Abstract
The antibacterial activity of propolis has long been of great interest, and the chemical composition of propolis is directly dependent on its source. We recently obtained a type of propolis from China with a red color. Firstly, the antibacterial properties of this unusual propolis were determined against Staphylococcus aureus and methicillin-resistant Staphylococcus aureus (MRSA). Studies on its composition identified and quantified 14 main polyphenols of Chinese red propolis extracts (RPE); quantification was carried out using liquid chromatography triple quadrupole tandem mass spectrometry (LC-QQQ-MS/MS) and RPE was found to be rich in pinobanksin, pinobanksin-3-acetate, and chrysin. In vitro investigations of its antibacterial activity revealed that its activity against S. aureus and MRSA is due to disruption of the cell wall and cell membrane, which then inhibits bacterial growth. Despite its similar antibacterial activities against S. aureus and MRSA, metabolomic analysis further revealed the effects of RPE on bacteria metabolism were different. The untargeted metabolomic results showed that a total of 7 metabolites in 12 metabolic pathways had significant changes (Fold change > 2, p < 0.05 *) after RPE treatment in S. aureus, while 11 metabolites in 9 metabolic pathways had significant changes (Fold change > 2, p < 0.05 *) after RPE treated on MRSA. Furthermore, RPE downregulated several specific genes related to bacterial biofilm formation, autolysis, cell wall synthesis, and bacterial virulence in MRSA. In conclusion, the data obtained indicate that RPE may be a promising therapeutic agent against S. aureus and MRSA.
1. Introduction
Propolis is a resinous substance collected by honeybees (Apis mellifera) when they collect natural plant shoots, resinous secretions, pollen, and soil and mix it with their own glandular secretions to repair the hive and protect it from external aggression [1]. The chemical composition of propolis is very complex and is directly related to the local plant source of the honeybees [2], mainly including polyphenols, flavonoids, terpenoids, quinones, minerals [3], etc. Depending on its botanical origin and color, propolis is classified as green, red, or brown in Brazil [4]. Baccharis dracunculifolia DC (Asteraceae) in southeastern Brazil is the main botanical source of green propolis [5]; red propolis is found in bee hives along the coast and mangroves of northeastern Brazil, and its botanical source is Dalbergia ecastophyllum (L.) Taub [6]. Most Chinese propolis is greenish-black or dark in color [7] and is typical of poplar propolis, whose main botanical source is poplar (Populus sp.) [8].
The biological activity of propolis is closely related to its chemical composition which in turn is linked to the botanical origin of the plants the honeybees visit [9]. Propolis has extensively reported wide ranging biological activity which in addition to antibacterial activity includes antioxidant, antitumor, immunomodulatory [10], and anti-inflammatory [11] activities.
The antibacterial activity of propolis has been of great interest to researchers, and the antibacterial activity of a variety of different propolis from all over the world has been studied. Couto et al. studied and demonstrated the in vitro inhibitory activity of Brazilian brown propolis against Enterococcus faecalis [12]. Silici and collegues studied the chemical composition and antibacterial activity of A. mellifera propolis from three different honeybee races. Forty-eight compounds were identified by GC/MS and the ethanolic extract of propolis was evaluated for antibacterial activity against Staphylococcus aureus, Escherichia coli, Pseudomonas aeruginosa, and Candida albicans. The results showed that propolis showed high antibacterial activity against Gram-positive bacteria and weaker activity against Gram-negative bacteria and yeasts [13]. Further studies on the antibacterial mechanism of action of propolis have postulated that propolis may reduce ATP production, increase membrane permeability, decrease bacterial motility, and/or disturb membrane potential [14].
Staphylococcus aureus (S. aureus) [15] and methicillin-resistant Staphylococcus aureus (MRSA) are two pathogens that pose a significant threat to public health worldwide [16]. S. aureus causes infections of the skin and soft tissues and with the extensive use of antibiotics, S. aureus is gradually acquiring mechanisms of resistance to various antibiotics [17], leading to the emergence of MRSA in 1961 [18]. The use of various antibiotics can also have negative effects on the body alongside their antibacterial action, via increasing the metabolic burden on the kidneys, inactivating surface activity in the lungs, and inhibiting bone marrow [19]. MRSA is of great scientific interest, with a recent study finding that an increased proportion of antibiotic resistance in MRSA was found in patients with significant pathogenicity and mortality [20]. Hence, there is an urgent need to find and develop natural antibacterial substances that can reduce the metabolic burden on the body whilst possessing antimicrobial effectiveness.
Metabolomics is the qualitative and quantitative study of small molecule metabolites (<1000–1500 Da), one of the main objectives of which is to identify biomarkers that have a direct impact on the metabolism or metabolic pathways of an organism [21]. Metabolomics applied to the study of antibacterial activity by analysis of changes in bacterial metabolites and metabolic pathways can reveal antibacterial mechanisms of action. Chen et al. used metabolomics to explore the antimicrobial mechanism of essential oil from the leaves of cinnamon tree (Cinnamomum camphora (Linn.) Presl using MRSA as a model, the results showed 74 differential metabolites—29 of which were upregulated and 45 downregulated—and a total of 7 pathways were enriched by common differential metabolites [22]. Non-targeted metabolomics was applied by He et al. to assess the mechanism of inhibition of Lysteria monocytogenes by linalool and found significant changes in metabolites involving amino acid, central carbon, lipid, and nucleic acid metabolisms [23].
Most reports on red propolis come from Cuba and northern Brazil [24]. In recent years, red propolis from China also has been discovered and investigated, researchers have reported that red propolis from China has strong antioxidant activity [25] and inhibits vascular endothelial growth factor-induced angiogenesis [26]. In this study, the main objective is to evaluate antibacterial activity of Chinese red propolis (Figure 1) against Staphylococcus aureus and MRSA and elucidate the mechanism of action. The effects of Chinese red propolis extracts (RPE) on the integrity of bacterial cell walls and cell membranes were observed, and the changes in metabolites after the action of RPE on S. aureus and methicillin-resistant S. aureus were also analyzed by LC-QTOF-MS/MS. We hope to better illustrate the inhibition mechanism of CRP on S. aureus and MRSA.
Figure 1.
Chinese red propolis extracts from Shandong, China. Left is Chinese red propolis extract, right is Chinese red propolis extract dissolved in methanol.
2. Results
2.1. Results of the Quantitative Analysis of Polyphenols in RPE
Quantification of the 14 polyphenols in RPE was achieved using Triple Quadrupole LC-MS/MS detection conditions for each of the 14 polyphenol standards MS/MS parameters were optimized with multiple reaction monitoring (MRM). The selected parameters including precursor, product ions, collision energy (CE), and quantification results for all polyphenols are listed in Table 1. MS/MS spectra of 14 polyphenols are shown in Supplementary Figure S1.

Table 1.
Quantitative results for 14 polyphenols in RPE.
2.2. RPE Has Anti-Bacterial Effect on S. aureus and MRSA
In this study, we utilized two strains of Staphylococcus aureus ATCC 25923 which is standard bacterial strain, and MRSA ATCC43300 strain. RPE was shown to have significant anti-bacterial effects against both bacterial strains. The zone of inhibition of RPE against S. aureus and MRSA using an agar-well diffusion test as previously described were assessed, methanol was used as a solvent control. Based on previous studies, the zone of inhibition diameter was used to ascribe descriptors of insensitive (≤8.0 mm), moderately sensitive (8.0 < diameter < 14.0 mm), sensitive (14.0 < diameter < 20.0 mm), and extremely sensitive (≥20.0 mm) [27]. Our study showed the inhibition zone of RPE against S. aureus to be 16.5 ± 0.5 mm, and the zone of inhibition of RPE against MRSA to be 19.3 ± 0.5 mm. No inhibition zone was found in the control group. The MIC and MBC of RPE against S. aureus and MRSA were determined by the microbroth dilution assay and the results are shown in Table 2. An MIC of 100 µg/mL for S. aureus and an MIC of 50 µg/mL for MRSA was determined, whilst the MBC of RPE for S. aureus and MRSA were found to be 300 µg/mL and 200 µg/mL respectively.

Table 2.
Antibacterial activity of RPE in terms of MIC and MBC against S. aureus and MRSA.
2.3. Time–Growth Curve Analysis
Following determination of the MIC and MBC of RPE against S. aureus and MRSA, we studied the growth curves of S. aureus and MRSA treated with MIC and 2 × MIC concentrations of RPE over 24 h, as shown in Figure 2. RPE showed significant inhibitory activities on S. aureus and MRSA in a concentration-dependent manner, the viability of S. aureus and MRSA were significantly (p < 0.05) inhibited by RPE in the 24 h following its administration.
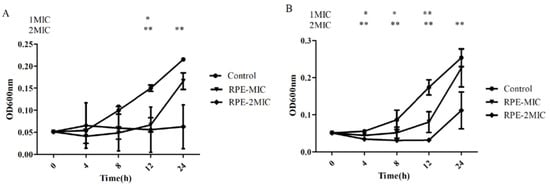
Figure 2.
Time-killing curve of RPE against S. aureus (A) and MRSA (B). (A) The time growth curve of RPE against S. aureus. (B) The time growth curve of RPE against MRSA. All data are presented as mean values ± SD, and n = 3 in each group. * p < 0.05, ** p < 0.01, vs. the solvent control.
2.4. Inhibitory Effect of RPE on Intracellular Protein and DNA Leakage Analysis
In this study, we further tested the effects of RPE treatment on the bacterial cell wall membrane integrity. S. aureus and MRSA cell wall membrane integrity were investigated by analyzing the absorbance of bacterial supernatant due to release of nucleic acid at 260 nm and protein at 280 nm. Our results on OD260 and OD280 assay (Figure 3), show RPE treatment (to concentrations of MIC and 2 × MIC) of S. aureus and MRSA significantly increased the OD260 and OD280 values (p < 0.05) in a concentration dependent manner, indicating that RPE can inhibit the synthesis of DNA and proteins in S. aureus and MRSA, and the integrity of the cell membrane is significantly impacted by RPE.
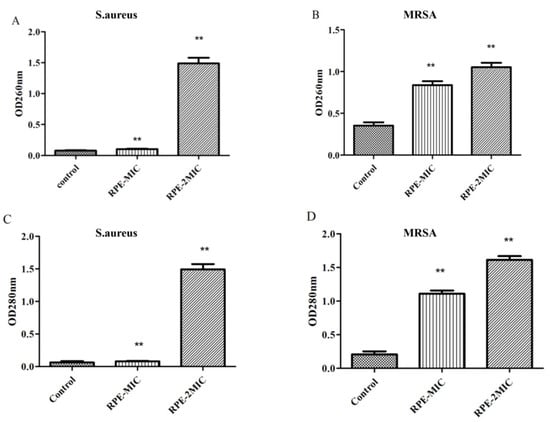
Figure 3.
260 nm absorbance values of S. aureus (A) and MRSA (B) treated with RPE, and 280 nm absorbance from S. aureus (C) and MRSA (D) treated with RPE. All data are presented as mean values ± SD, and n = 3 in each group. ** p < 0.01, vs. the solvent control.
2.5. Cell Wall Integrity Analysis
The effects of AKP leakage from bacteria are presented in Figure 4A and Figure 4B, compared to the control. The leakage of AKP from S. aureus (A) and MRSA (B) treated with RPE increased significantly in a dose-dependent manner. Figure 4C shows SEM images of S. aureus and MRSA treated with RPE alongside an untreated control. SEM images show cell wall damage in S. aureus and MRSA treated with 100 µg/mL of RPE. Untreated S. aureus and MRSA cells were spherical in shape, with a smooth surface and retained normal cell morphology. Following treatment with RPE, cells became irregular, exhibiting deformation of cell membranes in both S. aureus and MRSA groups.
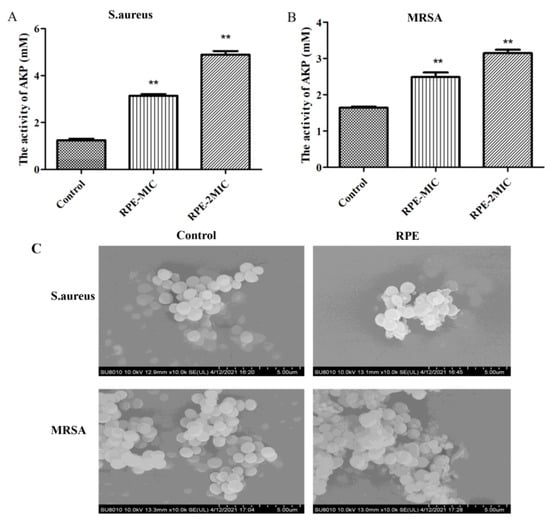
Figure 4.
Assessment of leakage of AKP from (A) S. aureus and (B) MRSA treated with RPE; and (C) scanning electron micrographs of S. aureus and MRSA treated with RPE. Scanning electron micrographs show cell wall integrity of S. aureus: (left) S. aureus control (magnification × 100,000); (right) S. aureus treated with the RPE at 100 µg/mL (magnification × 100,000); Scanning electron micrographs showing cell wall integrity of MRSA: (left) MRSA control (magnification × 100,000); (right) MRSA treated with the RPE at 100 µg/mL (magnification × 100,000). All data are presented as mean values ± SD, and n = 3 in each group. ** p < 0.01, vs. the solvent control.
2.6. Metabolomic Analysis of the Effect of RPE Treated on S. aureus and MRSA
The metabolic differences of S. aureus and MRSA following RPE treatment were investigated relative to untreated controls. After RPE treatment for 24 h, alterations in S. aureus and MRSA intracellular metabolites were analyzed. For S. aureus, a total of 984 metabolites were identified, and analyzed by ANOVA (p < 0.05) highlighting 428 differential metabolites. For MRSA, a total of 2511 metabolites were identified, including 161 differential metabolites (p < 0.05, analyzed by ANOVA). Orthogonal partial least-squares discriminant analysis (OPLS-DA) was applied for the recognition of the sample patterns, notably, the OPLS-DA spots (Figure 5) and showed the RPE treatment on S. aureus and MRSA group was significantly separated from relevant control groups. Enrichment analysis was carried out according to The Kyoto Encyclopedia of Genes and Genomes (KEGG) to categorize the differential metabolites into related pathways. The results indicated that 12 pathways were enriched after treatment with RPE treated on S. aureus. These included aminoacyl-tRNA biosynthesis; lysine biosynthesis; alanine, aspartate, and glutamate metabolism; glutathione metabolism; d-Arginine and d-ornithine metabolism; nitrogen metabolism; purine metabolism; glycine, serine, and threonine metabolism; riboflavin metabolism; benzoate degradation; arginine and proline metabolism; and arginine biosynthesis between the two groups (Figure 6A), these pathways contain metabolites in which xanthosine and 5-Amino-6-(1-d-ribitylamino)uracil are increased and l-Glutamate, l-Lysine, d-Ornithine, Betaine, and 3-Oxopimeloyl-CoA are decreased (Figure 6B).
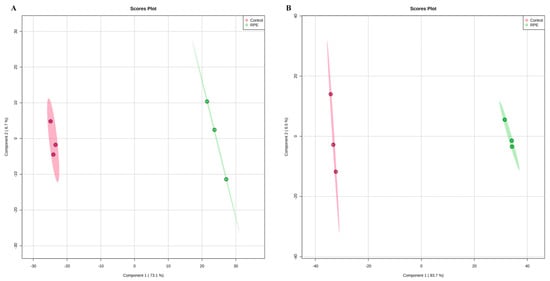
Figure 5.
OPLS-DA spot of RPE treated on S. aureus and MRSA compared with untreated respectively. (A) OPLS-DA analysis of effects of RPE treatment of S. aureus relative to control. Each point represents a duplicate sample in each group (n = 3). (B) OPLS-D analysis of effects of RPE treatment of MRSA versus control. Each dot in the figure represents a duplicate sample in each group (n = 3).
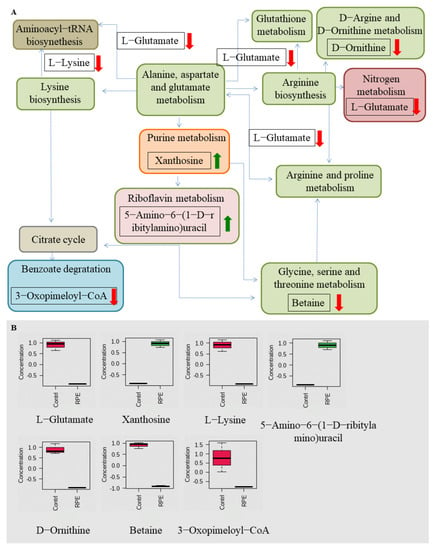
Figure 6.
Related metabolic pathways and metabolites with significant changes following RPE treatment in S. aureus compared to the untreated group. (A) Green and red arrows indicate an increase and decrease abundance of metabolites, respectively. Different colors in the pathway represent different types of metabolism, while the same color represents the same type of metabolism, e.g., green are all amino acid metabolism. (B) Box plots for metabolites with significant changes involved in related metabolic pathways.
Nine pathways were enriched after RPE treated on MRSA, including aminoacyl-tRNA biosynthesis; lysine biosynthesis; glycerophopholipid metabolism; purine metabolism; glycine, serine, and threonine metabolism; riboflavin metabolism; histidine metabolism; arginine and proline metabolism; and cysteine and methionine metabolism between the two groups (Figure 7A), pathways contain metabolites in which N2-Succinylglutamate, 5-Methylthio-d-ribose, xanthosine, and 5-Amino-6-(1-d-ribitylamino)uracil are increased, and Betaine, N-Formyl-l-asparfate, l-Lysine, phosphatidylethanolamine, and phosphatidylcholine (Lecithin) are decreased (Figure 7B).
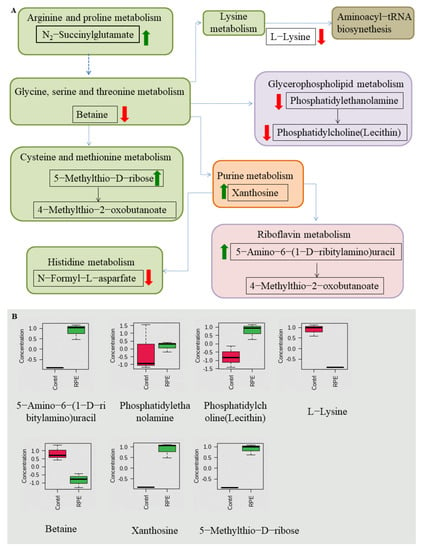
Figure 7.
Related metabolic pathways and metabolites with significant changes involved after treatment with RPE treated on MRSA compared to the untreated group. (A) Green and red arrows indicate increase and decrease abundance of metabolites, respectively. Different colors in the pathway represent different types of metabolism, while the same color represents the same type of metabolism, e.g., green are all amino acid metabolism. (B) Box plots for metabolites with significant changes involved in related metabolic pathways.
2.7. Effect of RPE on the Related Gene Expression of MRSA
Finally, we analyzed the gene expressions 11 related genes following exposure to RPE in MRSA. As shown in Figure 8 in all genes investigated, expression was downregulated compared to the control group; among them, the expression of icaC and ssaA was significantly downregulated compared to the control group (p < 0.05). The expression of icaR, MurE, and MurC were highly significantly downregulated compared to the control group (p < 0.01).
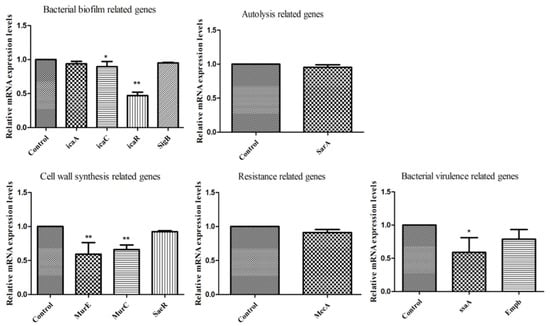
Figure 8.
Gene expression of MRSA after exposure to RPE by real-time quantitative reverse transcription PCR (bacterial biofilm-related genes: icaA, icaC, icaR, SigB; autolysis-related gene: SarA; cell wall synthesis-related genes: MurE, MurC, SaeR; resistance-related gene: MecA; bacterial virulence-related genes: ssaA, Empb). All data are presented as mean values ± SD, and n = 3 in each group. * p < 0.05, ** p < 0.01, vs. the solvent control.
3. Discussion
This study was carried out to assess the antibacterial activity and mechanism of RPE with a view to counteracting the harmful effects of S. aureus and MRSA. First, we quantitatively analyzed 14 main polyphenols in RPE and determined the antibacterial activity of RPE by means of the zone of inhibition, MIC, MBC, and time–growth curve. Subsequently, we investigated the inhibitory effect of RPE on the S. aureus and MRSA cell membrane and cell wall disruption by detecting protein and nucleic acid release, AKP release, and by performing SEM analysis. Finally, the metabolomic analysis showed that RPE inhibits S. aureus and MRSA by inducing changes in intracellular metabolite concentrations and related metabolic pathways. RPE downregulated genes expression related to bacterial biofilm formation, autolysis, cell wall synthesis, and bacterial virulence in MRSA.
Propolis in China is mainly yellowish brown and dark in color, with a chemical composition that is variable and complex, and dependent on source [28]. For red propolis from China, studies on its constituents and activities are still in the initial stages. It is well known that propolis has been reported for its antibacterial activity, however the mechanism of propolis antibacterial action is not well defined. Most studies suggesting that the antibacterial activity may be due to the synergistic effect of polyphenols contained in propolis [29]. In our study, we selected the 14 main polyphenols of RPE and subjected them to quantitative analysis by LC-QQQ. We found that the red propolis contained high levels of pinobanksin, pinobanksin-3-acetate, and chrysin. Darwish et al. investigated the antibacterial activity of Jordanian propolis of pine origin and oak origin. Three flavonoids: pinobanksin-3-O-acetate, pinocembrin, and chrysin were isolated from the propolis of pine origin and studied for antibacterial activity in vitro. Among the results, pinobanksin-3-O-acetate and pinocembrin showed antibacterial activity, especially against MRSA, while chrysin was active only against standard S. aureus [30]. Kopacz et al. tested morin; sodium salt of morin-5″-sulfonic acid (NaMSA); and new complexes of La (II), Gd (III), and Lu (III) with morin against Escherichia coli G (-), Klebsiella pneumoniae G (-), and S. aureus G (+). The results showed that morin was the most effective against E. coli and S. aureus [31]. Caffeic acid phenethyl ester showed higher antibacterial activity than 5-chlorogenic acid and caffeic acid against S. aureus, Bacillus subtilis, and Pseudomonas aeruginosa in an in vitro study by Kishimoto et al. [32]. Kharsany et al. studied the antibacterial activity of pinocembrin, galangin, and chrysin, the main compounds in South African propolis—both singularly and in combination—and found that the action of a single compound in propolis was not as effective as the synergistic action of the compounds in inhibiting bacteria [33].
In general, bacteria in the logarithmic growth phase are relatively stable, more resistant, and also more sensitive to external factors [34,35]. For a more accurate determination of the antibacterial activity of RPE against S. aureus and MRSA, the bacteria studied were all in the logarithmic growth phase. Gram-positive bacteria have a thick peptidoglycan layer in the cell wall, but cannot prevent polyphenols from entering the cell [36]. The antimicrobial activity of polyphenols depends on its chemical structure and environmental conditions [37]. The antibacterial activity of RPE against S. aureus and MRSA was evaluated and showed inhibitory effects, resulting in a MIC of 100 µg/mL for S. aureus and a MIC of 50 µg/mL for MRSA (Table 2). The MIC results of S. aureus are similar to those Bonvehiet et al. obtained from propolis, which ranged from 0.080 to 0.100 mg/mL [38], and lower than the MIC found by Rahman [39]. To further confirm the antibacterial activity of RPE against S. aureus and MRSA, time–kill curves of S. aureus and MRSA were plotted. Growth of S. aureus and MRSA was almost completely inhibited after 24 h of incubation at the RPE concentrations of 2 × MIC (Figure 2). These results demonstrated the dose dependency of RPE antibacterial effects.
Bacterial cell membranes are selectively permeable. The cell membrane protects the cell by preventing harmful substances from entering the cell, and also allowing substances that nourish the cell to enter [40], thus the integrity of the cell membrane can impact cell survival [15]. Small biomolecules, such as DNA, are permeable to move in the bacterial cell wall, and protein is important for maintaining the nutritional metabolism of the bacterial organism and for immune regulation [41]; in particular, soluble proteins play an important role in regulating cell permeability [42]. Damage to the integrity of the cell membrane can be determined by the release of proteins and nucleic acids [43]. In our study, the OD260 and OD280 of S. aureus and MRSA increased after treatment with increasing concentrations of RPE, suggesting that RPE damaged the cell membrane of S. aureus and MRSA (Figure 3). These results are in agreement with other reports of the action of antimicrobials on bacterial cells, thus suggesting that mechanism of action of RPE may be due to loss of membrane integrity [44].
The cell walls of bacteria are robust, and used to maintain the normal shape of bacterial cells, maintain normal pressure, and protect cells from the invasion by external substances [45]. AKP is found between the bacterial cell membrane and the cell wall; AKP is released when the cell wall is disrupted and as such can be used to determine the integrity of the bacterial cell wall [46]. AKP leakage from S. aureus (Figure 4A) and MRSA (Figure 4B) increased after treatment with increasing concentrations of RPE. SEM was used to study the bacteriostatic mechanism, with SEM results showing that S. aureus and MRSA all have a regular oval shape when untreated. However, following RPE treatment, the normal shape of the cells is disrupted and the contents are released (Figure 4C), suggesting that the action of RPE against S. aureus and MRSA is via damage to the cell wall—a theory further supported by the leakage of DNA and protein studies that we conducted. These results were in agreement with the conclusions of previous studies [47,48].
In addition to the above studies, changes in bacterial metabolism can also cause bacterial death [49], and antibacterial mechanisms can also be investigated by studying changes in the metabolic pathways of bacterial metabolites metabolic pathways [50]. By further studying the changes in metabolites and metabolic pathways following the action of RPE on S. aureus and MRSA, we hypothesize that new discoveries can be made regarding the antibacterial mechanism of RPE. In our study, the analysis of the metabolites in intracellular metabolites using a nontargeted metabolomics approach, the OPLS-DA spots (Figure 5) revealed the measurements from S. aureus and MRSA treated with RPE were significantly separated from the control group and clustered in defined ranges. In the results of the metabolic pathway analysis of S. aureus, a total of 7 metabolites in 12 metabolic pathways showed significant changes (Figure 6). Of these, 5 metabolites decreased in 10 metabolic pathways. The metabolite l-Glutamate decreased in aminoacyl-tRNA biosynthesis; alanine, aspartate, and glutamate metabolism; nitrogen metabolism; glutathione metabolism; arginine and proline metabolism; and arginine biosynthesis. l-Lysine decreased in aminoacyl-tRNA biosynthesis and lysine biosynthesis; d-Ornithine decreased in d-Arginine and d-ornithine metabolism; betaine decreased in glycine, serine, and threonine metabolism; and 3-Oxopimeloyl-CoA decreases benzoate degradation. Two metabolites increased in two metabolic pathways, the metabolite xanthosine is increased in purine metabolism, and 5-Amino-6-(1-d-ribitylamino)uracil is increased in riboflavin metabolism. Of the above metabolic pathways, all lysine biosynthesis; alanine, aspartate, and glutamate metabolism; glutathione metabolism; arginine and proline metabolism; glycine, serine, and threonine metabolism; arginine biosynthesis; and d-arginine and d-ornithine metabolism are involved in amino acid metabolism, indicating that amino acid metabolism plays a very important role in antibacterial activity, which is consistent with Chen’s [22] view. In previous studies, the metabolic pathways enriched by differential metabolites such as arginine and proline metabolism; alanine, aspartate, and glutamate metabolism; aminoacyl-tRNA biosynthesis [22]; purine metabolism [51]; glycine, serine, and threonine metabolism [52]; and nitrogen metabolism [53] were also detected.
In the results of the metabolic pathway analysis of MRSA, a total of 11 metabolites in the 9 metabolic pathways showed significant changes (Figure 7). Of these, five metabolites decreased in five metabolic pathways. The metabolite l-Lysine was decreased in aminoacyl-tRNA biosynthesis and lysine biosynthesis; betaine is decreased in glycine, serine, and threonine metabolism; n-Formyl-l-asparfate is decreased in histidine metabolism, phosphatidylethanolamine; and phosphatidylcholine (lecithin) is decreased in glycerophopholipid metabolism. Four metabolites increased in four metabolic pathways. The metabolite N2-succinylglutamate increases in arginine and proline metabolism, 5-Methylthio-d-ribose increases in cysteine and methionine metabolism, xanthosine increases in purine metabolism, and 5-Amino-6-(1-d-ribitylamino) uracil increases in riboflavin metabolism. Of the above metabolic pathways, all arginine and proline metabolism; cysteine and methionine metabolism; glycine, serine, and threonine metabolism; lysine biosynthesis; and histidine metabolism are involved in amino acid metabolism, also indicating that amino acid metabolism plays a very important role in antibacterial activity in MRSA. In previous studies, the metabolic pathways enriched by differential metabolites—such as arginine and proline metabolism; purine metabolism [53]; glycine, serine, and threonine metabolism [52]; aminoacyl-tRNA biosynthesis; and cysteine and methionine metabolism [22]—glycerophopholipid metabolism and histidine metabolism [54] were also detected.
We further determined the changes in expression of relevant genes of MRSA following treatment with RPE. A very important factor affecting MRSA resistance is the formation of biofilms; biofilm formation can effectively prevent infection. Biofilm formation is associated with many cell surface and secreted virulence factors [55]. The formation of a bacterial biofilm begins with the attachment of bacteria to an external surface, followed by the formation of a complex membrane structure, a process that critically depends critically on the synthesis of polysaccharide intercellular adhesion (PIA) [56], which is regulated by the ica operon, which contains icaA, icaB, icaC, icaD, and icaR [57]. In the present study, we detected that the bacterial biofilm-related gene (icaA, icaC, icaR, and SigB) expression levels, as shown in Figure 8, were downregulated compared with control group, suggesting that RPE may prevent biofilm formation through the altering the PIA pathway. Bacterial virulence-related genes, including ssaA and Empb, were downregulated compared with control group. ssaA was significantly downregulated (p < 0.05), indicating that RPE treatment weakened the secretion of bacterial virulence factors. The autolysis-related gene—sarA, also associated with the release of virulence factors and the formation of biofilms [58]—was also downregulated, a result consistent with a previous study [59]. Expression of the cell wall synthesis-related genes, MurE and MurC, was significantly inhibited compared with control group (p < 0.01), which was consistent with AKP leakage and SEM results, suggesting that RPE can kill MRSA by inhibiting and disrupting cell wall synthesis.
4. Materials and Methods
4.1. Extraction of CRP
CRP samples were collected from Shandong, China, and the samples were extracted with 95% v/v ethanol as solvent with the ultrasound assisted method [60], and the extracts were concentrated by rotary evaporation (Switzerland Buchi, R-300) to obtain constant weight CRP. The RPE was then dissolved in methanol to obtain a stock solution at a concentration of 50 mg/mL, which was stored at 4 °C
4.2. Calibration and Quantification of the Main Polyphenols in RPE
The RPE stock solution was diluted to a concentration of 1 mg/mL with methanol. Fourteen standards—pinobanksin-3-acetate, morin, ferulic acid, caffeic acid, caffeic acid phenethyl ester, pinobanksin, galangin, quercetin, p-coumaric acid, apigenin, kaempferol, chrysin, pinocembrin, and naringenin—were prepared at a concentration of 100 mg/mL in 100 µL of methanol. The standard stock solutions were then mixed in equal parts to maintain a concentration of each standard of 1 mg/mL and were then diluted to the following concentrations in methanol: 100, 200, 400, 600, and 800 µg/mL, a calibration curve was constructed by injecting each standard solution at each concentration level. The RPE and mixed standard solutions of different concentrations were filtered through a 0.22 µm membrane filter into a LC bottle ready to be assayed.
Agilent 6495 Triple Quadrupole LC/MS was applied to quantify the 14 major polyphenols in RPE. Analysis was performed on an Agilent Infinity Lab Poroshell SB-C18 (3.0 × 100 mm, 2.7 µm) with the column temperature set at 35 °C, mobile phase A was 0.1% v/v formic acid-water and phase B was methanol, with an injection volume of 2 µL and a flow rate of 0.25 mL/min. The gradient elution procedure was as follows: 5–55% B in 1–6 min; 55–95% B in 6–20 min; 95–5% B in 20–21.5 min, post-run time was 5 min. The electrospray ionization source [15] was used for mass spectrometry, and the samples were detected in negative ion mode with the following parameters: gas temp 270 °C, carrier gas (N2) flow rate 10 L/min, atomization pressure 25 psi, sheath gas temperature 350 °C, sheath gas flow rate 12 L/min, capillary voltage (Vcap) 4000 V, and nozzle voltage 1500 V.
4.3. Bacterial Strains and Growth Conditions
S. aureus (ATCC 25923) and MRSA (ATCC 43300) strains were obtained from China Center of Industrial Culture Collection. It is usually acknowledged that the logarithmic phase cells are more sensitive to external stress factors, while stationary phase cells are more resistant [33,34]. The bacteria strains were cultured overnight to the logarithmic phase of growth at 37 °C (approximately 16 h) in Luria-Bertani broth culture medium (AOBOX Biotechnology, Beijing, China). Both inoculums were measured at OD600 and adjusted to the concentration of 5 × 107 CFU/mL using the McFarland standard. Culture purity was examined by streaking each culture on plates of LB agar and nutrient agar for S. aureus and MRSA and incubating at 37 °C overnight.
4.4. Determination of Antibacterial Activity of RPE
RPE antimicrobial activity was assessed using the agar disk diffusion method [61]. Bacterial cultures of S. aureus and MRSA were spread evenly on LB plates with applicator sticks at a concentration of 5 × 107 CFU/mL. Wells of 6 mm in diameter were bored into LB solid agar and each well filled with 100 µL RPE with a concentration of 100 µg/mL or control (methanol) separately. Following incubation overnight at 37 °C the zone of inhibition was measured and reported as diameter in mm.
4.5. Determination of Minimal Inhibitory Concentration and Minimum Bactericidal Concentration (MBC)
The MIC and MBC of RPE against S. aureus and MRSA were determined following the Clinical and Laboratory Standards Institute 2017 (CLSI 2017) by the gradient dilution assay [52]. RPE was dissolved in methanol and prepared in LB broth medium ranging from 50 µg/mL to 600 µg/mL, methanol was used as a solvent control. After incubation at 37 °C for 24 h, 50 µL of the cultures were spread evenly on LB plates with applicator sticks (5 × 107 CFU/mL) and incubated at 37 °C for 16 h to identify the lowest RPE concentration that inhibited bacterial growth as MIC and the bacterial concentration that showed no bacterial growth as MBC.
4.6. Time–Growth Curve Assay
The time–growth analysis of RPE against S. aureus and MRSA were performed using previous methods with slight modifications [62], RPE was tested at a concentration of MIC and 2 × MIC was added to logarithmic phase of growth S. aureus and MRSA suspensions (5 × 107 CFU/mL), while methanol was used as a solvent control. After incubation at 37 °C for 0, 4, 8, 12, and 24 h, 200 µL of suspension transferred into a 96-well plate and OD600 nm was measured. Growth curves of S. aureus and MRSA were constructed by plotting OD600 against time.
4.7. Intracellular Protein and Nucleic Acid Leakage Assay
The effects RPE treatment on the leakage of the intracellular contents of S. aureus and MRSA was evaluated, according to reported procedures with some modifications [63,64]. RPE was added to an overnight culture of S. aureus and MRSA suspensions (5 × 107 CFU/mL), respectively both in logarithmic phase of growth at 37 °C. Tests were performed at final concentrations of RPE of MIC and 2 × MIC, whilst bacterial culture exposed to equivalent methanol concentration as solvent control. Bacterial solutions were incubated at 37 °C for 24 h, then centrifuged at 8000× g for 10 min. The optical density of the supernatant was then measured at 260 and 280 nm using an UV spectrophotometer (JING HUA) in order to determine the release of intracellular nucleic acid and protein respectively.
4.8. Alkaline Phosphatase (AKP) Release Determination and Scanning Electron Microscopy (SEM)
To examine the mechanisms of action of RPE against cell wall of S. aureus and MRSA, AKP release was evaluated according to the instruction of AKP assay kit (Beyotime Biotechnology) [65], SEM studies were carried out as previously reported with some modifications [66]. Logarithmic growth phase cells of S. aureus and MRSA (each approximately 5 × 107 CFU/mL) were treated with RPE at the same concentration (100 µg/mL). The control and RPE treated S. aureus and MRSA were incubation at 37 °C for 24 h. After incubation, cells were collected by centrifugation for 10 min at 8000× g and washed twice with 0.1 M phosphate buffer solution (PBS, PH = 7.0), then were resuspended in 2.5% glutaraldehyde and kept overnight at −4 °C to fix the cells. After centrifugation, the cells were further dehydrated in ethanol with increasing concentrations respectively (35%, 50%, 75%, 90%, and 100% v/v). The dried samples were observed using Hitachi S-750 scanning electron microscopy (Hitachi Company, Tokyo, Japan).
4.9. Metabolomic Analysis
Bacterial metabolites of S. aureus and MRSA treated with RPE were determined using metabolomic analysis using an Agilent 6545 LC-QTOF-MS/MS. First, RPE was co-cultured with S. aureus and MRSA separately at a concentration of 2 × MIC for 24 h (37 °C) in LB broth medium with an initial concentration of 5 × 107 CFU/mL of bacterial solution, methanol in the bacterial culture broth was used as a solvent control. The culture broth was removed from the incubator, centrifuged at 8000× g for 10 min, and the supernatant removed. The remaining pellet was washed with PBS and transferred to a 1.5 mL centrifuge tube and then quenched in liquid nitrogen. Samples were removed from liquid nitrogen and 1 mL of a methanol/acetonitrile/water solvent mixture (2:2:1, v/v) was added and vortexed, followed by repeated freeze–thawing cycle between liquid nitrogen and a 37 °C water bath three times then the samples were left to stand for 1 h at −20 °C and centrifuged at 13,000 rpm for 15 min at 4 °C. The supernatant was taken and spun dry using a rotary evaporator. 100 µL of water/acetonitrile (1:1, v/v) was added again and sonicated, and the supernatant was centrifuged through a 0.22 µm into an LC bottle ready for assay.
4.10. Real-Time PCR (RT-PCR) Analysis Related Gene Expressions of MRSA
Bacterial biofilm-related genes (icaA, icaC, icaR, SigB), autolysis-related gene (SarA), cell wall synthesis-related genes (MurE, MurC, SaeR), resistance-related gene (MecA), and bacterial virulence-related genes (ssaA, Empb) were selected for quantitative analysis by RT-PCR. MRSA bacteria was centrifuged after incubation in LB broth medium for 12 h at 37 °C and resuspended in TRIzol regent containing 100 µg/mL lysostaphin. Total RNA was extracted using RNasy mini kit (Qiagen, Düsseldorf, Germany) and then converted to cDNA using PrimeScript RT reagent kit (TaKaRa) according to the manufacturer’s instructions. The RT-PCR reaction using cDNA templates were performed with TaKaRa TB Green™ Premix Ex Taq™ II (Tli RNaseH Plus; RR820A) on QuantStudio1 applied biosystems. The cycle threshold values of all tested genes were normalized using GAPDH as reference gene, gene expressions were calculated by 2(−ΔΔCT) method [67]. The primers [68] used are listed in Table S1.
4.11. Statistical Analysis
A completely randomized design with three replications was performed and the data was expressed as the mean ± standard deviation. One-way analysis of variance (ANOVA) using SPSS20.0 was used to identify any significant differences between the means using a 95% confidence interval (p < 0.05).
5. Conclusions
On basis of these results, it may be concluded that RPE was rich in pinobanksin, pinobanksin-3-acetate, and chrysin and possessed considerable antibacterial activities against S. aureus and MRSA. RPE was found to inhibit S. aureus and MRSA by disrupting the cell wall, cell membrane, and induce changes in cell morphology. In addition, metabolomic analysis further revealed differences in bacterial metabolism following RPE treatment in S. aureus and MRSA. Finally, RPE downregulated gene expression related to bacterial biofilm formation, autolysis, cell wall synthesis, and bacterial virulence of MRSA. Thus, our study provides novel mechanistic insight to understand RPE action against S. aureus and MRSA. The findings of this study suggest RPE may be a promising therapeutic agent for use against S. aureus and MRSA.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/molecules27051693/s1. Figure S1: MS/MS spectra of 14 polyphenol; Table S1: Primer sequence information for MRSA-related genes.
Author Contributions
Conceptualization, K.W. and W.Z.; Methodology, D.W. and W.Y.; Resources, X.X. and L.W.; Data curation, S.Y. and S.Q.; Writing—original draft preparation, W.Z.; Writing—review and editing, G.E.M. and W.Z.; Project administration, L.W.; Funding acquisition, K.W. and L.W. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by National Natural Science Foundation of China (32172791) and the Special Project of National Bee Industry Technical System (CARS-44).
Conflicts of Interest
The authors declare no conflict of interest.
Sample Availability
Chinese red bee propolis samples, 14 polyphenol standardsare available from the authors.
References
- Silva, J.C.; Rodrigues, S.S.Q.; Feás, X.; Estevinho, M.L.M.F. Antimicrobial activity, phenolic profile and role in the inflammation of propolis. Food Chem. Toxicol. 2012, 50, 1790–1795. [Google Scholar] [CrossRef] [PubMed]
- Rufatto, L.C.; Luchtenberg, P.; Garcia, C.; Thomassigny, C.; Bouttier, S.; Henriques, J.A.P.; Roesch-Ely, M.; Dumas, F.; Moura, S. Brazilian red propolis: Chemical composition and antibacterial activity determined using bioguided fractionation. Microbiol. Res. 2018, 214, 74–82. [Google Scholar] [CrossRef] [PubMed]
- Tosic, S.; Stojanovic, G.; Mitic, S.; Pavlovic, A.; Alagic, S. Mineral Composition of Selected Serbian Propolis Samples. J. Apic. Sci. 2017, 61, 5–15. [Google Scholar] [CrossRef]
- Machado, B.A.S.; Silva, R.P.D.; Barreto, G.; Costa, S.S.; da Silva, D.F.; Brandão, H.N.; Da Rocha, J.L.C.; Dellagostin, O.A.; Henriques, J.A.P.; Umsza-Guez, M.A.; et al. Chemical Composition and Biological Activity of Extracts Obtained by Supercritical Extraction and Ethanolic Extraction of Brown, Green and Red Propolis Derived from Different Geographic Regions in Brazil. PLoS ONE 2016, 11, e0145954. [Google Scholar] [CrossRef]
- Teixeira, É.W.; Negri, G.; Meira, R.M.; Salatino, A. Plant Origin of Green Propolis: Bee Behavior, Plant Anatomy and Chemistry. Evid.-Based Complement. Altern. Med. 2005, 2, 85–92. [Google Scholar] [CrossRef]
- Silva, B.B.; Rosalen, P.L.; Cury, J.A.; Ikegaki, M.; Souza, V.C.; Esteves, A.; Alencar, S.M. Chemical Composition and Botanical Origin of Red Propolis, a New Type of Brazilian Propolis. Evid.-Based Complement. Altern. Med. 2008, 5, 313–316. [Google Scholar] [CrossRef]
- Gong, S.; Luo, L.; Gong, W.; Gao, Y.; Xie, M. Multivariate analyses of element concentrations revealed the groupings of propolis from different regions in China. Food Chem. 2012, 134, 583–588. [Google Scholar] [CrossRef]
- Wu, Y.-W.; Sun, S.-Q.; Zhao, J.; Li, Y.; Zhou, Q. Rapid discrimination of extracts of Chinese propolis and poplar buds by FT-IR and 2D IR correlation spectroscopy. J. Mol. Struct. 2008, 883, 48–54. [Google Scholar] [CrossRef]
- Jiang, X.; Tian, J.; Zheng, Y.; Zhang, Y.; Wu, Y.; Zhang, C.; Zheng, H.; Hu, F. A New Propolis Type from Changbai Mountains in North-east China: Chemical Composition, Botanical Origin and Biological Activity. Molecules 2019, 24, 1369. [Google Scholar] [CrossRef]
- Wolska, K.; Gorska, A.; Antosik, K.; Lugowska, K. Immunomodulatory Effects of Propolis and its Components on Basic Immune Cell Functions. Indian J. Pharm. Sci. 2019, 81, 575–588. [Google Scholar] [CrossRef]
- Anjum, S.I.; Ullah, A.; Khan, K.A.; Attaullah, M.; Khan, H.; Ali, H.; Bashir, M.A.; Tahir, M.; Ansari, M.J.; Ghramh, H.A.; et al. Composition and functional properties of propolis (bee glue): A review. Saudi J. Biol. Sci. 2019, 26, 1695–1703. [Google Scholar] [CrossRef]
- Pimenta, H.C.; Violante, I.M.P.; De Musis, C.R.; Borges, Á.H.; Aranha, A.M.F. In vitro effectiveness of Brazilian brown propolis against Enterococcus faecalis. Braz. Oral Res. 2015, 29, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Silici, S.; Kutluca, S. Chemical composition and antibacterial activity of propolis collected by three different races of honeybees in the same region. J. Ethnopharmacol. 2005, 99, 69–73. [Google Scholar] [CrossRef] [PubMed]
- Przybyłek, I.; Karpiński, T.M. Antibacterial Properties of Propolis. Molecules 2019, 24, 2047. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Z.; Ouyang, P.; Gu, K.; Rehman, T.; Zhang, T.; Yin, Z.; Fu, H.; Lin, J.; He, C.; Shu, G.; et al. The antibacterial mechanism of oridonin against methicillin-resistant Staphylococcus aureus (MRSA). Pharm. Biol. 2019, 57, 710–716. [Google Scholar] [CrossRef] [PubMed]
- Ferrera-Suanzes, M.; Prieto, V.; Medina-Olivera, A.J.; Botubol-Ares, J.M.; Galán-Sánchez, F.; Rodríguez-Iglesias, M.A.; Hernández-Galán, R.; Durán-Peña, M.J. Synthesis of Degraded Limonoid Analogs as New Antibacterial Scaffolds against Staphylococcus aureus. Antibiotics 2020, 9, 488. [Google Scholar] [CrossRef] [PubMed]
- Periasamy, H.; Iswarya, S.; Pavithra, N.; Senthilnathan, S.; Gnanamani, A. In vitro antibacterial activity of plumbagin isolated from Plumbago zeylanica L. against methicillin-resistant Staphylococcus aureus. Lett. Appl. Microbiol. 2019, 69, 41–49. [Google Scholar] [CrossRef]
- DeLeo, F.R.; Chambers, H.F. Reemergence of antibiotic-resistant Staphylococcus aureus in the genomics era. J. Clin. Investig. 2009, 119, 2464–2474. [Google Scholar] [CrossRef]
- Nguyen, H.M.; Graber, C.J. Limitations of antibiotic options for invasive infections caused by methicillin-resistant Staphylococcus aureus: Is combination therapy the answer? J. Antimicrob. Chemother. 2010, 65, 24–36. [Google Scholar] [CrossRef]
- Iwata, Y.; Sakai, N.; Yoneda, I.; Satou, K.; Furuichi, K.; Senda, Y.; Sakai-Takemori, Y.; Wada, T.; Fujita, S.; Ogura, H.; et al. The increased frequency of methicillin-resistant Staphylococcus aureus with low MIC of beta-lactam antibiotics isolated from hospitalized patients. J. Infect. Chemother. 2020, 26, 604–610. [Google Scholar] [CrossRef]
- Zhang, W.; Qi, S.; Xue, X.; Al Naggar, Y.; Wu, L.; Wang, K. Understanding the Gastrointestinal Protective Effects of Polyphenols using Foodomics-Based Approaches. Front. Immunol. 2021, 12, 671150. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Tang, C.; Zhang, R.; Ye, S.; Zhao, Z.; Huang, Y.; Xu, X.; Lan, W.; Yang, D. Metabolomics analysis to evaluate the antibacterial activity of the essential oil from the leaves of Cinnamomum camphora (Linn.) Presl. J. Ethnopharmacol. 2020, 253, 112652. [Google Scholar] [CrossRef] [PubMed]
- He, R.; Chen, W.; Zhang, M.; Zhong, Q.; Zhang, H.; Chen, H.; Chen, W. Antibacterial mechanism of linalool against L. monocytogenes, a metabolomic study. Food Control 2021, 132, 108533. [Google Scholar] [CrossRef]
- Alencar, S.; Oldoni, T.; Castro, M.; Cabral, I.; Costa-Neto, C.; Cury, J.; Rosalen, P.L.; Ikegaki, M. Chemical composition and biological activity of a new type of Brazilian propolis: Red propolis. J. Ethnopharmacol. 2007, 113, 278–283. [Google Scholar] [CrossRef]
- Hatano, A.; Nonaka, T.; Yoshino, M.; Ahn, M.-R.; Tazawa, S.; Araki, Y.; Kumazawa, S. Antioxidant Activity and Phenolic Constituents of Red Propolis from Shandong, China. Food Sci. Technol. Res. 2012, 18, 577–584. [Google Scholar] [CrossRef]
- Izuta, H.; Shimazawa, M.; Tsuruma, K.; Araki, Y.; Mishima, S.; Hara, H. Bee products prevent VEGF-induced angiogenesis in human umbilical vein endothelial cells. BMC Complement. Altern. Med. 2009, 9, 45. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.-H.; Cai, M.; Liu, Y.-S.; Sun, P.L.; Luo, S.-L. Antibacterial Activity and Mechanisms of Essential Oil from Citrus medica L. var. sarcodactylis. Molecules 2019, 24, 1577. [Google Scholar] [CrossRef]
- Hu, F.L.; Zhu, W.; Li, Y.J.; Zheng, H.Q. Study on Chemical and Biological Active Diversity of Propolis. J. Bee 2006, 1. [Google Scholar]
- Bankova, V.; Popova, M.; Trusheva, B. Propolis volatile compounds: Chemical diversity and biological activity: A review. Chem. Cent. J. 2014, 8, 28. [Google Scholar] [CrossRef]
- Darwish, R.M.; Abu Fares, R.J.; Abu Zarga, M.H.; Nazer, I.K. Antibacterial effect of Jordanian propolis and isolated flavonoids against human pathogenic bacteria. Afr. J. Biotechnol. 2010, 9, 5966–5974. [Google Scholar] [CrossRef]
- Kopacz, M.; Woźnicka, E.; Gruszecka, J. Antibacterial activity of morin and its complexes with La(III), Gd(III) and Lu(III) ions. Acta Pol. Pharm.-Drug Res. 2005, 62, 65–67. [Google Scholar]
- Kishimoto, N.; Kakino, Y.; Iwai, K.; Mochida, K.; Fujita, T. In Vitro Antibacterial, Antimutagenic and Anti-Influenza Virus Activity of Caffeic Acid Phenethyl Esters. Biocontrol. Sci. 2005, 10, 155–161. [Google Scholar] [CrossRef]
- Kharsany, K. The Antimicrobial Properties of The Major Compounds Found in South African Propolis; University of the Witwatersrand, Faculty of Health Sciences: Johannesburg, South Africa, 2019. [Google Scholar]
- Kheadr, E.; Bernoussi, N.; Lacroix, C.; Fliss, I. Comparison of the sensitivity of commercial strains and infant isolates of bifidobacteria to antibiotics and bacteriocins. Int. Dairy J. 2004, 14, 1041–1053. [Google Scholar] [CrossRef]
- Phillips, C.; Duggan, J. The effect of temperature and citric acid, alone, and in combination with nisin, on the growth of Arcobacter butzleri in culture. Food Control. 2002, 13, 463–468. [Google Scholar] [CrossRef]
- Huber, B.; Eberl, L.; Feucht, W.; Polster, J. Influence of Polyphenols on Bacterial Biofilm Formation and Quorum-sensing. Z. Für Nat. C 2003, 58, 879–884. [Google Scholar] [CrossRef]
- Almajano, M.; Carbó, R.; Delgado, M.; Gordon, M. Effect of pH on the Antimicrobial Activity and Oxidative Stability of Oil-in-Water Emulsions Containing Caffeic Acid. J. Food Sci. 2007, 72, C258–C263. [Google Scholar] [CrossRef]
- Bankova, V.; Marcucci, M.; Simova, S.; Nikolova, N.; Kujumgiev, A.; Popov, S. Antibacterial Diterpenic Acids from Brazilian Propolis. Z. Für Nat. C 1996, 51, 277–280. [Google Scholar] [CrossRef]
- Rahman, M.M.; Richardson, A.; Sofian-Azirun, M. Antibacterial activity of propolis and honey against Staphylococcus aureus and Escherichia coli. Afr. J. Microbiol. Res. 2010, 4, 1872. [Google Scholar] [CrossRef]
- Strahl, H.; Errington, J. Bacterial Membranes: Structure, Domains, and Function. Annu. Rev. Microbiol. 2017, 71, 519–538. [Google Scholar] [CrossRef]
- Typas, A.; Sourjik, V. Bacterial protein networks: Properties and functions. Nat. Rev. Genet. 2015, 13, 559–572. [Google Scholar] [CrossRef]
- Setlow, B.; Setlow, P. Binding of small, acid-soluble spore proteins to DNA plays a significant role in the resistance of Bacillus subtilis spores to hydrogen peroxide. Appl. Environ. Microb. 1993, 59, 3418–3423. [Google Scholar] [CrossRef] [PubMed]
- Diao, W.-R.; Hu, Q.-P.; Zhang, H.; Xu, J.-G. Chemical composition, antibacterial activity and mechanism of action of essential oil from seeds of fennel (Foeniculum vulgare Mill.). Food Control. 2014, 35, 109–116. [Google Scholar] [CrossRef]
- Zhou, D.; Liu, Z.-H.; Wang, D.-M.; Li, D.-W.; Yang, L.-N.; Wang, W. Chemical composition, antibacterial activity and related mechanism of valonia and shell from Quercus variabilis Blume (Fagaceae) against Salmonella paratyphi a and Staphylococcus aureus. BMC Complement. Altern. Med. 2019, 19, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Wei, F.; Song, C.; Jiang, B.; Tian, S.; Yi, J.; Yu, C.; Song, Z.; Sun, L.; Bao, Y.; et al. Dodartia orientalis L. essential oil exerts antibacterial activity by mechanisms of disrupting cell structure and resisting biofilm. Ind. Crop. Prod. 2017, 109, 358–366. [Google Scholar] [CrossRef]
- Yuan, L.-H.; Hu, Q.-P.; Xu, J.-G. Antibacterial Mechanism of The Ehyl Acetate Extracts From Naked Oat Again Bacillus subtilis. Aceh Int. J. Sci. Technol. 2014, 4. [Google Scholar] [CrossRef]
- Wang, F.; Liu, H.; Li, J.; Zhang, W.; Jiang, B.; Xuan, H. Australian propolis ethanol extract exerts antibacterial activity against methicillin-resistant Staphylococcus aureus by mechanisms of disrupting cell structure, reversing resistance, and resisting biofilm. Braz. J. Microbiol. 2021, 52, 1651–1664. [Google Scholar] [CrossRef]
- Bryan, J.; Redden, P.; Traba, C. The mechanism of action of Russian propolis ethanol extracts against two antibiotic-resistant biofilm-forming bacteria. Lett. Appl. Microbiol. 2016, 62, 192–198. [Google Scholar] [CrossRef]
- He, F.; Li, K.; Zhang, X.; Yang, Y.; Fang, Y.; Xiang, F. Components and antibacterial activity of a novel essential oil from the nutrient broth of Eremothecium ashbyii H4565. LWT 2018, 101, 389–394. [Google Scholar] [CrossRef]
- An, P.; Yang, X.; Yu, J.; Qi, J.; Ren, X.; Kong, Q. α-terpineol and terpene-4-ol, the critical components of tea tree oil, exert antifungal activities in vitro and in vivo against Aspergillus niger in grapes by inducing morphous damage and metabolic changes of fungus. Food Control 2019, 98, 42–53. [Google Scholar] [CrossRef]
- Belenky, P.; Ye, J.D.; Porter, C.B.; Cohen, N.R.; Lobritz, M.A.; Ferrante, T.; Jain, S.; Korry, B.J.; Schwarz, E.G.; Walker, G.C.; et al. Bactericidal Antibiotics Induce Toxic Metabolic Perturbations that Lead to Cellular Damage. Cell Rep. 2015, 13, 968–980. [Google Scholar] [CrossRef]
- Schelli, K.; Zhong, F.; Zhu, J. Comparative metabolomics revealing Staphylococcus aureus metabolic response to different antibiotics. Microb. Biotechnol. 2017, 10, 1764–1774. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Feng, M.; Yang, K.; Cao, Y.; Zhang, J.; Xu, J.; Hernández, S.H.; Wei, X.; Fan, M. Transcriptomic and metabolomic analyses reveal antibacterial mechanism of astringent persimmon tannin against Methicillin-resistant Staphylococcus aureus isolated from pork. Food Chem. 2020, 309, 125692. [Google Scholar] [CrossRef]
- Hussein, M.; Karas, J.A.; Schneider-Futschik, E.K.; Chen, F.; Swarbrick, J.; Paulin, O.K.A.; Hoyer, D.; Baker, M.; Zhu, Y.; Li, J.; et al. The Killing Mechanism of Teixobactin against Methicillin-Resistant Staphylococcus aureus: An Untargeted Metabolomics Study. MSystems 2020, 5, 00077-20. [Google Scholar] [CrossRef] [PubMed]
- Selvaraj, A.; Jayasree, T.; Valliammai, A.; Pandian, S.K. Myrtenol Attenuates MRSA Biofilm and Virulence by Suppressing sarA Expression Dynamism. Front. Microbiol. 2019, 10, 2027. [Google Scholar] [CrossRef] [PubMed]
- Arciola, C.R.; Campoccia, D.; Baldassarri, L.; Donati, M.E.; Pirini, V.; Gamberini, S.; Montanaro, L. Detection of biofilm formation inStaphylococcus epidermidis from implant infections. Comparison of a PCR-method that recognizes the presence ofica genes with two classic phenotypic methods. J. Biomed. Mater. Res. Part A 2005, 76, 425–430. [Google Scholar] [CrossRef] [PubMed]
- Ahmadrajabi, R.; Layegh-Khavidaki, S.; Kalantar-Neyestanaki, D.; Fasihi, Y. Molecular analysis of immune evasion cluster (IEC) genes and intercellular adhesion gene cluster (ICA) among methicillin-resistant and methicillin-sensitive isolates of Staphylococcus aureus. J. Prev. Med. Hyg. 2017, 58, E308–E314. [Google Scholar] [CrossRef] [PubMed]
- Bronner, S.; Monteil, H.; Prévost, G. Regulation of virulence determinants in Staphylococcus aureus: Complexity and applications. FEMS Microbiol. Rev. 2004, 28, 183–200. [Google Scholar] [CrossRef]
- Bai, J.; Wu, Y.; Liu, X.; Zhong, K.; Huang, Y.; Gao, H. Antibacterial Activity of Shikimic Acid from Pine Needles of Cedrus deodara against Staphylococcus aureus through Damage to Cell Membrane. Int. J. Mol. Sci. 2015, 16, 27145–27155. [Google Scholar] [CrossRef]
- Wang, K.; Zhang, J.; Ping, S.; Ma, Q.; Chen, X.; Xuan, H.; Shi, J.; Zhang, C.; Hu, F. Anti-inflammatory effects of ethanol extracts of Chinese propolis and buds from poplar (Populus × canadensis). J. Ethnopharmacol. 2014, 155, 300–311. [Google Scholar] [CrossRef]
- Liu, X.; Zhong, S.; Wu, K.; Yu, H.; Chai, X. Antimicrobial activity in the vapour phase of a combination of cinnamon and clove essential oils. Food Chem. 2009, 116, 982–989. [Google Scholar]
- Elnakady, Y.A.; Chatterjee, I.; Bischoff, M.; Rohde, M.; Josten, M.; Sahl, H.-G.; Herrmann, M.; Müller, R. Investigations to the Antibacterial Mechanism of Action of Kendomycin. PLoS ONE 2016, 11, e0146165. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Li, J.; Yang, L.; Shi, F.; Yang, L.; Ye, M. Antibacterial activity and a membrane damage mechanism of Lachnum YM30 melanin against Vibrio parahaemolyticus and Staphylococcus aureus. Food Control 2017, 73, 1445–1451. [Google Scholar] [CrossRef]
- Sannasiddappa, T.H.; Lund, P.A.; Clarke, S.R.; Sannasiddappa, T. In Vitro Antibacterial Activity of Unconjugated and Conjugated Bile Salts on Staphylococcus aureus. Front. Microbiol. 2017, 8, 1581. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Li, C.; Dai, J.; Cui, H.; Lin, L. Antibacterial activity and mechanism of Litsea cubeba essential oil against methicillin-resistant Staphylococcus aureus (MRSA). Ind. Crop. Prod. 2019, 130, 34–41. [Google Scholar] [CrossRef]
- Tang, C.; Chen, J.; Zhang, L.; Zhang, R.; Zhang, S.; Ye, S.; Zhao, Z.; Yang, D. Exploring the antibacterial mechanism of essential oils by membrane permeability, apoptosis and biofilm formation combination with proteomics analysis against methicillin-resistant staphylococcus aureus. Int. J. Med. Microbiol. 2020, 310, 151435. [Google Scholar] [CrossRef] [PubMed]
- Kot, B.; Sytykiewicz, H.; Sprawka, I. Expression of the Biofilm-Associated Genes in Methicillin-Resistant Staphylococcus aureus in Biofilm and Planktonic Conditions. Int. J. Mol. Sci. 2018, 19, 3487. [Google Scholar] [CrossRef]
- Qu, X.; Yang, H.; Jia, B.; Yu, Z.; Zheng, Y.; Dai, K. Biodegradable Zn–Cu alloys show antibacterial activity against MRSA bone infection by inhibiting pathogen adhesion and biofilm formation. Acta Biomater. 2020, 117, 400–417. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).