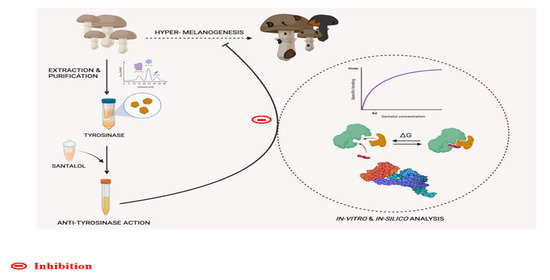
Elucidating the Role of Santalol as a Potent Inhibitor of Tyrosinase: In Vitro and In Silico Approaches
Abstract
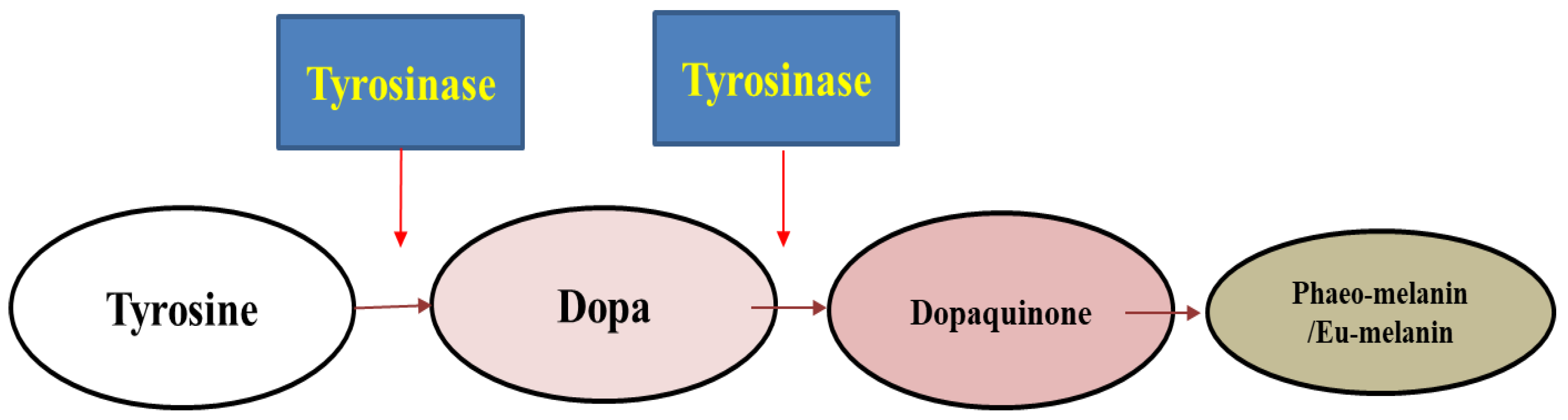
1. Introduction
2. Results and Discussion
2.1. Extraction of Enzyme Tyrosinase and Ammonium Sulphate Precipitation
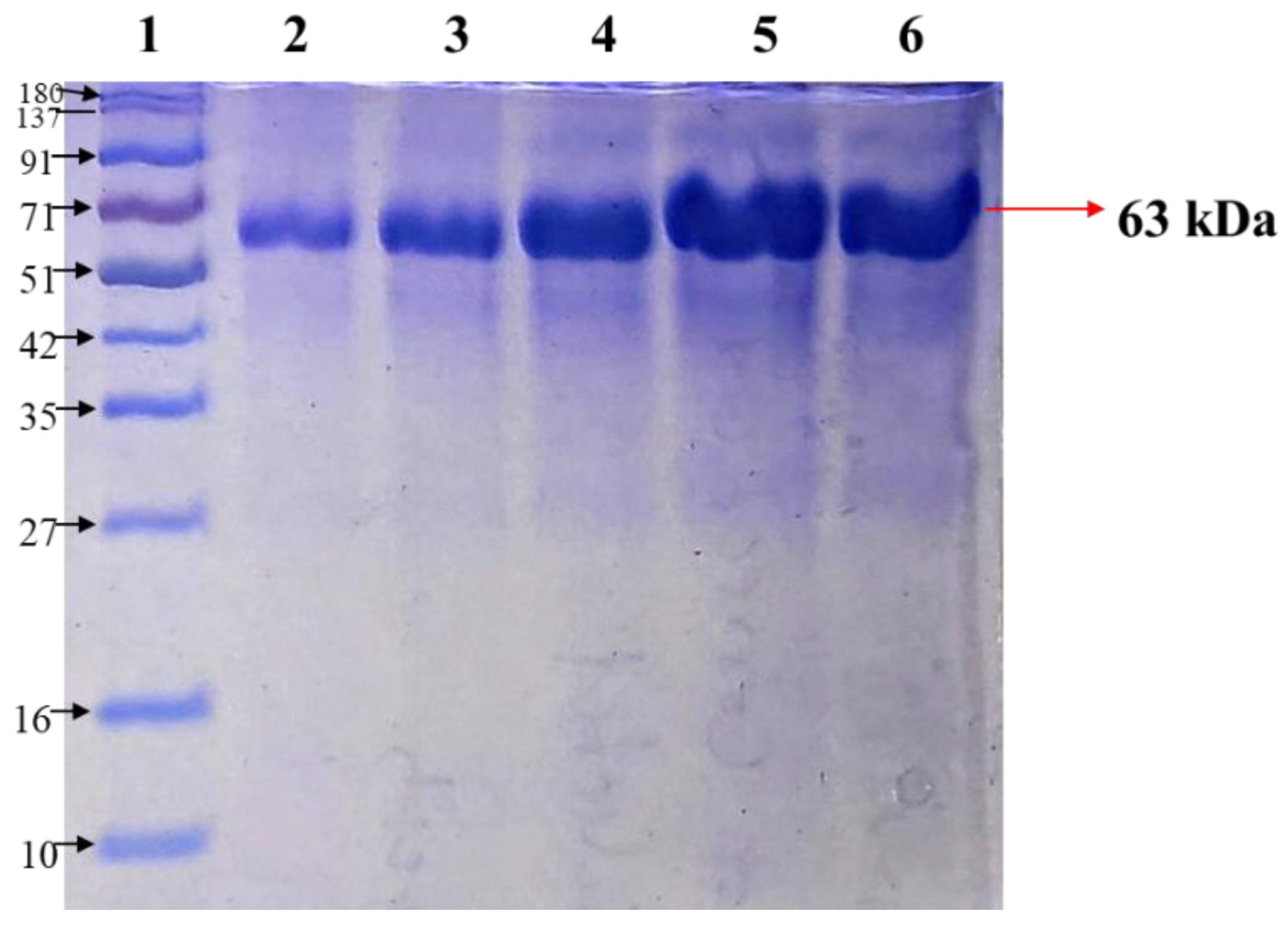
2.2. Purification of Enzyme Tyrosinase Using Chromatography
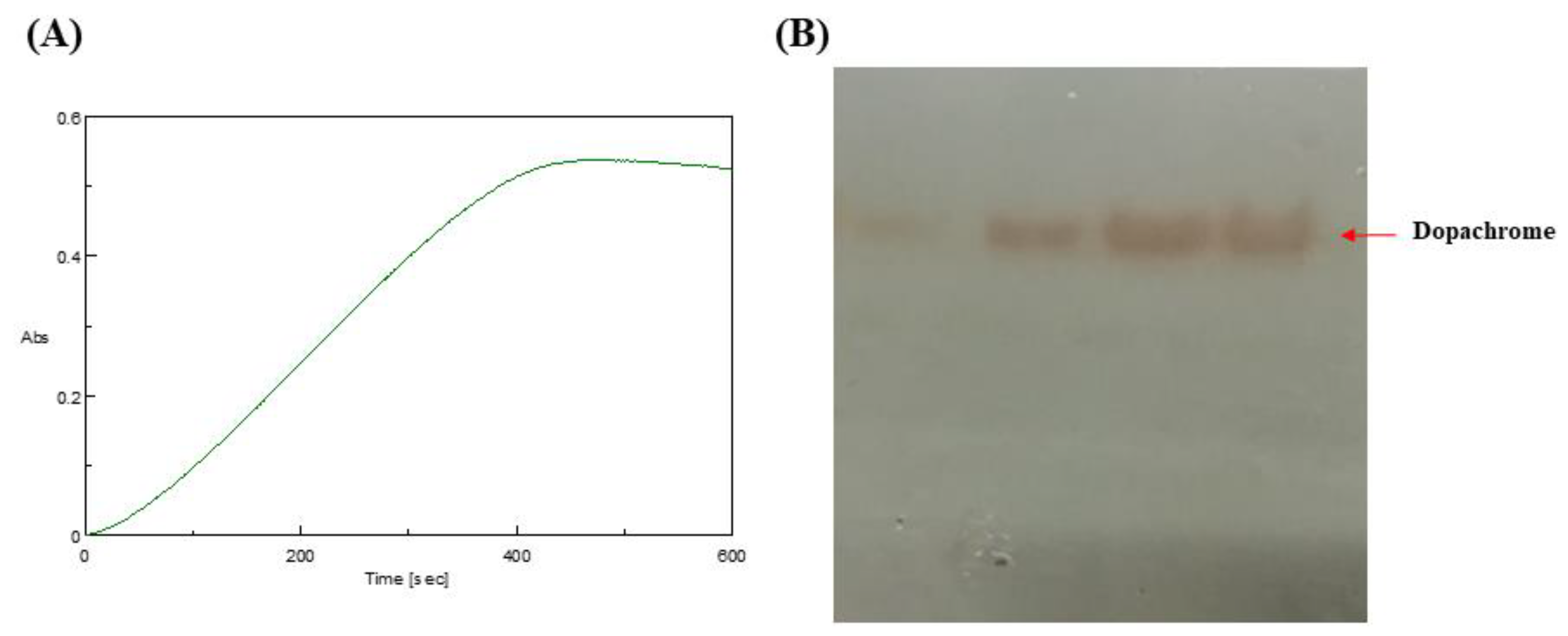
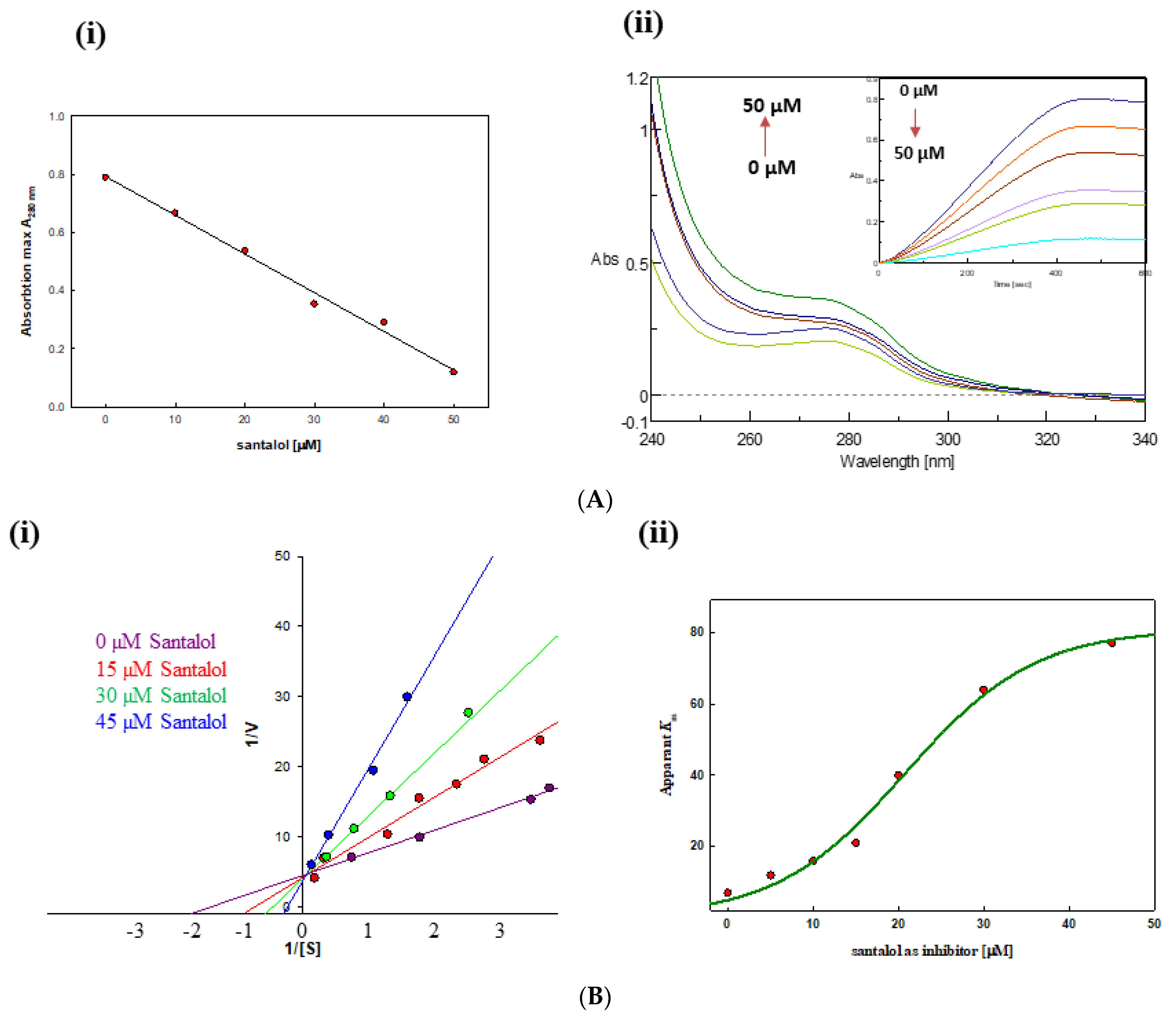
2.3. Tyrosinase Activity Assay
2.4. Tyrosinase Inhibition Kinetics
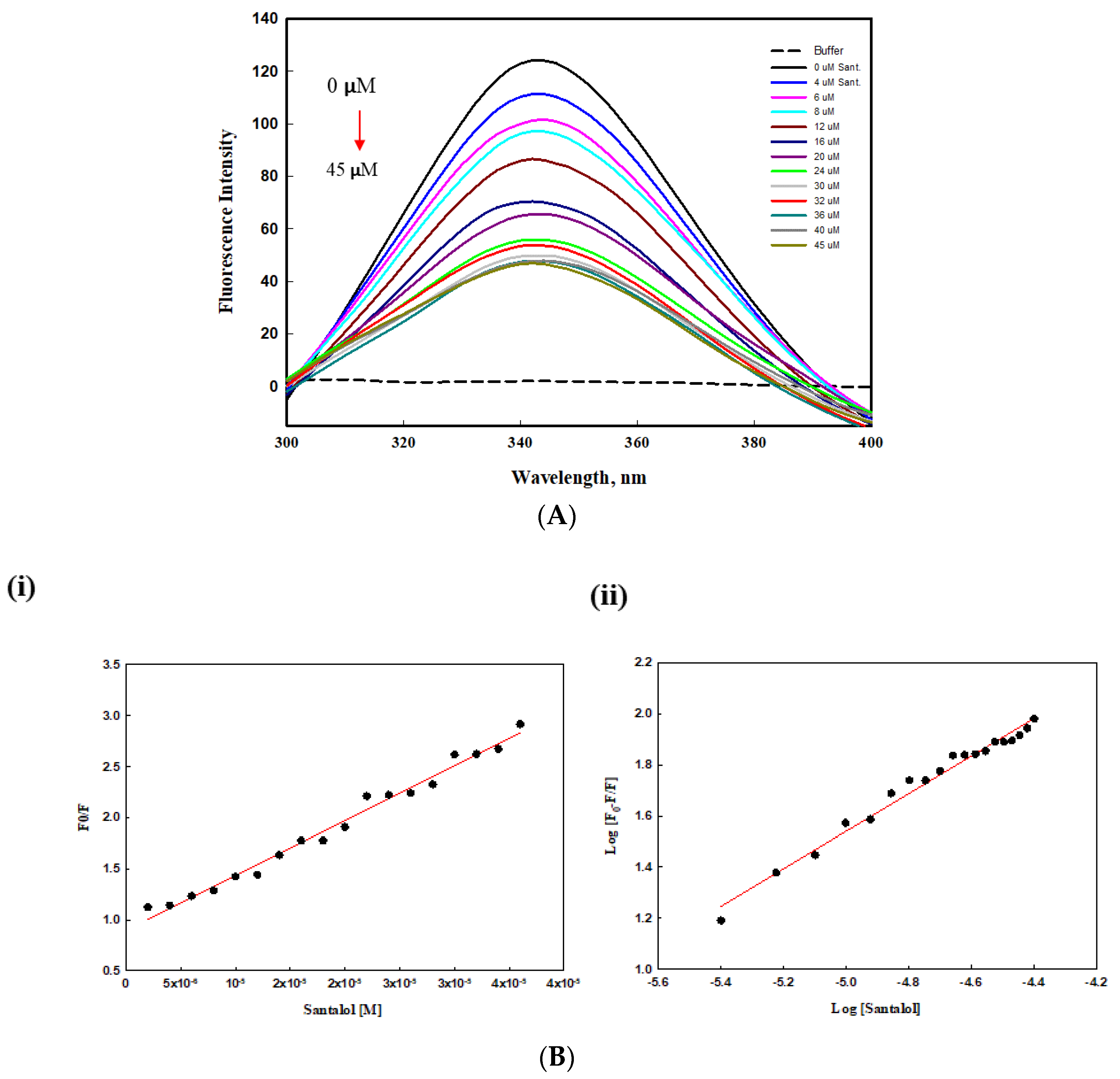
2.5. Intrinsic Fluorescence Binding Study
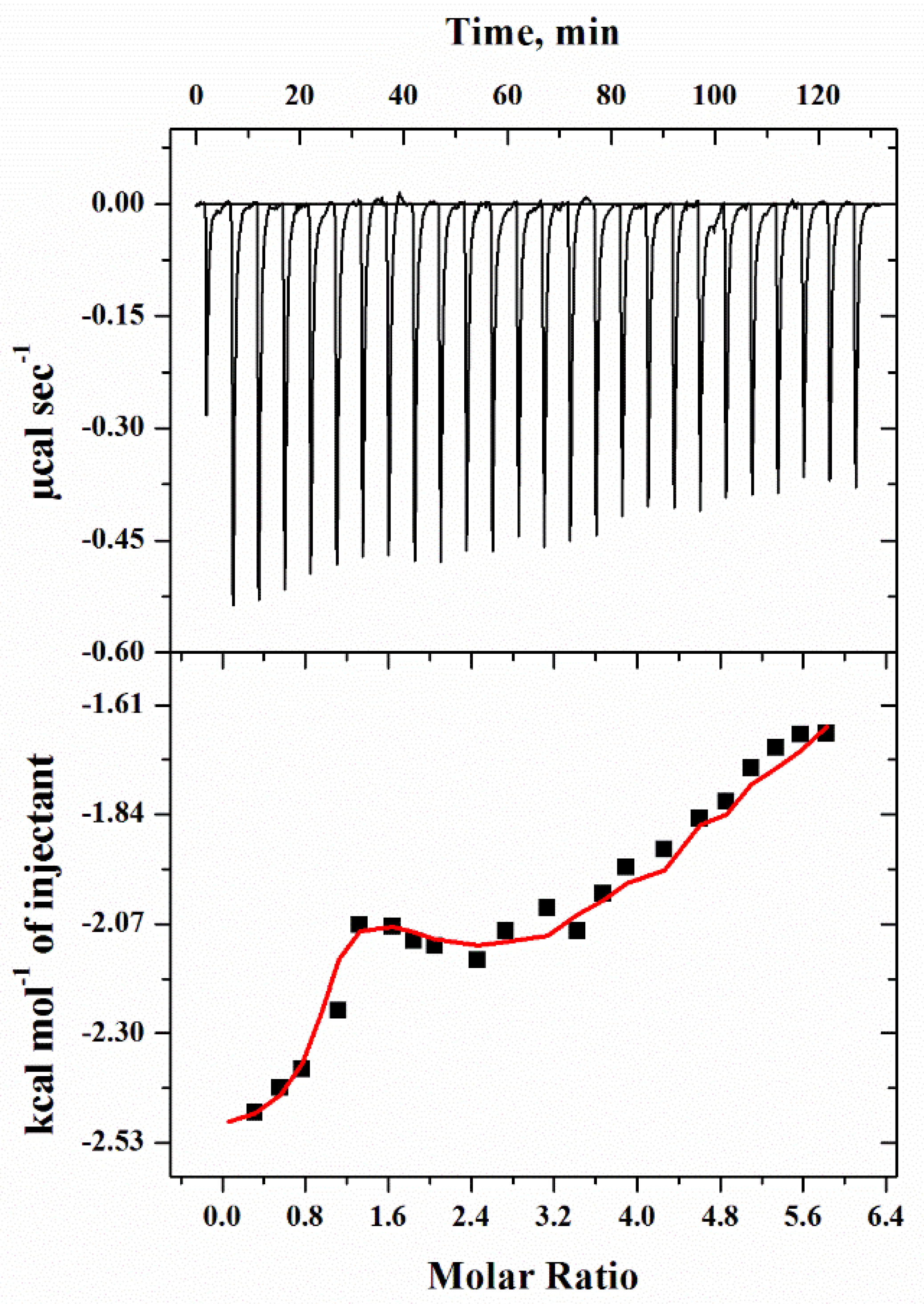
2.6. Isothermal Titration Calorimetry (ITC)
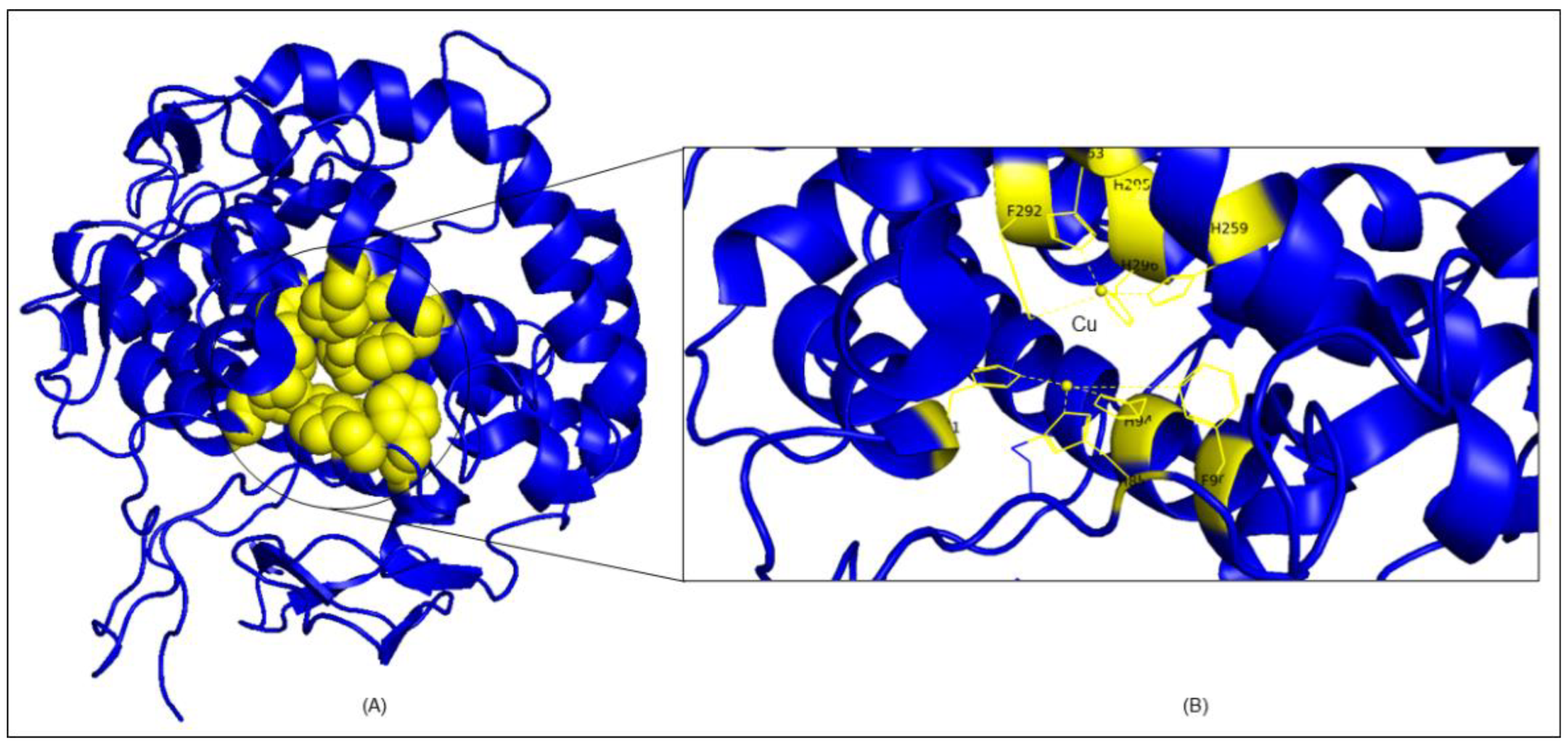
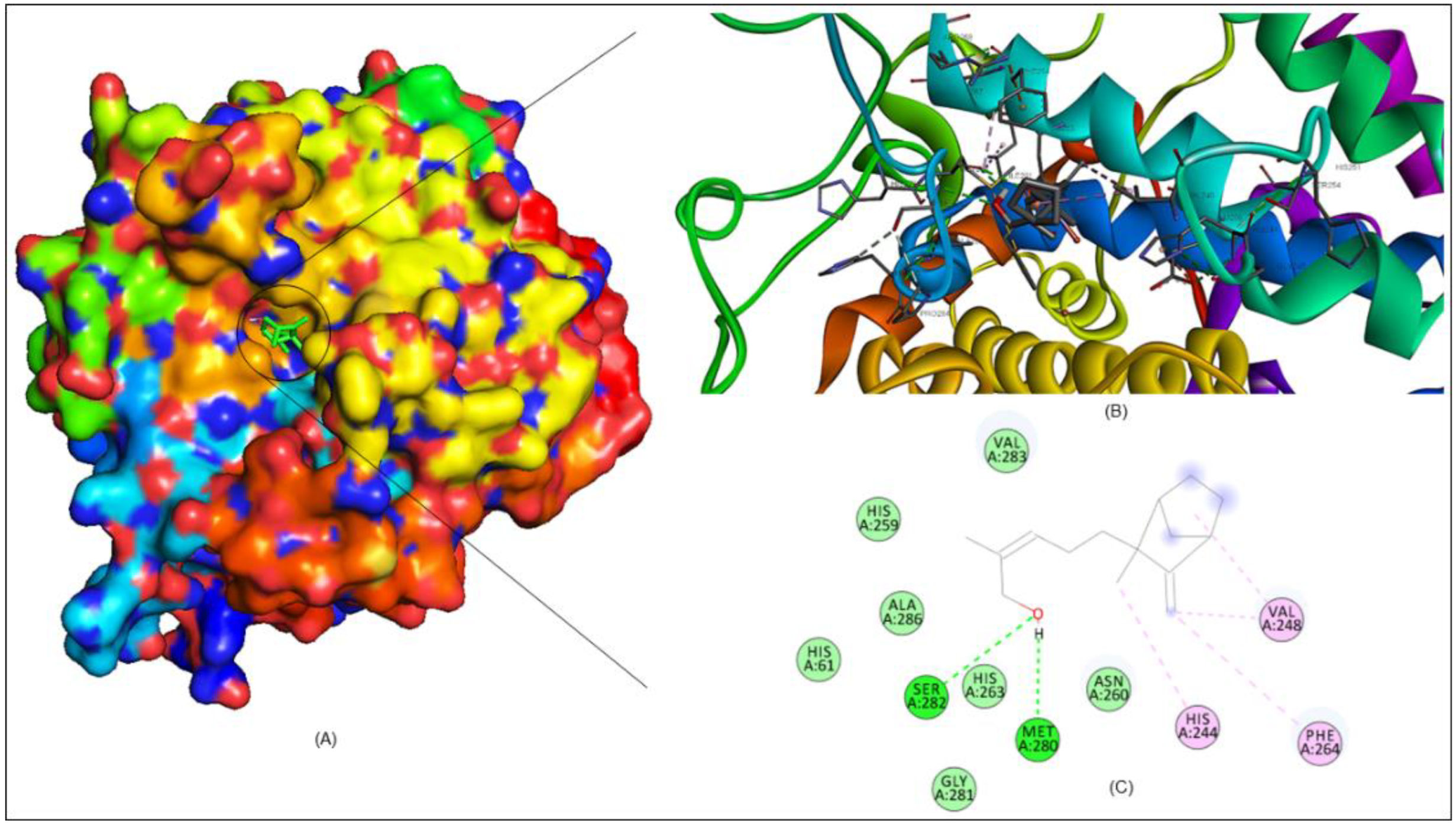
2.7. Molecular Docking Analysis
3. Materials and Methods
3.1. Materials
3.2. Methods
3.2.1. Extraction of the Enzyme Tyrosinase
3.2.2. Ammonium Sulphate Precipitation with Dialysis
3.2.3. Purification of Enzyme Tyrosinase Using Chromatography
3.2.4. Tyrosinase Activity Assay
3.2.5. Tyrosinase Inhibition Kinetics and UV-Visible Spectral Measurements
3.2.6. Intrinsic Fluorescence Binding Study
3.2.7. Isothermal Titration Calorimetry (ITC)
3.2.8. Molecular Docking Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kamilah, S.N.; Muslim, C.; Astuti, Y.D.; Pasaribu, M.D. Phenotypic Variation in Pigmentation of Persons with Albinism in Rejang Lebong, Bengkulu. In 3rd KOBI Congress, International and National Conferences (KOBICINC 2020); Atlantis Press: Paris, France, 2021; pp. 391–394. [Google Scholar]
- Battistella, C.; McCallum, N.C.; Vanthournout, B.; Forman, C.J.; Ni, Q.Z.; La Clair, J.J.; Burkart, M.D.; Shawkey, M.D.; Gianneschi, N.C. Bioinspired chemoenzymatic route to artificial melanin for hair pigmentation. Chem. Mater. 2020, 32, 9201–9210. [Google Scholar] [CrossRef]
- Solano, F. Photoprotection and skin pigmentation: Melanin-related molecules and some other new agents obtained from natural sources. Molecules 2020, 25, 1537. [Google Scholar] [CrossRef] [PubMed]
- Azumi, J.; Takeda, T.; Shimada, Y.; Aso, H.; Nakamura, T. The organogermanium compound THGP suppresses melanin synthesis via complex formation with L-DOPA on mushroom tyrosinase and in B16 4A5 melanoma cells. Int. J. Mol. Sci. 2019, 20, 4785. [Google Scholar] [CrossRef] [PubMed]
- Hexsel, D.; Lacerda, D.A.; Cavalcante, A.S.; Filho, C.A.M.; Kalil, C.L.P.; Ayres, E.L.; Azulay-Abulafia, L.; Weber, M.B.; Serra, M.S.; Lopes, N.F. Epidemiology of melasma in B razilian patients: A multicenter study. Int. J. Dermatol. 2014, 53, 440–444. [Google Scholar] [CrossRef]
- Suh, A.; Pham, A.; Cress, M.J.; Pincelli, T.; TerKonda, S.P.; Bruce, A.J.; Zubair, A.C.; Wolfram, J.; Shapiro, S.A. Adipose-derived cellular and cell-derived regenerative therapies in dermatology and aesthetic rejuvenation. Ageing Res. Rev. 2019, 54, 100933. [Google Scholar] [CrossRef]
- Hongbo, Y.; Thomas, C.L.; Harrison, M.A.; Salek, M.S.; Finlay, A.Y. Translating the science of quality of life into practice: What do dermatology life quality index scores mean? J. Investig. Dermatol. 2005, 125, 659–664. [Google Scholar] [CrossRef]
- Khalilzadeh Ganjalikhani, M.; Tirgari, B.; Roudi Rashtabadi, O.; Shahesmaeili, A. Studying the effect of structured ostomy care training on quality of life and anxiety of patients with permanent ostomy. Int. Wound J. 2019, 16, 1383–1390. [Google Scholar] [CrossRef]
- Wang, D.; Wang, P.; Liu, D.; Zhou, Z. Fluorometric atrazine assay based on the use of nitrogen-doped graphene quantum dots and on inhibition of the activity of tyrosinase. Microchim. Acta 2019, 186, 527. [Google Scholar] [CrossRef]
- Ogawa, T.; Fujii, S.; Kuya, K.; Kitao, S.-i.; Shinohara, Y.; Ishibashi, M.; Tanabe, Y. Role of neuroimaging on differentiation of Parkinson’s disease and its related diseases. Yonago Acta Med. 2018, 61, 145–155. [Google Scholar] [CrossRef]
- Zhou, X.; Xiao, Y.; Meng, X.; Liu, B. Full inhibition of Whangkeumbae pear polyphenol oxidase enzymatic browning reaction by L-cysteine. Food Chem. 2018, 266, 1–8. [Google Scholar] [CrossRef]
- Chen, C.-Y.; Lin, L.-C.; Yang, W.-F.; Bordon, J.; D Wang, H.-M. An updated organic classification of tyrosinase inhibitors on melanin biosynthesis. Curr. Org. Chem. 2015, 19, 4–18. [Google Scholar] [CrossRef]
- Hearing, V.J. Determination of melanin synthetic pathways. J. Investig. Dermatol. 2011, 131, E8. [Google Scholar] [CrossRef] [PubMed]
- Upadhyay, S.; Xu, X.; Lowry, D.; Jackson, J.C.; Roberson, R.W.; Lin, X. Subcellular compartmentalization and trafficking of the biosynthetic machinery for fungal melanin. Cell Rep. 2016, 14, 2511–2518. [Google Scholar] [CrossRef] [PubMed]
- Ismaya, W.T.; Rozeboom, H.J.; Schurink, M.; Boeriu, C.G.; Wichers, H.; Dijkstra, B.W. Crystallization and preliminary X-ray crystallographic analysis of tyrosinase from the mushroom Agaricus bisporus. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2011, 67, 575–578. [Google Scholar] [CrossRef] [PubMed]
- Ismaya, W.T.; Rozeboom, H.J.; Weijn, A.; Mes, J.J.; Fusetti, F.; Wichers, H.J.; Dijkstra, B.W. Crystal structure of Agaricus bisporus mushroom tyrosinase: Identity of the tetramer subunits and interaction with tropolone. Biochemistry 2011, 50, 5477–5486. [Google Scholar] [CrossRef]
- You, Z.; Wang, Y.-Q.; He, Q.-X. Homology Modeling of Human Tyrosinase. J. Southwest Univ. 2020, 42, 42–48. [Google Scholar]
- Peng, Z.; Feng, L.; Tan, L.; Chen, L.; Shi, Q.; Zeng, Q.-H.; Liu, H.; Wang, J.J.; Zhao, Y. Anti-tyrosinase, antioxidant and antibacterial activities of gallic acid-benzylidenehydrazine hybrids and their application in preservation of fresh-cut apples and shrimps. Food Chem. 2022, 378, 132127. [Google Scholar] [CrossRef]
- Pavan, M.E.; López, N.I.; Pettinari, M.J. Melanin biosynthesis in bacteria, regulation and production perspectives. Appl. Microbiol. Biotechnol. 2020, 104, 1357–1370. [Google Scholar] [CrossRef]
- Zolghadri, S.; Bahrami, A.; Hassan Khan, M.T.; Munoz-Munoz, J.; Garcia-Molina, F.; Garcia-Canovas, F.; Saboury, A.A. A comprehensive review on tyrosinase inhibitors. J. Enzym. Inhib. Med. Chem. 2019, 34, 279–309. [Google Scholar] [CrossRef]
- Saeedi, M.; Khezri, K.; Seyed Zakaryaei, A.; Mohammadamini, H. A comprehensive review of the therapeutic potential of α-arbutin. Phytother. Res. 2021, 35, 4136–4154. [Google Scholar] [CrossRef]
- Kim, Y.-J.; Uyama, H. Tyrosinase inhibitors from natural and synthetic sources: Structure, inhibition mechanism and perspective for the future. Cell. Mol. Life Sci. CMLS 2005, 62, 1707–1723. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.-J.; Chung, J.E.; Kurisawa, M.; Uyama, H.; Kobayashi, S. New tyrosinase inhibitors,(+)-catechin− aldehyde polycondensates. Biomacromolecules 2004, 5, 474–479. [Google Scholar] [CrossRef] [PubMed]
- Parvez, S.; Kang, M.; Chung, H.S.; Bae, H. Naturally occurring tyrosinase inhibitors: Mechanism and applications in skin health, cosmetics and agriculture industries. Phytother. Res. Int. J. Devoted Pharmacol. Toxicol. Eval. Nat. Prod. Deriv. 2007, 21, 805–816. [Google Scholar] [CrossRef] [PubMed]
- Bommareddy, A.; Brozena, S.; Steigerwalt, J.; Landis, T.; Hughes, S.; Mabry, E.; Knopp, A.; VanWert, A.L.; Dwivedi, C. Medicinal properties of alpha-santalol, a naturally occurring constituent of sandalwood oil. Nat. Prod. Res. 2019, 33, 527–543. [Google Scholar] [CrossRef] [PubMed]
- Misra, B.B.; Dey, S. Evaluation of in vivo anti-hyperglycemic and antioxidant potentials of α-santalol and sandalwood oil. Phytomedicine 2013, 20, 409–416. [Google Scholar] [CrossRef] [PubMed]
- Burdock, G.A.; Carabin, I.G. Safety assessment of sandalwood oil (Santalum album L.). Food Chem. Toxicol. 2008, 46, 421–432. [Google Scholar] [CrossRef] [PubMed]
- Dubnicka, M.; Cromwell, B.; Levine, M. Investigation of the Adulteration of Essential Oils by GC-MS. Curr. Anal. Chem. 2020, 16, 965–969. [Google Scholar] [CrossRef]
- Jain, R.; Nair, S. Sandalwood oil for the chemoprevention of skin cancer: Mechanistic insights, anti-inflammatory, and in vivo anticancer potential. Curr. Pharmacol. Rep. 2019, 5, 345–358. [Google Scholar] [CrossRef]
- Duong-Ly, K.C.; Gabelli, S.B. Salting out of proteins using ammonium sulfate precipitation. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 2014; pp. 85–94. [Google Scholar]
- Moringo, N.A.; Bishop, L.D.; Shen, H.; Misiura, A.; Carrejo, N.C.; Baiyasi, R.; Wang, W.; Ye, F.; Robinson, J.T.; Landes, C.F. A mechanistic examination of salting out in protein–polymer membrane interactions. Proc. Natl. Acad. Sci. USA 2019, 116, 22938–22945. [Google Scholar] [CrossRef]
- Hagel, L. Gel-filtration chromatography. Curr. Protoc. Mol. Biol. 1998, 44, 10.9. 1–10.9. 2. [Google Scholar] [CrossRef]
- Lopez-Tejedor, D.; Palomo, J.M. Efficient purification of a highly active H-subunit of tyrosinase from Agaricus bisporus. Protein Expr. Purif. 2018, 145, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Ali, N.; Aiman, A.; Shamsi, A.; Hassan, I.; Shahid, M.; Gaur, N.A.; Islam, A. Identification of Thermostable Xylose Reductase from Thermothelomyces thermophilus: A Biochemical Characterization Approach to Meet Biofuel Challenges. ACS Omega 2022. [Google Scholar] [CrossRef] [PubMed]
- Kielkopf, C.L.; Bauer, W.; Urbatsch, I.L. Bradford assay for determining protein concentration. Cold Spring Harb. Protoc. 2020, 2020, pdb-prot102269. [Google Scholar] [CrossRef] [PubMed]
- Zeng, H.-j.; Liu, Z.; Hu, G.-z.; Qu, L.-b.; Yang, R. Investigation on the binding of aloe-emodin with tyrosinase by spectral analysis and molecular docking. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2019, 211, 79–85. [Google Scholar] [CrossRef] [PubMed]
- Mohammad, T.; Mathur, Y.; Hassan, M.I. InstaDock: A single-click graphical user interface for molecular docking-based virtual high-throughput screening. Brief. Bioinform. 2021, 22, bbaa279. [Google Scholar] [CrossRef] [PubMed]
- Tisserand, R.; Young, R. Essential Oil Safety: A Guide for Health Care Professionals; Elsevier Health Sciences: Amsterdam, The Netherlands, 2013. [Google Scholar]
- Pires, D.E.; Blundell, T.L.; Ascher, D.B. pkCSM: Predicting small-molecule pharmacokinetic and toxicity properties using graph-based signatures. J. Med. Chem. 2015, 58, 4066–4072. [Google Scholar] [CrossRef]
- Haghbeen, K.; Tan, E.W. Direct spectrophotometric assay of monooxygenase and oxidase activities of mushroom tyrosinase in the presence of synthetic and natural substrates. Anal. Biochem. 2003, 312, 23–32. [Google Scholar] [CrossRef]
- Andrew, S.M.; Titus, J.A.; Zumstein, L. Dialysis and concentration of protein solutions. Curr. Protoc. Toxicol. 2001, 10, A. 3H. 1–A. 3H. 5. [Google Scholar] [CrossRef]
- Wingfield, P. Protein precipitation using ammonium sulfate. Curr. Protoc. Protein Sci. 1998, 13, A. 3F. 1–A. 3F. 8. [Google Scholar]
- Gao, Z.; Shen, P.; Lan, Y.; Cui, L.; Ohm, J.-B.; Chen, B.; Rao, J. Effect of alkaline extraction pH on structure properties, solubility, and beany flavor of yellow pea protein isolate. Food Res. Int. 2020, 131, 109045. [Google Scholar] [CrossRef]
- Pugh, M.E.; Schultz, E. Assessment of the purification of a protein by ion exchange and gel permeation chromatography. Biochem. Mol. Biol. Educ. 2002, 30, 179–183. [Google Scholar] [CrossRef]
- Schägger, H. Tricine–sds-page. Nat. Protoc. 2006, 1, 16–22. [Google Scholar] [CrossRef] [PubMed]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Hassan Sajedi, R.; Bagheri Kalmarzi, M.; Asadolahi, E.; Mahmoodi, N.; Mahdavi, A.; Haji Hosseini, R. Enzymatic browning, Inhibition, Nitroanilines, Tyrosinase. Modares J. Biotechnol. 2012, 3, 67–80. [Google Scholar]
- Wilkesman, J.; Kurz, L. Zymography principles. Zymography 2017, 1626, 3–10. [Google Scholar]
- Cui, Y.; Hu, Y.-H.; Yu, F.; Zheng, J.; Chen, L.-S.; Chen, Q.-X.; Wang, Q. Inhibition kinetics and molecular simulation of p-substituted cinnamic acid derivatives on tyrosinase. Int. J. Biol. Macromol. 2017, 95, 1289–1297. [Google Scholar] [CrossRef]
- Larsson, L.K. Computational and Structural Characterization of Engineered Type-3-Copper Variants; Norwegian University of Life Sciences: Ås, Norway, 2021. [Google Scholar]
- Kim, D.; Park, J.; Kim, J.; Han, C.; Yoon, J.; Kim, N.; Seo, J.; Lee, C. Flavonoids as mushroom tyrosinase inhibitors: A fluorescence quenching study. J. Agric. Food Chem. 2006, 54, 935–941. [Google Scholar] [CrossRef]
- Suryawanshi, V.D.; Walekar, L.S.; Gore, A.H.; Anbhule, P.V.; Kolekar, G.B. Spectroscopic analysis on the binding interaction of biologically active pyrimidine derivative with bovine serum albumin. J. Pharm. Anal. 2016, 6, 56–63. [Google Scholar] [CrossRef]
- Parray, Z.A.; Ahmad, F.; Alajmi, M.F.; Hussain, A.; Hassan, M.I.; Islam, A. Formation of molten globule state in horse heart cytochrome c under physiological conditions: Importance of soft interactions and spectroscopic approach in crowded milieu. Int. J. Biol. Macromol. 2020, 148, 192–200. [Google Scholar] [CrossRef]
- Parray, Z.A.; Ahmad, F.; Alajmi, M.F.; Hussain, A.; Hassan, M.I.; Islam, A. Interaction of polyethylene glycol with cytochrome c investigated via in vitro and in silico approaches. Sci. Rep. 2021, 11, 6475. [Google Scholar] [CrossRef]
- Parray, Z.A.; Ahmad, F.; Hassan, M.I.; Hasan, I.; Islam, A. Effects of Ethylene Glycol on the Structure and Stability of Myoglobin Using Spectroscopic, Interaction, and In Silico Approaches: Monomer Is Different from Those of Its Polymers. ACS Omega 2020, 5, 13840–13850. [Google Scholar] [CrossRef] [PubMed]
| S. No. | Purification Steps | Fraction Volume (mL) | Protein Conc. (mg/mL) b | Total Amt. of Protein (mg) | Total Activity (Units) | Specific Activity (Unit/mg) | Yields (%) | Purification Fold |
|---|---|---|---|---|---|---|---|---|
| 1 | Crude extract a | 310.0 | 1.250 | 387.5 | 4121 | 10.6 | 100 | 1 |
| 2 | Ammonium sulphate precipitation 30% cut-off | 120 | 0.853 | 102.36 | 3172 | 30.9 | 26.42 | 2.91 |
| 3 | Ion-exchange chromatography, DEAE-Sepharose column | 65 | 0.552 | 35.88 | 2627.5 | 73.25 | 9.25 | 7.07 |
| 4 | Gel filtration chromatography, using Superdex 200 column | 45 | 0.15 | 7 | 2256.2 | 322.3 | 1.8 | 32.14 |
| Thermodynamic Parameters (Units) | Step 1 | Step 2 | Step 3 |
|---|---|---|---|
| Ka (M−1) | 7.89 × 105 ± 8.5 × 104 | 1.32 × 104 ± 1.7 × 103 | 2.49 × 103 ± 2.8 × 102 |
| ∆H (cal mol−1) | −2.595 × 103 | −6.497 × 103 | −4.16 × 104 |
| ∆S (cal mol−1deg−1) | 18.3 | −2.94 | −124 |
| ∆G° (cal mol−1) | −8.048 × 103 ± 90.0 | −5.62 × 103 ± 7.87 × 102 | 4.16 × 103 ± 90.0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ali, N.; Zehra, Z.; Shamsi, A.; Beg, M.A.; Parray, Z.A.; Israil; Imam, M.A.; Gaur, N.A.; Hassan, M.I.; Chaudhary, A.A.; et al. Elucidating the Role of Santalol as a Potent Inhibitor of Tyrosinase: In Vitro and In Silico Approaches. Molecules 2022, 27, 8915. https://doi.org/10.3390/molecules27248915
Ali N, Zehra Z, Shamsi A, Beg MA, Parray ZA, Israil, Imam MA, Gaur NA, Hassan MI, Chaudhary AA, et al. Elucidating the Role of Santalol as a Potent Inhibitor of Tyrosinase: In Vitro and In Silico Approaches. Molecules. 2022; 27(24):8915. https://doi.org/10.3390/molecules27248915
Chicago/Turabian StyleAli, Nabeel, Zainy Zehra, Anas Shamsi, Md. Amjad Beg, Zahoor Ahmad Parray, Israil, Md. Ali Imam, Naseem A. Gaur, Md. Imtaiyaz Hassan, Anis Ahmad Chaudhary, and et al. 2022. "Elucidating the Role of Santalol as a Potent Inhibitor of Tyrosinase: In Vitro and In Silico Approaches" Molecules 27, no. 24: 8915. https://doi.org/10.3390/molecules27248915
APA StyleAli, N., Zehra, Z., Shamsi, A., Beg, M. A., Parray, Z. A., Israil, Imam, M. A., Gaur, N. A., Hassan, M. I., Chaudhary, A. A., Rudayni, H. A., Alghonaim, M. I., Alsalamah, S. A., & Islam, A. (2022). Elucidating the Role of Santalol as a Potent Inhibitor of Tyrosinase: In Vitro and In Silico Approaches. Molecules, 27(24), 8915. https://doi.org/10.3390/molecules27248915