Automated Microbial Library Generation Using the Bioinformatics Platform IDBac
Abstract
1. Introduction
2. Results and Discussion
2.1. Collection and Processing of Environmental Samples
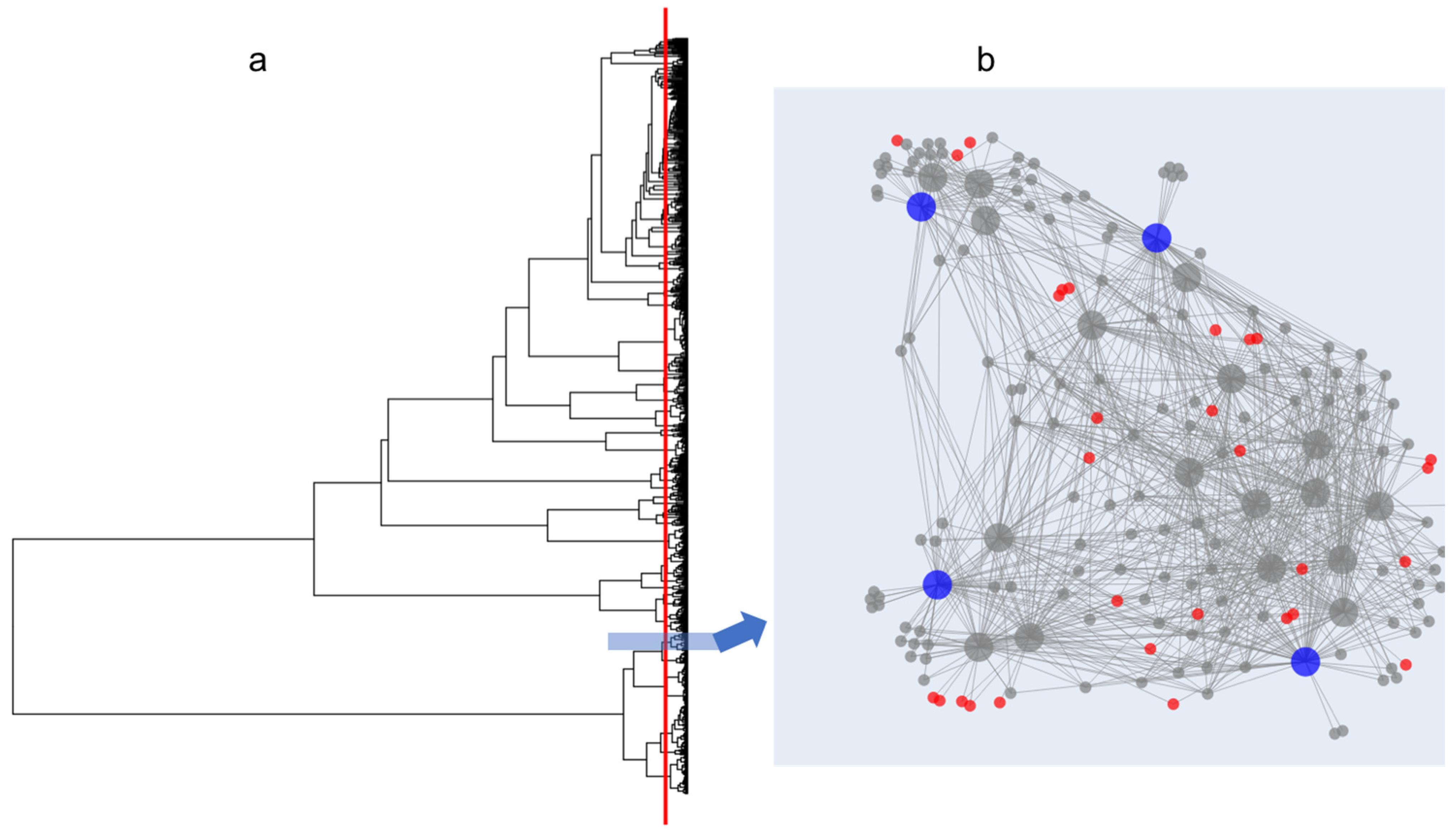
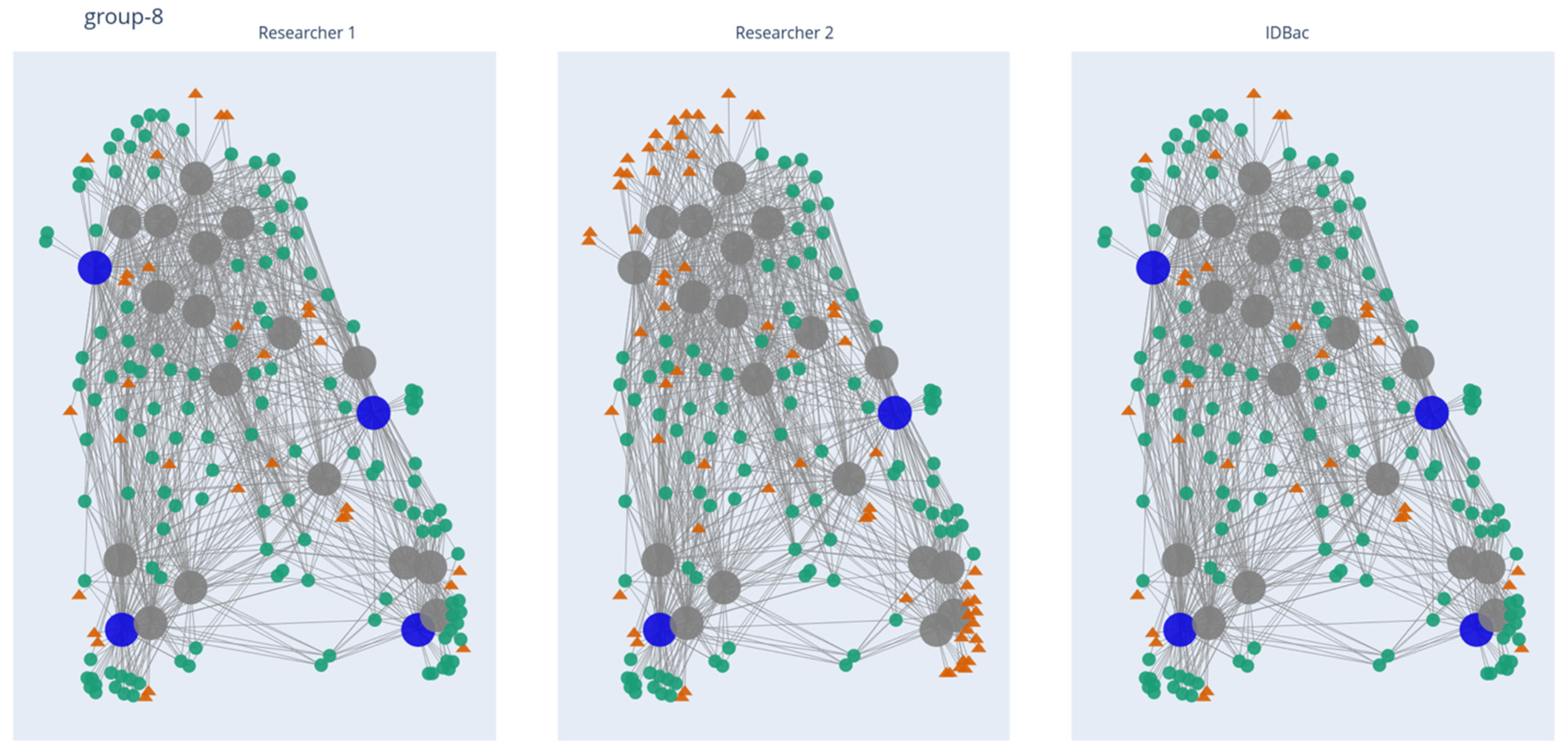
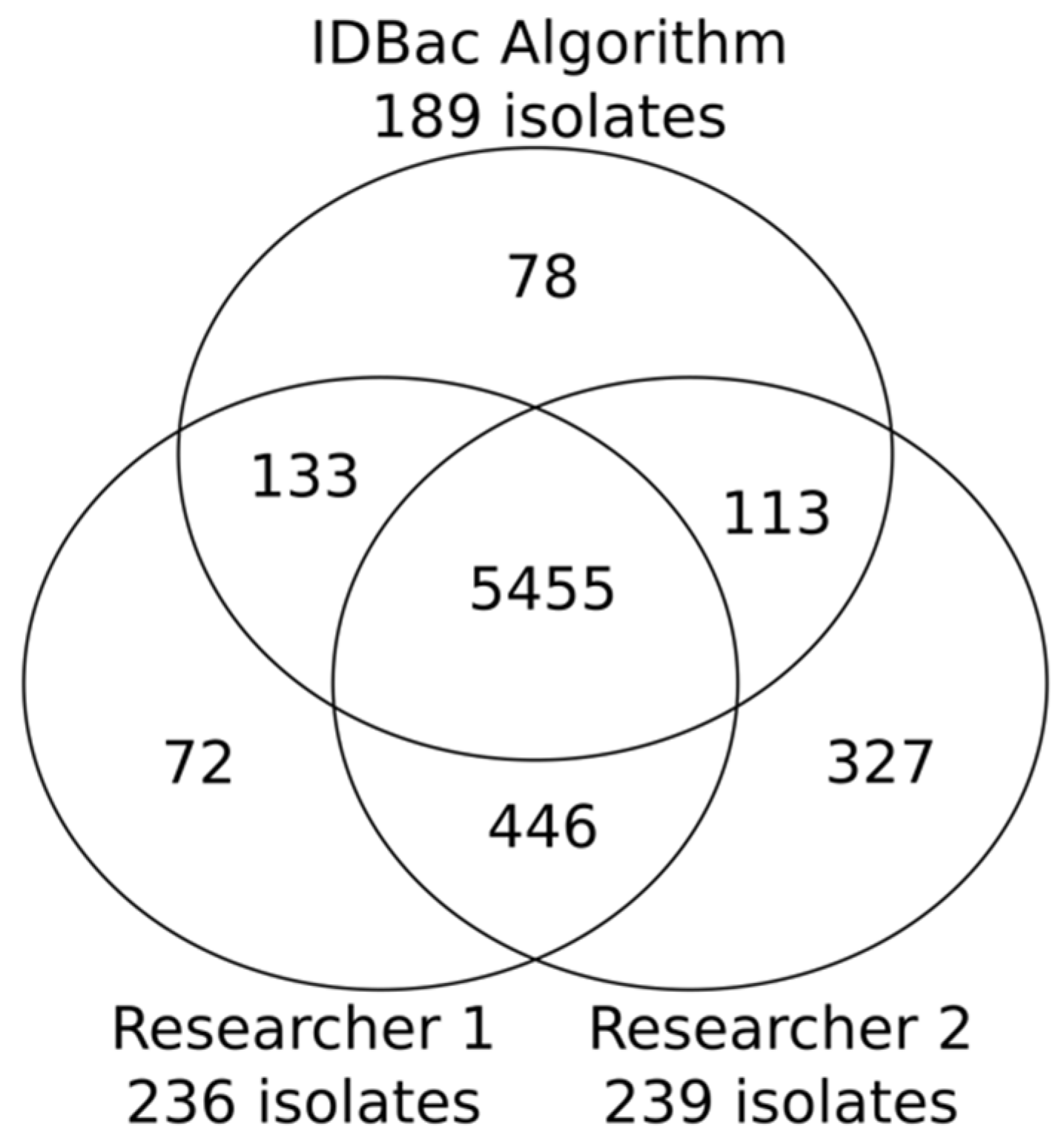
2.2. Application of the Isolate Prioritization Algorithm to a Collection of Unknown Environmental Bacterial Isolates
2.3. Study Limitations and Perspectives
3. Conclusions
4. Experimental Section
Sample Collection and Processing MALDI-TOF MS
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bouslimani, A.; Sanchez, L.M.; Garg, N.; Dorrestein, P.C. Mass spectrometry of natural products: Current, emerging and future technologies. Nat. Prod. Rep. 2014, 31, 718–729. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; Braun, D.R.; Michel, C.R.; Klassen, J.L.; Adnani, N.; Wyche, T.P.; Bugni, T.S. Microbial strain prioritization using metabolomics tools for the discovery of natural products. Anal. Chem. 2012, 84, 4277–4283. [Google Scholar] [PubMed]
- Yang, J.Y.; Sanchez, L.M.; Rath, C.M.; Liu, X.; Boudreau, P.D.; Bruns, N.; Glukhov, E.; Wodtke, A.; de Felicio, R.; Fenner, A.; et al. Molecular networking as a dereplication strategy. J. Nat. Prod. 2013, 76, 1686–1699. [Google Scholar] [CrossRef] [PubMed]
- Molinski, T.F. Microscale methodology for structure elucidation of natural products. Curr. Opin. Biotechnol. 2010, 21, 819–826. [Google Scholar] [PubMed]
- Bauman, K.D.; Butler, K.S.; Moore, B.S.; Chekan, J.R. Genome mining methods to discover bioactive natural products. Nat. Prod. Rep. 2021, 38, 2100–2129. [Google Scholar]
- Wilson, B.A.P.; Thornburg, C.C.; Henrich, C.J.; Grkovic, T.; O’Keefe, B.R. Creating and screening natural product libraries. Nat. Prod. Rep. 2020, 37, 893–918. [Google Scholar]
- Potts, M.B.; Kim, H.S.; Fisher, K.W.; Hu, Y.; Carrasco, Y.P.; Bulut, G.B.; Ou, Y.-H.; Herrera-Herrera, M.L.; Cubillos, F.; Mendiratta, S.; et al. Using functional signature ontology (FUSION) to identify mechanisms of action for natural products. Sci. Signal. 2013, 6, ra90. [Google Scholar] [CrossRef]
- Hernandez, A.; Nguyen, L.T.; Dhakal, R.; Murphy, B.T. The need to innovate sample collection and library generation in microbial drug discovery: A focus on academia. Nat. Prod. Rep. 2021, 38, 292–300. [Google Scholar] [CrossRef]
- Liu, X.; Ashforth, E.; Ren, B.; Song, F.; Dai, H.; Liu, M.; Wang, J.; Xie, Q.; Zhang, L. Bioprospecting microbial natural product libraries from the marine environment for drug discovery. J. Antibiot. 2010, 63, 415–422. [Google Scholar] [CrossRef]
- Knight, V.; Sanglier, J.-J.; DiTullio, D.; Braccili, S.; Bonner, P.; Waters, J.; Hughes, D.; Zhang, L. Diversifying microbial natural products for drug discovery. Appl. Microbiol. Biotechnol. 2003, 62, 446–458. [Google Scholar] [CrossRef]
- Okuda, T.; Ando, K.; Bills, G. Fungal germplasm for drug discovery and industrial applications. In Handbook of Industrial Mycology; An, Z., Ed.; CRC Press: Boca Raton, FL, USA, 2004; pp. 123–166. [Google Scholar] [CrossRef]
- Fujimori, F.; Okuda, T. Application of the random amplified polymorphic DNA using the polymerase chain reaction for efficient elimination of duplicate strains in microbial screening. J. Antibiot. 1994, 47, 173–182. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ziemert, N.; Lechner, A.; Wietz, M.; Millán-Aguiñaga, N.; Chavarria, K.L.; Jensen, P.R. Diversity and evolution of secondary metabolism in the marine actinomycete genus Salinispora. Proc. Natl. Acad. Sci. USA 2014, 111, E1130-9. [Google Scholar] [CrossRef] [PubMed]
- Chevrette, M.G.; Carlos-Shanley, C.; Louie, K.B.; Bowen, B.P.; Northen, T.R.; Currie, C.R. Taxonomic and metabolic incongruence in the ancient genus Streptomyces. Front. Microbiol. 2019, 10, 2170. [Google Scholar] [CrossRef] [PubMed]
- Baltz, R.H. Marcel Faber Roundtable: Is our antibiotic pipeline unproductive because of starvation, constipation or lack of inspiration? J. Ind. Microbiol. Biotechnol. 2006, 33, 507–513. [Google Scholar] [CrossRef] [PubMed]
- Pye, C.R.; Bertin, M.J.; Lokey, R.S.; Gerwick, W.H.; Linington, R.G. Retrospective analysis of natural products provides insights for future discovery trends. Proc. Natl. Acad. Sci. USA 2017, 114, 5601–5606. [Google Scholar] [CrossRef]
- Clark, C.M.; Costa, M.S.; Sanchez, L.M.; Murphy, B.T. Coupling MALDI-TOF mass spectrometry protein and specialized metabolite analyses to rapidly discriminate bacterial function. Proc. Natl. Acad. Sci. USA 2018, 115, 4981–4986. [Google Scholar] [CrossRef]
- Clark, C.M.; Costa, M.S.; Conley, E.; Li, E.; Sanchez, L.M.; Murphy, B.T. Using the open-source MALDI TOF-MS IDBac pipeline for analysis of microbial protein and specialized metabolite data. J. Vis. Exp. 2019, 147, e59219. [Google Scholar] [CrossRef]
- Costa, M.S.; Clark, C.M.; Ómarsdóttir, S.; Sanchez, L.M.; Murphy, B.T. Minimizing taxonomic and natural product redundancy in microbial libraries using MALDI-TOF MS and the bioinformatics pipeline IDBac. J. Nat. Prod. 2019, 82, 2167–2173. [Google Scholar] [CrossRef]
- Strejcek, M.; Smrhova, T.; Junkova, P.; Uhlik, O. Whole-cell MALDI-TOF MS versus 16S rRNA gene analysis for identification and dereplication of recurrent bacterial isolates. Front. Microbiol. 2018, 9, 1294. [Google Scholar] [CrossRef]
- Dumolin, C.; Aerts, M.; Verheyde, B.; Schellaert, S.; Vandamme, T.; Van der Jeugt, F.; De Canck, E.; Cnockaert, M.; Wieme, A.D.; Cleenwerck, I.; et al. Introducing SPeDE: High-throughput dereplication and accurate determination of microbial diversity from matrix-assisted laser desorption-ionization time of flight mass spectrometry data. mSystems 2019, 4, e00437-19. [Google Scholar] [CrossRef]
- Chudejova, K.; Bohac, M.; Skalova, A.; Rotova, V.; Papagiannitsis, C.C.; Hanzlickova, J.; Bergerova, T.; Hrabák, J. Validation of a novel automatic deposition of bacteria and yeasts on MALDI target for MALDI-TOF MS-based identification using MALDI Colonyst robot. PLoS ONE 2017, 12, e0190038. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Clark, C.M.; Nguyen, L.; Pham, V.C.; Sanchez, L.M.; Murphy, B.T. Automated Microbial Library Generation Using the Bioinformatics Platform IDBac. Molecules 2022, 27, 2038. https://doi.org/10.3390/molecules27072038
Clark CM, Nguyen L, Pham VC, Sanchez LM, Murphy BT. Automated Microbial Library Generation Using the Bioinformatics Platform IDBac. Molecules. 2022; 27(7):2038. https://doi.org/10.3390/molecules27072038
Chicago/Turabian StyleClark, Chase M., Linh Nguyen, Van Cuong Pham, Laura M. Sanchez, and Brian T. Murphy. 2022. "Automated Microbial Library Generation Using the Bioinformatics Platform IDBac" Molecules 27, no. 7: 2038. https://doi.org/10.3390/molecules27072038
APA StyleClark, C. M., Nguyen, L., Pham, V. C., Sanchez, L. M., & Murphy, B. T. (2022). Automated Microbial Library Generation Using the Bioinformatics Platform IDBac. Molecules, 27(7), 2038. https://doi.org/10.3390/molecules27072038





