies for ZM241385 is −4.82 and −4.88 kcal mol−1, respectively. Hence, the presence of an ethyl group in the 9-position of adenine appears beneficial to achieve affinity at the analyzed AR subtypes, while its absence appears to provide some selectivity for the A2AAR.
3. Materials and Methods
3.1. Chemical Synthesis
General Methods
Melting points were determined with a Büchi apparatus and are uncorrected. 1H NMR and 13C NMR spectra were obtained with a Bruker Ascend 500 MHz spectrometer; δ values are in ppm, J values are in Hz. All exchangeable protons were confirmed by the addition of D2O. 13C NMR spectra are reported in the Supplementary Materials. Mass spectra were recorded on an HP 1100-MSD series instrument. Thin-layer chromatography (TLC) was carried out on pre-coated TLC plates with silica gel 60 F254 (Fluka). For column chromatography, silica gel 60 (Merck) was used. Elemental analyses were determined on Fisons Instruments Model EA 1108 CHNS-O model analyzer and are within 0.4% of theoretical values. Purity of the compounds is ≥98% according to elemental analysis data.
9-Ethyl-N2-(4-methoxyphenethyl)-9H-purine-2,6-diamine (7): 6 (287 mg; 1.45 mmol) was suspended in 4 mL of 4-methoxyphenethylamine in a sealed vessel and was left to react at 130 °C for 24 h. Volatiles were removed under vacuum and the crude was purified by gravimetric chromatography eluted with EtOAc-cHex-MeOH (60:36:4). The desired compound 7 (387 mg, 1.24 mmol; yield 86%) was obtained as white powder after crystallization by MeCN-MeOH. M.p.: 151–153 °C; 1H-NMR (DMSO-d6) δ: 1.38 (t, 3H, J = 7.3 Hz, CH2CH3), 2.78 (t, 2H, J = 7.2 Hz, CH2Ph), 3.41 (m, 2H, CH2NH), 3.73 (s, 3H, CH3O), 4.02 (q, 2H, J = 7.2 Hz, CH2CH3), 6.22 (t, 1H, NH), 6.65 (s, 2H, NH2), 6.87 (d, 2H, J = 8.3 Hz, H-Ph), 7.19 (d, 2H, J = 8.3 Hz, H-Ph), 7.73 (s, 1H, H-8). ESI-MS: positive mode m/z: 313.0 ([M+H]+), 335.0 ([M+Na]+). Elemental analysis calcd for C16H20N6O: C 61.52, H 6.45, N 26.90; found C 61.28, H 6.69, N 26.80.
8-Bromo-9-ethyl-N2-(4-methoxyphenethyl)-9H-purine-2,6-diamine (8): 7 (220 mg; 0.71 mmol) was dissolved in 5 mL of dry DMF and a solution of NBS (190 mg; 1.07 mmol) in 5 mL of dry DMF was added dropwise over 30 min in which there was the total consumption of the starting material. The mixture was evaporated to dryness under vacuum, neutralized with 1N NaOH, and extracted with ethyl acetate (3 × 20 mL). The organic phases were combined and placed under stirring with anhydrous sodium sulphate. The suspension was filtered, the filtrate was brought to dryness, and the residue purified via flash column chromatography eluting with EtOAc-cHex (60:40). 8 (122 mg, 0.312 mmol, yield 44%,) was obtained as white powder after crystallization with MeCN. M.p.: 155–157 °C; 1H-NMR (DMSO-d6) δ: 1.30 (t, 3H, J = 7.1 Hz, CH2CH3), 2.77 (t, 2H, J = 8.1 Hz, CH2Ph), 3.41 (m, 2H, CH2NH), 3.73 (s, 3H, OCH3), 4.02 (q, 2H, J = 7.3 Hz, CH2CH3), 6.42 (bs, 1H, NH), 6.86 (m, 4H, H-Ph and NH2), 7.18 (d, 2H, J = 8.6 Hz, H-Ph). ESI-MS: positive mode m/z: 390.9 ([M+H]+), 414.9 ([M+Na]+). Elemental analysis calcd for C16H19BrN6O: C 49.12, H 4.89, N 21.48; found C 49.46, H 4.55, N 21.32.
4-(2-((6-Amino-9-ethyl-8-bromo-9H-purin-2-yl)amino)ethyl)phenol (9): 8 (120 mg, 0.307 mmol) was suspended in 4 mL of HBr 48% and heated at 100 °C for 2 h. The reaction was quenched with ice, neutralized at 0 °C with NH4OH 33%, followed by extraction with EtOAc (3 × 20 mL). The organic phases were dried with anhydrous sodium sulphate and filtered. The filtrate was brought to dryness and the residue purified via flash column chromatography eluting with EtOAc-cHex-MeOH (60:39:1) to obtain 9 (44 mg, 0.12 mmol; yield 23%), 10 (19 mg, 0.06 mmol; yield 10%), and 11 (52 mg, 0.17 mmol; yield 32%).
9: M.p.: 221–223 °C. 1H-NMR (DMSO-d6) δ: 1.19 (t, 3H, J = 6.6 Hz, CH2CH3), 2.74 (t, 2H, J = 7.5 Hz, CH2Ph), 3.32 (q, 2H, J = 7.0 Hz, CH2NH), 3.67 (q, 2H, J = 7.0 Hz, CH2CH3), 5.96 (bs, 2H, NH2), 6.25 (bs, 1H, NH), 6.80 (d, 2H, J = 8.4 Hz, H-Ph), 7.05 (d, 2H, J = 8.4 Hz, H-Ph), 9.58 (bs, 1H, OH). ESI-MS: positive mode m/z: 378.9 ([M+H]+), 400.9 ([M+Na]+). Elemental analysis calcd for C15H17BrN6O: C 47.76, H 4.54, N 22.28; found C 47.82, H 4.3, N 22.38.
10: m.p.: 204–205 °C. 1H-NMR (DMSO-d6) δ: 1.19 (t, 3H, J = 7.0 Hz, CH2CH3), 2.74 (t, 2H, J = 7.5 Hz, CH2Ph), 3.32 (q, 2H, J = 7.0 Hz, CH2NH), 3.67 (q, 2H, J = 7.0 Hz, CH2CH3), 3.72 (s, 3H, OCH3), 5.98 (bs, 2H, NH2), 6.21 (bs, 1H, NH), 6.85 (d, 2H, J = 8.4 Hz, H-Ph), 7.16 (d, 2H, J = 8.4 Hz, H-Ph), 9.53 (bs, 1H, OH). ESI-MS: positive mode m/z: 329.0 ([M+H]+), 351.0 ([M+Na]+). Elemental analysis calcd for C16H21N6O2: C 58.52, H 6.14, N 25.59; found C 58.23, H 6.43, N 25.57.
11: m.p.: 220 °C dec. 1H-NMR (DMSO-d6) δ: 1.19 (t, 3H, J = 7.0 Hz, CH2CH3), 2.69 (t, 2H, J = 7.5 Hz, CH2Ph), 3.67 (m, 2H, CH2NH), 4.04 (q, 2H, J = 7.1 Hz, CH2CH3), 5.99 (bs, 2H, NH2), 6.19 (bs, 1H, NH), 6.67 (d, 2H, J = 8.4 Hz, H-Ph), 7.02 (d, 2H, J = 8.4 Hz, H-Ph), 9.13 (bs, 1H, OH), 9.53 (bs, 1H, OH). ESI-MS: positive mode m/z: 315.0 ([M+H]+), 336.9 ([M+Na]+). Elemental analysis calcd for C15H18N6O2: C 57.31, H 5.77, N 26.74; found C 57.10, H 5.66, N 26.95.
General procedure for the synthesis of 4-(2-((6-Amino-9-ethyl-8-(furan-2-yl)-9H-purin-2-yl)amino)ethyl)phenol (13) and 9-ethyl-8-(furan-2-yl)-N2-(4-methoxyphenethyl)-9H-purine-2,6-diamine (12): 2-Tributylstannylfuran (104 µL, 0.33 mmol) and bis(triphenylphosphine)-palladium dichloride (3 mg, 0.004 mmol) were, in turn, added to a solution of 9 (25 mg, 0.066 mmol) or 8 (26 mg, 0.066 mmol) in anhydrous THF (330 µL). The mixtures were refluxed for 4 h and 6 h, respectively, after which volatiles were evaporated to dryness and the crude mixtures purified through a flash column silica gel eluting with DCM-MeOH (100:0 to 99:1) to afford 13 (16 mg, 0.044 mmol, yield 67%) and with DCM-MeOH (97:3) to obtain 12 (9.1 mg, 0.023 mmol, yield 35%) as white solids after crystallization from MeCN.
13: m.p.: 150–151 °C. 1H-NMR (DMSO-d6) δ: 1.32 (t, J = 7.0 Hz, 3H, CH2CH3), 2.79 (t, J = 7.5 Hz, 2H, CH2Ph), 3.44 (q, J = 6.9 Hz, 2H, CH2NH), 3.72 (s, 3H, OCH3), 4.28 (q, J = 7.0 Hz, 2H, CH2CH3), 6.38 (bs, 1H, NH), 6.71 (m, 1H, H-4′), 6.80 (bs, 2H, NH2), 6.86 (d, J = 8.4 Hz, 2H, H-Ph), 6.98 (d, J = 3.3 Hz, 1H, H-3′), 7.18 (d, J = 8.4 Hz, 2H, H-Ph), 7.89 (m, 1H, H-5′); ESI-MS: positive mode m/z: 379.2 ([M+H]+), 391.2 ([M+Na]+). Elemental analysis calcd for C20H22N6O2: C 63.48, H 5.86, N 22.21; found C 63.76, H 5.58, N 22.33.
12: m. p.: 166–168 °C; 1H-NMR (DMSO-d6) δ: 1.29 (t, J = 7.2 Hz, 3H, CH2CH3), 2.71 (t, J = 6.8 Hz, 2H, CH2Ph), 3.39 (q, J = 6.0 Hz, 2H, CH2NH), 4.26 (q, J = 7.2 Hz, 2H, CH2CH3), 6.34 (m, 1H, NH), 6.66 (d, J = 8.0 Hz, 2H, H-Ph), 6.68 (m, 1H, H-4′), 6.79 (bs, 2H, NH2), 6.95 (d, J = 3.2 Hz, 1H, H-3′), 7.03 (d, J = 8.4 Hz, 2H, H-Ph), 7.87 (m, 1H, H-5′), 9.13 (s, 1H, OH); ESI-MS: positive mode m/z: 365.2 ([M+H]+), 387.2 ([M+Na]+); negative mode m/z 363.1 ([M−H]−), 409.1 ([M-Cl]−). Elemental analysis calcd for C19H20N6O2: C 62.62, H 5.53, N 23.06; found C 62.86, H 5.40, N 23.20).
5-(Methylthio)-1H-1,2,4-triazol-3-amine (15): Dimethyl N-cyanodithioimino-carbonate (14, 1.008 g, 6.89 mmol) was dissolved in toluene (3 mL) and reacted with hydrazine hydrate (228 μL, 10.33 mmol) under stirring in an ice bath under an inert atmosphere. The mixture was left at 0 °C for 1 h and then at r. t. for 4 h; after that it was filtered, and the residue was washed with cHex. The organic layers were dried over sodium sulfate, then filtered and evaporated and the residue was purified by flash column chromatography eluting with DCM-MeOH (97.5:2.5) to obtain the desired compound 15 as a white solid (741 mg; 5.70 mmol; yield 83%). M.p.: 136–139 °C. 1H-NMR (DMSO-d6) δ: 2.38 (s, 3H, CH3), 5.99 (bs, 2H, NH2), 11.91 ppm (bs, 1H, NH). ESI-MS: positive mode m/z: 131.0 ([M+H]+), 153.0 ([M+Na]+). Elemental analysis calcd for C3H6N4S: C 27.68, H 4.65, N 43.04; found C 27.86, H 4.55, N 43.18.
5-(Methylsulfonyl)-1H-1,2,4-triazol-3-amine (16): Compound 16 was obtained by reacting the intermediate 15 (460 mg, 3.53 mmol) with m-CPBA (2.434 g, 14.14 mmol), both dissolved in DCM (32 mL) at 0 °C for 4 h and subsequently at r. t. for 20 h. After removal of the solvent under vacuum, compound 16 was obtained by crystallization with EtOAc, from the crude mixture, as a white solid (457 mg, 2.82 mmol, yield 80%). M.p.: 235–237 °C. 1H-NMR (DMSO-d6) δ: 3.18 (s, 3H, SCH3), 6,55 (s, 2H, NH2), 12.86 ppm (s, 1H, NH). ESI-MS: positive mode m/z: 185.0 ([M+Na]+), 346.9 ([2M+Na]+); negative mode m/z: 161.1 ([M−H]−), 197.1 ([M+Cl]−), 323.0 ([2M−H]−), 358.9 ([2M+Cl]−). Elemental analysis calcd for C3H6N4O2S: C 22.22, H 3.73, N 34.55; found C 22.48, H 3.47, N 34.50).
2-(Methylsulfonyl)-5-(methylthio)-[1,2,4]triazolo [1,5-a][1,3,5]triazin-7-amine (17): The intermediate 16 (1.114 g, 6.87 mmol) was reacted with dimethyl N-cyanodithioiminocarbonate (14, 2.009 g, 13.74 mmol) in a metal container hermetically sealed and placed at 170 ° C for 6.5 h. The product becomes a sticky yellow paste which was purified by flash column chromatography eluting with DCM-MeOH (99.5:0.5) to obtain the desired product 17 as a white solid (391 mg, 1.5 mmol; yield 22%). M.p.: 250 °C (dec). 1H-NMR (DMSO-d6) δ: 2.48 (s, 3H, SCH3), 3.40 (s, 3H, SO2CH3), 9.15 ppm (bs, 2H, NH2). ESI-MS: positive mode m/z: 282.9 ([M+Na]+), 542.6 ([2M+Na]+); negative mode m/z: 259.1 ([M−H]−). Elemental analysis calcd for C6H8N6O2S2: C 27.69, H 3.10, N 32.29; found C 27.36, H 3.23, N 32.49).
2-Ethoxy-5-(methylthio)-[1,2,4]triazolo[1,5-a][1,3,5]triazin-7-amine (18) and 2,5-diethoxy-[1,2,4]triazolo[1,5-a][1,3,5]triazin-7-amine (19): 3 mL of ethanol are reacted with sodium hydride (252 mg, 6.45 mmol) for 2 h at room temperature under nitrogen flow. Then compound 17 (150 mg, 0.61 mmol) was added and the reaction mixture was left at 50 °C for 28 h. Then, the reaction was neutralized with an aqueous solution of 2N HCl and brought to dryness under vacuum. Water was added, and the product was extracted with ethyl acetate (3 × 20 mL). The organic phases were dried with anhydrous sodium sulphate and filtered. The filtrate is brought to dryness and the residue purified via flash column chromatography eluting with DCM-MeOH (99.5: 0.5) to obtain the desired compound 18 (44 mg, 0.19 mmol; yield 32%) and a secondary product 19 as white solid (7 mg, 0.031 mmol; 5%).
18: m.p.: 220 °C (dec). 1H-NMR (DMSO-d6) δ: 1.34 (t, 3H, J = 7.1 Hz, CH3), 2.45 (s, 3H, SCH3), 4.36 (q, 2H, J = 7.0 Hz, OCH2), 8.33 (bs, 1H, HNH), 8.75 ppm (bs, 1H, HNH). ESI-MS: positive mode m/z: 227.1 ([M+H]+), 249.1 ([M+Na]+), 475.0 ([2M+Na]+); negative mode m/z: 225.3 ([M−H]−). elemental analysis calcd for C7H10N6OS: C 7.16, H 4.45, N 37.14; found C 7.46, H 4.24, N 37.00.
19: m.p.: 180–181 °C. 1H-NMR (DMSO-d6) δ: 1.27 (t, 3H, J = 6.8 Hz, CH3), 1.33 (t, 3H, J = 6.8 Hz, CH3), 4.28 (q, 2H, J = 7.2 Hz, OCH2), 4.34 (q, 2H, J = 7.2 Hz, OCH2), 8.42 (bs, 2H, NH2). ESI-MS: positive mode m/z: 225.1 ([M+H]+), 247.1 ([M+Na]+), 471.0 ([2M+Na]+); negative mode m/z: 223.2 ([M−H]−). Elemental analysis calcd for C8H12N6O2: C 42.85, H 5.39, N 37.48; found C 42.75, H 5.51, N 37.40.
2-Ethoxy-[1,2,4]triazolo[1,5-a][1,3,5]triazin-7-amine (21): Compound 18 (35 mg, 0.15 mmol) was suspended in tetrahydrofuran (THF, 7 mL) and Pd/C 10% (8 mg, 0.076 mmol) was added. The mixture was placed at 0 °C and then triethylsilane (159 mg, 1.37 mmol) was added. The reaction mixture was left at 0 °C for 30 min and then at r. t. for 7.5 h. The formed suspension was filtered, the filtrate was evaporated to dryness under vacuum, and the residue was crystallized from a mixture of DCM-MeOH (9:1) to obtain the desired product (12 mg, 0.066 mmol; yield 44%). M.p.: 212–213 °C. 1H-NMR (DMSO-d6) δ: 1.35 (t, 3H, J = 6.8 Hz, CH2CH3), 4.40 (q, 2H, J = 6.8 Hz, CH2CH3), 8.21 (s, 1H, H-5), 8.40 (bs, 1H, HNH), 8.85 (s, 1H, HNH). ESI-MS: positive mode m/z: 181.2 ([M+H]+), 203.1 ([M+Na]+), 383.0 ([2M+Na]+); negative mode m/z: 179.3 ([M−H]−), 259.2 ([2M−H]−). Elemental analysis calcd for C14H17N7O2: C 53.32, H 5.43, N 31.09; found C 53.16, H 5.59, N 31.20).
4-(2-((7-amino-2-ethoxy-[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino)ethyl)phenol (20): To a suspension of 18 (80 mg, 0.35 mmol) in THF, m-CPBA (88 mg, 0.51 mmol) was added at r. t. and left to react for 2 h. Then, to the reaction mixture, a solution of tyramine (77 mg, 0.35 mmol) in THF was added. The temperature was brought to 40 °C and left for 24 h. To the reaction mixture, 2N NaHCO3 was added to neutralize the pH and extracted with EtOAc (3 × 20 mL). The organic phases were dried with anhydrous sodium sulphate and filtered. The filtrate was brought to dryness and the residue purified via flash column chromatography eluting with DCM-MeOH (98:2). The compound was obtained by crystallization with MeOH-EtOAc-Petroleum ether (9:1:1) to obtain 20 as a white powder (4 mg, mmol; yield 10%). 1H-NMR (DMSO-d6) δ: 1.31 (t, 3H, CH3), 2.66 (m, 2H, CH2Ph), 3.32 (m, CH2NH2), 4.29 (q, 2H, OCH2), 6.98 (d, 2H, J = 7.0 Hz, H-Ph), 6.65 (d, 2H, H-Ph), 7.28 (m, 1H, NH), 9.15 (s, 1H, HO-Ph). ESI-MS: positive mode m/z: 316.1 ([M+H]+), 337.9 ([M+Na]+), 653.0 ([2M+Na]+). Elemental analysis calcd for C6H8N6O: C 40.00, H 4.48, N 46.65; found C 40.24, H 4.44, N 46.42.
3.2. Crystallographic Study
The crystallographic data for complex 17 were collected on a Bruker APEX II single-crystal diffractometer working with monochromatic Mo-Kα radiation and equipped with an area detector. The structure was solved by direct methods and refined against F2 with SHELXL-2014/7 with anisotropic thermal parameters for all non-hydrogen atoms [62,63]. Idealized geometries were assigned to the hydrogen atoms. Crystallographic data were deposited with the Cambridge Crystallographic Data Centre as a supplementary publication (reference code CCDC 2133489). Copies of the data can be obtained free of charge upon application to the CCDC, 12 Union Road, Cambridge CB2 1EZ, UK (fax, (+44) 1223 336033; e-mail, deposit@ccdc.cam.ac.uk).
3.3. Biological Assays
3.3.1. Binding Evaluation at ARs
Cell culture: The Chinese hamster ovary cells (CHO) stably transfected with the desired ARs were grown in DMEM/F12 medium containing 10% FBS (fetal bovine serum), 100 U/mL penicillin, 100 µg/mL streptomycin, 2.5 μg/mL amphotericin B, 1 mM sodium pyruvate, 0.1 mg/mL genetecina (G418), and incubated at 37 °C in a gaseous mixture of 5% CO2: 95% O2.
Preparation of membranes: Cell membranes for binding assays were prepared mechanically detaching the cells from the petri and suspending them in a cold hypotonic buffer (5 mM Tris/HCl, 2 mM EDTA, pH 7.4). The cell suspension was homogenized (Ultra-Turrax, 2 × 15 s at maximum speed) and the homogenate centrifuged for 10 min (4 °C) to 1000× g. The supernatant was subsequently centrifuged for 30 min (4 °C) at 100,000× g and the pellet containing the membrane proteins was re-suspended in a specific buffer for each receptor subtype (A1: 50 mM Tris/HCl, pH 7.4; A2A: 50 mM Tris/HCl, 10 mM MgCl2, pH 7.4; A3: 50 mM Tris/HCl, 10 mM MgCl2, 1 mM EDTA, pH 8.25). The amount of the protein suspension was determined according to the Bradford method using the BCA Protein Assay Kit (Pierce), frozen in liquid nitrogen, and stored at −80 °C.
Binding assays: The dissociation constants of the radioligand (KD) were calculated through saturation assays, while the dissociation constants of the compounds under study (Ki) were determined by competition experiments. All binding data were calculated by nonlinear regression curves using the software GraphPad Prism 5 (GraphPad Software, San Diego, CA, USA). The increasing concentrations of the radioligand saturation binding ([3H]-CCPA for the A1 receptor subtype, [3H]-NECA for the A2A receptor subtype, and [3H]-HEMADO for the A3 receptor) subtype were incubated in a total volume of 250 μL containing 0.2 U/mL of Ado deaminase (ADA) and 12 micrograms of membrane proteins in the specific buffer. In competition experiments for the A1 receptor subtype, each compound was added at increasing concentrations, to the well containing 1 nM of [3H]-CCPA, to a final volume of 200 μL. The samples were left to incubate for 3 h at room temperature, filtered in a filter 96-well plate (UniFilter GF/C, Perkin-Elmer Life and Analytical Science, Boston, MA) using the FilterMate Cell Harvester (Perkin-Elmer), and washed 3 times with cold distilled water. After drying the filter plate at 40 °C for 30′, they 20 μL of scintillating liquid (Microscint-20, Perkin-Elmer) was added to each well, and then the radioactivity was quantized with the MicroBeta2 Plate Counter (Perkin-Elmer). The conditions for binding experiments at A2A and A3 receptor subtypes were essentially the same as for the A1AR using [3H]-NECA (N-ethylcarboxyamideAdo) at a concentration of 10 nM and [3H]-HEMADO (2-(1-hexyne)-N6-methylAdo) at a concentration of 1 nM as radioligands, respectively. The samples were incubated with 12 micrograms of protein for 3 h at room temperature and filtered as described above.
The non-specific binding was determined in the presence of 1 mM theophylline for the A1AR, or 100 µM R-PIA (N6-(1-methyl-2-phenylethyl)Ado) for the A2A and A3 receptors.
3.3.2. Anti-Inflammatory Activity Assays
Cell Cultures and Treatments: BV-2 murine microglial cells (a gift from Prof. Elisabetta Blasi of the University of Modena and Reggio Emilia, Modena, Italy) were maintained at 37 °C and 5% CO2 in a humidified incubator and grown in DMEM supplemented with 10% (v/v) low-endotoxin fetal bovine serum (Euroclone—Milan, Italy), 2 mM L-glutamine, 50 U/mL penicillin, and 50 μg/mL streptomycin (Sigma-Aldrich–Merck, Milan, Italy). Cells were seeded at 50,000 cells/mL in 96-well tissue culture plates for the viability assay, in 6-well tissue culture plates for the Griess assay and RT-PCR assay. BV-2 cells were treated for 24 h with ANR 94 or 13 at different concentrations and then exposed to LPS (100 ng/mL; Sigma-Aldrich–Merck) for a further 24 h.
MTT Viability Assay: MTT assay was performed to quantify the viability of BV-2 cells as previously reported [52]. At the end of the experiments, a cell culture medium was replaced with 0.5 mg/mL MTT solution (Sigma-Aldrich–Merck), and after 30 min the formed formazan crystals were solubilized, replacing the MTT solution with DMSO. The absorbance was measured at 595 nm using a microplate spectrophotometer (VICTOR3 V Multilabel Counter; PerkinElmer, Wellesley, MA, USA). Data are expressed as a percentage of the control cells, which is considered 100% cell viability.
Measurement of NO Production: The amount of nitrite accumulation in the culture medium of BV-2 cells was measured using Griess reagent. 50 µL of culture medium from each sample was mixed with the same volume of the Griess reagent (Sigma-Aldrich-Merck). The NO concentration was evaluated by measuring the absorbance at 540 nm with a 96-well microplate spectrophotometer (VICTOR3 V Multilabel Counter; PerkinElmer). Concentration of nitrites released in the medium was determined from the standard curve generated with known concentrations of sodium nitrite. Results are expressed as mean concentration of nitrites (μM) ± SEM, from three different samples.
Real-Time Polymerase Chain Reaction (PCR): At the end of the experiments, total RNA was extracted by BV-2 cells using RNeasy Mini Kit (QIAGEN GmbH, Hilden, Germany). RNA concentraion and quality were measured on a NanoVue Spectrophotometer (GE Healthcare, Milano, Italy). iScript cDNA Synthesis Kit (Bio-Rad, Hercules, CA, USA) was used to synthesize cDNA starting from the extracted RNA, following the supplier’s instructions. PCR was performed by adding 2.5 μL (12.5 ng) of cDNA, 5 μL SsoAdvanced Universal SYBR Green Supermix (Bio-Rad), and 0.5 μL (500 nM) of each primer (Table 2) to a PCR tube. cDNA amplification was started at 95 °C for 30 s to activate the polymerase, followed by 40 cycles of 5 s at 95 °C and 30 s at 60 °C. Normalized expression levels were calculated relative to the control cells according to the 2−ΔΔCT method.

Table 2.
Reports the primers used (Sigma-Aldrich–Merck). GAPDH was used as the reference gene.
3.4. Molecular Modeling
Receptor refinement and energy minimization tasks were carried out using MOE. Docking experiments were performed with CCDC Gold [57].
A2AAR and A1AR crystal structures refinement: The crystal structures of the human A2AAR and A1AR in complex with ZM241385 and the antagonist PSB36, respectively, were downloaded by the Protein Data Bank webpage (http://www.rcsb.org accessed on 7 January 2022; pdb code: 5NM4; 1.7-Å resolution [26], and pdb code: 5N2S; 3.6-Å resolution [56], respectively). The two structures were checked into MOE by restoring missing loops and the wild-type receptor sequences, and by adding all the hydrogen atoms. The protein structures were then energetically minimized with MOE using the AMBER99 force field until the RMS gradient of the potential energy was less than 0.05 kJ mol−1 Å−1.
Homology modeling of the human A3AR structure: A homology model of the human A3AR was built using the above cited X-ray structure of the antagonist-bound A1AR as a template (pdb code: 5N2S). A multiple alignment of the AR primary sequences was built within MOE as the preliminary step. The Homology Modeling tool of MOE was employed for this task. The obtained A3AR model was then energetically minimized with the same protocol of the other receptors (see above).
Molecular docking analysis: Docking analyses were performed by using CCDC Gold [57], which was set with default efficiency settings by selecting ChemScore as the scoring function and generating 50 poses for each ligand.
Post Docking analysis. Residue interaction analysis: The interactions between the ligands and the AR receptors binding sites were analyzed by using the IF-E 6.0 tool retrievable at the SVL exchange service (Chemical Computing Group, Inc. SVL exchange: http://svl.chemcomp.com accessed on 7 January 2022). The program calculates and displays the atomic and residue interaction forces as 3D vectors. It also calculates the per-residue interaction energies, where negative and positive energy values are associated to favorable and unfavorable interactions, respectively. For each AR structure, a shell of residues contained within a 10 Å distance from the ligand were considered for this analysis.
4. Conclusions
On the base of ZM241385 and ANR 94 structures, two series of new compounds endowed with the triazolotriazine, and purine scaffolds were designed and synthesized. Radioligand binding and functional assays at human recombinant ARs showed that the 9-ethyl-8-furyl-2-(4-hydroxyphenyl)-2-ethylaminoadenine (13) and its 4-methoxyphenyl analogue 12 showed high affinity (Ki in the low nM range) and a different degree of selectivity at the A2AAR. Furthermore, 13 was found to exert anti-inflammatory properties in a microglial model of neuroinflammation, since it was able to reduce pro-inflammatory mediators and to increase anti-inflammatory cytokines in microglia cells.
Comparing the ARs binding mode of selected compounds belonging to the two series, molecular modeling studies support the hypothesis that the triazolotriazine and the purine scaffolds are interchangeable only when the 2 and 5 positions of the triazolotriazine moiety (corresponding to the purine 2- and 8-positions) are substituted. Taken together, the results of our experiments reveal that the novel A2AAR antagonist 13 is a potential tool for studying neuroinflammation and neuroprotective processes and could be a useful anti-PD agent suitable for further investigations.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/molecules27082386/s1, 1H NMR and 13C NMR spectra of compounds 7–13 and 17–21.
Author Contributions
Conceptualization, R.V., G.M.; methodology, A.S., C.L., M.B., M.C.B., S.H., D.D.B., C.G., K.-N.K.; software, D.D.B.; data curation, C.A., G.M.; writing—original draft, C.L.; visualization, A.M.; supervision, R.V., C.A., G.M., D.D.B.; funding acquisition, S.K.T., C.L., C.A., M.U. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the University of Camerino, grant number FPI000065 FAR 2019.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
Authors declare no conflict of interest.
References
- Fredholm, B.B.; IJzerman, A.P.; Jacobson, K.A.; Linden, J.; Muller, C.E. International Union of Basic and Clinical Pharmacology. LXXXI. Nomenclature and classification of adenosine receptors-An update. Pharm. Rev 2011, 63, 1–34. [Google Scholar] [CrossRef] [PubMed]
- Wilson, C.N.; Mustafa, S.J. Adenosine Receptors in Health and Disease; Springer: Berlin/Heidelberg, Germany, 2009; Volume 193. [Google Scholar]
- Borea, P.A.; Gessi, S.; Merighi, S.; Vincenzi, F.; Varani, K. Pharmacology of Adenosine Receptors: The State of the Art. Physiol. Rev. 2018, 98, 1591–1625. [Google Scholar] [CrossRef] [PubMed]
- Hasko, G.; Cronstein, B. Regulation of inflammation by adenosine. Front. Immunol 2013, 4, 85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Antonioli, L.; Pacher, P.; Hasko, G. Adenosine and inflammation: It’s time to (re)solve the problem. Trends Pharmacol. Sci. 2022, 43, 43–55. [Google Scholar] [CrossRef] [PubMed]
- Haddad, M. Impact of Adenosine A2 Receptor Ligands on BCL2 Expression in Skeletal Muscle Cells. Appl. Sci. 2021, 11, 2272. [Google Scholar] [CrossRef]
- Rebola, N.; Simoes, A.P.; Canas, P.M.; Tome, A.R.; Andrade, G.M.; Barry, C.E.; Agostinho, P.M.; Lynch, M.A.; Cunha, R.A. Adenosine A2A receptors control neuroinflammation and consequent hippocampal neuronal dysfunction. J. Neurochem. 2011, 117, 100–111. [Google Scholar] [CrossRef]
- Pinna, A.; Tronci, E.; Schintu, N.; Simola, N.; Volpini, R.; Pontis, S.; Cristalli, G.; Morelli, M. A new ethyladenine antagonist of adenosine A2A receptors: Behavioral and biochemical characterization as an antiparkinsonian drug. Neuropharmacology 2010, 58, 613–623. [Google Scholar] [CrossRef]
- Marti Navia, A.; Dal Ben, D.; Lambertucci, C.; Spinaci, A.; Volpini, R.; Marques-Morgado, I.; Coelho, J.E.; Lopes, L.V.; Marucci, G.; Buccioni, M. Adenosine Receptors as Neuroinflammation Modulators: Role of A1 Agonists and A2A Antagonists. Cells 2020, 9, 1739. [Google Scholar] [CrossRef]
- Allan, S.M.; Tyrrell, P.J.; Rothwell, N.J. Interleukin-1 and neuronal injury. Nat. Rev. Immunol. 2005, 5, 629–640. [Google Scholar] [CrossRef]
- Lai, T.T.; Kim, Y.J.; Ma, H.I.; Kim, Y.E. Evidence of Inflammation in Parkinson’s Disease and Its Contribution to Synucleinopathy. J. Mov. Disord. 2022, 15, 1–14. [Google Scholar] [CrossRef]
- Jenner, P.; Mori, A.; Hauser, R.; Morelli, M.; Fredholm, B.B.; Chen, J.F. Adenosine, adenosine A2A antagonists, and Parkinson’s disease. Parkinsonism Relat. Disord. 2009, 15, 406–413. [Google Scholar] [CrossRef] [PubMed]
- Pinna, A.; Serra, M.; Morelli, M.; Simola, N. Role of adenosine A2A receptors in motor control: Relevance to Parkinson’s disease and dyskinesia. J. Neural Transm. 2018, 125, 1273–1286. [Google Scholar] [CrossRef] [PubMed]
- Shang, P.; Baker, M.; Banks, S.; Hong, S.I.; Chol, D.S. Emerging Nondopaminergic Medications for Parkinson’s Disease: Focusing on A2A Receptor Antagonists and GLP1 Receptor Agonists. J. Mov. Disord. 2021, 14, 193–203. [Google Scholar] [CrossRef]
- Hauser, R.A.; Hattori, N.; Fernandez, H.; Isaacson, S.H.; Mochizuki, H.; Rascol, O.; Stocchi, F.; Li, J.; Mori, A.; Nakajima, Y.; et al. Efficacy of Istradefylline, an Adenosine A2A Receptor Antagonist, as Adjunctive Therapy to Levodopa in Parkinson’s Disease: A Pooled Analysis of 8 Phase 2b/3 Trials. J. Parkinson’s Dis. 2021, 11, 1663–1675. [Google Scholar] [CrossRef] [PubMed]
- Jenner, P.; Mori, A.; Aradi, S.D.; Hauser, R.A. Istradefylline-A first generation adenosine A2A antagonist for the treatment of Parkinson’s disease. Expert Rev. Neurother. 2021, 21, 317–333. [Google Scholar] [CrossRef]
- Chen, J.F.; Cunha, R.A. The belated US FDA approval of the adenosine A2A receptor antagonist istradefylline for treatment of Parkinson’s disease. Purinergic Signal. 2020, 16, 167–174. [Google Scholar] [CrossRef]
- FDA Approves New Add-On Drug to Treat off Episodes in Adults with Parkinson’s Disease. Available online: https://www.fda.gov/news-events/press-announcements/fda-approves-new-add-drug-treat-episodes-adults-parkinsons-disease (accessed on 22 February 2022).
- Muller, C.E.; Jacobson, K.A. Recent developments in adenosine receptor ligands and their potential as novel drugs. Biochim. Biophys. Acta 2011, 1808, 1290–1308. [Google Scholar] [CrossRef] [Green Version]
- Muller, C.E.; Jacobson, K.A. Xanthines as adenosine receptor antagonists. Handb. Exp. Pharm. 2011, 200, 151–199. [Google Scholar]
- Jacobson, K.A.; IJzerman, A.P.; Muller, C.E. Medicinal chemistry of P2 and adenosine receptors: Common scaffolds adapted for multiple targets. Biochem. Pharmacol. 2021, 187, 114311. [Google Scholar] [CrossRef]
- Williams, M.; Francis, J.; Ghai, G.; Braunwalder, A.; Psychoyos, S.; Stone, G.A.; Cash, W.D. Biochemical characterization of the triazoloquinazoline, CGS 15943, a novel, non-xanthine adenosine antagonist. J. Pharmacol. Exp. Ther. 1987, 241, 415–420. [Google Scholar]
- LeWitt, P.A.; Aradi, S.D.; Hauser, R.A.; Rascol, O. The challenge of developing adenosine A2A antagonists for Parkinson disease: Istradefylline, preladenant, and tozadenant. Parkinsonism Relat. Disord. 2020, 80, S54–S63. [Google Scholar] [CrossRef] [PubMed]
- Merck Provides Update on Phase III Clinical Program for Preladenant, the Company’s Investigational Parkinson’s Disease Medicine. Available online: www.merck.com/news/merck-provides-update-on-phase-iii-clinical-program-for-preladenant-the-companys-investigational-parkinsons-disease-medicine (accessed on 24 February 2022).
- Hauser, R.A.; Stocchi, F.; Rascol, O.; Huyck, S.B.; Capece, R.; Ho, T.W.; Sklar, P.; Lines, C.; Michelson, D.; Hewitt, D. Preladenant as an Adjunctive Therapy with Levodopa in Parkinson Disease Two Randomized Clinical Trials and Lessons Learned. JAMA Neurol. 2015, 72, 1491–1500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weinert, T.; Olieric, N.; Cheng, R.; Brunle, S.; James, D.; Ozerov, D.; Gashi, D.; Vera, L.; Marsh, M.; Jaeger, K.; et al. Serial millisecond crystallography for routine room-temperature structure determination at synchrotrons. Nat. Commun. 2017, 8, 542. [Google Scholar] [CrossRef] [PubMed]
- Carpenter, B.; Lebon, G. Human Adenosine A2A Receptor: Molecular Mechanism of Ligand Binding and Activation. Front. Pharm. 2017, 8, 898. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jaakola, V.P.; Griffith, M.T.; Hanson, M.A.; Cherezov, V.; Chien, E.Y.; Lane, J.R.; Ijzerman, A.P.; Stevens, R.C. The 2.6 angstrom crystal structure of a human A2A adenosine receptor bound to an antagonist. Science 2008, 322, 1211–1217. [Google Scholar] [CrossRef] [Green Version]
- Lambertucci, C.; Spinaci, A.; Buccioni, M.; Dal Ben, D.; Ngouadjeu Ngnintedem, M.A.; Kachler, S.; Marucci, G.; Klotz, K.N.; Volpini, R. New A2A adenosine receptor antagonists: A structure-based upside-down interaction in the receptor cavity. Bioorg. Chem. 2019, 92, 103183. [Google Scholar] [CrossRef]
- Lambertucci, C.; Marucci, G.; Catarzi, D.; Colotta, V.; Francucci, B.; Spinaci, A.; Varano, F.; Volpini, R. A2A Adenosine Receptor Antagonists and their Potential in Neurological Disorders. Curr. Med. Chem. 2022. accepted. [Google Scholar] [CrossRef]
- Lambertucci, C.; Antonini, I.; Buccioni, M.; Dal Ben, D.; Kachare, D.D.; Volpini, R.; Klotz, K.N.; Cristalli, G. 8-Bromo-9-alkyl adenine derivatives as tools for developing new adenosine A2A and A2B receptors ligands. Bioorg. Med. Chem. 2009, 17, 2812–2822. [Google Scholar] [CrossRef]
- Camaioni, E.; Costanzi, S.; Vittori, S.; Volpini, R.; Klotz, K.N.; Cristalli, G. New substituted 9-alkylpurines as adenosine receptor ligands. Bioorg. Med. Chem. 1998, 6, 523–533. [Google Scholar] [CrossRef]
- Jorg, M.; Agostino, M.; Yuriev, E.; Mak, F.S.; Miller, N.D.; White, J.M.; Scammells, P.J.; Capuano, B. Synthesis, molecular structure, NMR spectroscopic and computational analysis of a selective adenosine A2A antagonist, ZM241385. Struct. Chem. 2013, 24, 1241–1251. [Google Scholar] [CrossRef]
- Caulkett, P.W.R.; Jones, G.; McPartlin, M.; Renshaw, N.D.; Stewart, S.K.; Wright, B. Adenine isosteres with bridgehead nitrogen. Part 1. Two independent syntheses of the [1,2,4]triazolo[1,5-a][1,3,5]triazine ring system leading to a range of substituents in the 2, 5 and 7 positions. J. Chem. Soc. Perkin Trans. 1995, 1, 801–808. [Google Scholar] [CrossRef]
- Klotz, K.N.; Falgner, N.; Kachler, S.; Lambertucci, C.; Vittori, S.; Volpini, R.; Cristalli, G. [3H]HEMADO-a novel tritiated agonist selective for the human adenosine A3 receptor. Eur. J. Pharmacol. 2007, 556, 14–18. [Google Scholar] [CrossRef] [PubMed]
- Falsini, M.; Ceni, C.; Catarzi, D.; Varano, F.; Dal Ben, D.; Marucci, G.; Buccioni, M.; Marti Navia, A.; Volpini, R.; Colotta, V. New 8-amino-1,2,4-triazolo[4,3-a]pyrazin-3-one derivatives. Evaluation of different moieties on the 6-aryl ring to obtain potent and selective human A2A adenosine receptor antagonists. Bioorg. Med. Chem. Lett. 2020, 30, 127126. [Google Scholar] [CrossRef] [PubMed]
- Buccioni, M.; Marucci, G.; Dal Ben, D.; Giacobbe, D.; Lambertucci, C.; Soverchia, L.; Thomas, A.; Volpini, R.; Cristalli, G. Innovative functional cAMP assay for studying G protein-coupled receptors: Application to the pharmacological characterization of GPR17. Purinergic Signal. 2011, 7, 463–468. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jacobson, K.A.; Gao, Z.G. Adenosine receptors as therapeutic targets. Nat. Rev. Drug Discov. 2006, 5, 247–264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klotz, K.N.; Kachler, S.; Lambertucci, C.; Vittori, S.; Volpini, R.; Cristalli, G. 9-Ethyladenine derivatives as adenosine receptor antagonists: 2- and 8-substitution results in distinct selectivities. Naunyn Schmiedebergs Arch. Pharm. 2003, 367, 629–634. [Google Scholar] [CrossRef]
- He, J.; Zhu, G.; Wang, G.; Zhang, F. Oxidative Stress and Neuroinflammation Potentiate Each Other to Promote Progression of Dopamine Neurodegeneration. Oxid. Med. Cell. Longev. 2020, 2020, 6137521. [Google Scholar] [CrossRef]
- Wang, Y.; Li, L.; Wu, Y.; Zhang, S.; Ju, Q.; Yang, Y.; Jin, Y.; Shi, H.; Sun, C. CD44 deficiency represses neuroinflammation and rescues dopaminergic neurons in a mouse model of Parkinson’s disease. Pharmacol. Res. 2022, 177, 106133. [Google Scholar] [CrossRef]
- Badanjak, K.; Fixemer, S.; Smajic, S.; Skupin, A.; Grunewald, A. The Contribution of Microglia to Neuroinflammation in Parkinson’s Disease. Int. J. Mol. Sci. 2021, 22, 4676. [Google Scholar] [CrossRef]
- Tansey, M.G.; McCoy, M.K.; Frank-Cannon, T.C. Neuroinflammatory mechanisms in Parkinson’s disease: Potential environmental triggers, pathways, and targets for early therapeutic intervention. Exp. Neurol. 2007, 208, 1–25. [Google Scholar] [CrossRef] [Green Version]
- Gyoneva, S.; Shapiro, L.; Lazo, C.; Garnier-Amblard, E.; Smith, Y.; Miller, G.W.; Traynelis, S.F. Adenosine A2A receptor antagonism reverses inflammation-induced impairment of microglial process extension in a model of Parkinson’s disease. Neurobiol. Dis. 2014, 67, 191–202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moehle, M.S.; West, A.B. M1 and M2 immune activation in Parkinson’s Disease: Foe and ally? Neuroscience 2015, 302, 59–73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sica, A.; Mantovani, A. Macrophage plasticity and polarization: In vivo veritas. J. Clin. Investig. 2012, 122, 787–795. [Google Scholar] [CrossRef] [PubMed]
- Gustin, A.; Kirchmeyer, M.; Koncina, E.; Felten, P.; Losciuto, S.; Heurtaux, T.; Tardivel, A.; Heuschling, P.; Dostert, C. NLRP3 Inflammasome Is Expressed and Functional in Mouse Brain Microglia but Not in Astrocytes. PLoS ONE 2015, 10, e0130624. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, H.M.; Jiang, J.; Wilson, B.; Zhang, W.; Hong, J.S.; Liu, B. Microglial activation-mediated delayed and progressive degeneration of rat nigral dopaminergic neurons: Relevance to Parkinson’s disease. J. Neurochem. 2002, 81, 1285–1297. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, S.; Malovic, E.; Harishchandra, D.S.; Ghaisas, S.; Panicker, N.; Charli, A.; Palanisamy, B.N.; Rokad, D.; Jin, H.; Anantharam, V.; et al. Mitochondrial impairment in microglia amplifies NLRP3 inflammasome proinflammatory signaling in cell culture and animal models of Parkinson’s disease. NPJ Parkinson’s Dis. 2017, 3, 30. [Google Scholar] [CrossRef]
- Ding, R.R.; Chen, W.; Guo, C.Y.; Liao, W.T.; Yang, X.; Liao, F.E.; Lin, J.M.; Mei, H.F.; Zeng, Y. Dangguishaoyao-San attenuates LPS-induced neuroinflammation via the TLRs/NF-kappaB signaling pathway. Biomed. Pharmacother. 2018, 105, 187–194. [Google Scholar] [CrossRef]
- Lee, D.G.; Nam, B.R.; Huh, J.W.; Lee, D.S. Isoliquiritigenin Reduces LPS-Induced Inflammation by Preventing Mitochondrial Fission in BV-2 Microglial Cells. Inflammation 2021, 44, 714–724. [Google Scholar] [CrossRef]
- Angeloni, S.; Freschi, M.; Marrazzo, P.; Hrelia, S.; Beghelli, D.; Juan-Garcia, A.; Juan, C.; Caprioli, G.; Sagratini, G.; Angeloni, C. Antioxidant and Anti-Inflammatory Profiles of Spent Coffee Ground Extracts for the Treatment of Neurodegeneration. Oxid. Med. Cell. Longev. 2021, 2021, 6620913. [Google Scholar] [CrossRef]
- Henn, A.; Lund, S.; Hedtjarn, M.; Schrattenholz, A.; Porzgen, P.; Leist, M. The suitability of BV2 cells as alternative model system for primary microglia cultures or for animal experiments examining brain inflammation. ALTEX 2009, 26, 83–94. [Google Scholar] [CrossRef] [Green Version]
- Bauernfeind, F.G.; Horvath, G.; Stutz, A.; Alnemri, E.S.; MacDonald, K.; Speert, D.; Fernandes-Alnemri, T.; Wu, J.; Monks, B.G.; Fitzgerald, K.A.; et al. Cutting edge: NF-kappaB activating pattern recognition and cytokine receptors license NLRP3 inflammasome activation by regulating NLRP3 expression. J. Immunol. 2009, 183, 787–791. [Google Scholar] [CrossRef] [PubMed]
- Oishi, S.; Takano, R.; Tamura, S.; Tani, S.; Iwaizumi, M.; Hamaya, Y.; Takagaki, K.; Nagata, T.; Seto, S.; Horii, T.; et al. M2 polarization of murine peritoneal macrophages induces regulatory cytokine production and suppresses T-cell proliferation. Immunology 2016, 149, 320–328. [Google Scholar] [CrossRef] [PubMed]
- Cheng, R.K.Y.; Segala, E.; Robertson, N.; Deflorian, F.; Dore, A.S.; Errey, J.C.; Fiez-Vandal, C.; Marshall, F.H.; Cooke, R.M. Structures of Human A1 and A2A Adenosine Receptors with Xanthines Reveal Determinants of Selectivity. Structure 2017, 25, 1275–1285.e4. [Google Scholar] [CrossRef] [PubMed]
- Jones, G.; Willett, P.; Glen, R.C.; Leach, A.R.; Taylor, R. Development and validation of a genetic algorithm for flexible docking. J. Mol. Biol. 1997, 267, 727–748. [Google Scholar] [CrossRef] [Green Version]
- Molecular Operating Environment (MOE 2020.09). Chemical Computing Group, Inc.: Montreal, QC, Canada 1255 University St.: Suite 1600, H3B 3X3. Available online: https://www.chemcomp.com/Products.htm (accessed on 7 January 2022).
- Dal Ben, D.; Buccioni, M.; Lambertucci, C.; Marucci, G.; Santinelli, C.; Spinaci, A.; Thomas, A.; Volpini, R. Simulation and Comparative Analysis of Different Binding Modes of Non-nucleoside Agonists at the A2A Adenosine Receptor. Mol. Inform. 2016, 35, 403–413. [Google Scholar] [CrossRef] [PubMed]
- Dal Ben, D.; Buccioni, M.; Lambertucci, C.; Marucci, G.; Thomas, A.; Volpini, R.; Cristalli, G. Molecular modeling study on potent and selective adenosine A3 receptor agonists. Bioorg. Med. Chem. 2010, 18, 7923–7930. [Google Scholar] [CrossRef]
- Dal Ben, D.; Buccioni, M.; Lambertucci, C.; Thomas, A.; Volpini, R. Simulation and comparative analysis of binding modes of nucleoside and non-nucleoside agonists at the A2B adenosine receptor. In Silico Pharmacol. 2013, 1, 24. [Google Scholar] [CrossRef] [Green Version]
- Sheldrick, G.M. Crystal structure refinement with SHELXL. Acta Crystallogr. C Struct. Chem. 2015, 71, 3–8. [Google Scholar] [CrossRef]
- Farrugia, L.J. WinGX and ORTEP for Windows: An Update. J. Appl. Crystallogr. 2012, 45, 849–854. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
