Abstract
Signal transducer and activator of transcription 3 (STAT3) and nuclear factor erythroid-derived 2-like 2 (NRF2, also known as NFE2L2), are two of the most complicated transcription regulators, which participate in a variety of physiological processes. Numerous studies have shown that they are overactivated in multiple types of tumors. Interestingly, STAT3 and NRF2 can also interact with each other to regulate tumor progression. Hence, these two important transcription factors are considered key targets for developing a new class of antitumor drugs. This review summarizes the pivotal roles of the two transcription regulators and their interactions in the tumor microenvironment to identify potential antitumor drug targets and, ultimately, improve patients’ health and survival.
1. Introduction
Signal transducer and activator of transcription 3 (STAT3) belongs to the STAT family, which includes STAT1, STAT2, STAT3, STAT4, STAT5a, STAT5b, and STAT6, and mediates signal transduction from the cell membrane to the nucleus in multiple intracellular and extracellular activities [1,2]. As a crucial transcription factor, STAT3 exerts a vital role on all STAT proteins. As it is essential for early development, it gets involved in regulating the transcription of a good many crucial genes related to cell proliferation, differentiation, apoptosis, survival, angiogenesis, inflammation, immunity, and metastasis, thereby participating in various physiological and pathological processes [3,4,5]. There is mounting evidence showing that STAT3 plays a crucial role in various diseases, such as cancer [6,7,8], cerebrovascular diseases [9,10], cardiovascular diseases [11,12], and obesity [13].
The STAT3 gene is located on chromosome 17q21 [14,15]. The STAT3 protein consists of 770 amino acids and has six conserved domains (Figure 1). The amino-terminal domain (NTD) of STAT3 performs multiple functions, including protein–protein interactions, cooperative DNA binding, and nuclear translocation [16]. STAT3 interacts with other transcription factors and regulatory proteins via coiled-coil domain (CCD) [17]. The DNA-binding domain (DBD) facilitates STAT3 interactions with target genes. STAT3 dimerization is formed via the Src homology-2 (SH2) domain by identifying phosphorylated Tyr-705 of another STAT monomer [17]. The phosphorylation of serine sites on the C-terminal transcription activation domain (TAD) promotes the assembly of STAT3 with other transcriptional activators [17,18,19]. The structure of the STAT3 protein determines its special functions, which lays the foundation for signal transduction.
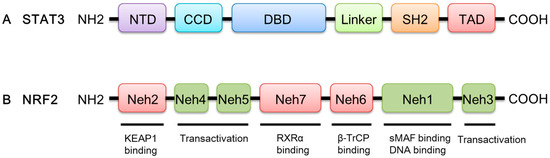
Figure 1.
A schematic diagram of the domain structures of STAT3 and NRF2. (A) STAT3 has six functional domains, including NTD, CCD, DBD, Linker, SH2, and TAD. (B) NRF2 has seven conserved domains labeled Neh1–Neh7.
Nuclear factor erythroid-derived 2-like 2 (NRF2) possesses a unique Cap ‘n’ Collar (CNC) motif followed by a basic leucine zipper (bZip), which belongs to the CNC transcription factor family. Human NRF2 contains seven highly conserved domains, namely NRF2-ECH (erythroid cell-derived protein with CNC homology) homology and (Neh) 1–Neh7; each domain has its own unique function (Figure 1). A bZip DNA-binding domain and a heterodimerization domain together form the Neh1 domain, which enables DNA-binding to the antioxidant response element (ARE) and the dimerization of NRF2 with small musculoaponeurotic fibrosarcoma (sMAF) proteins [20]. Neh2 is a negative regulatory domain of NRF2, which is crucial for Kelch-like ECH-associated protein 1 (KEAP1)-mediated repression of NRF2 [21]. The C-terminal Neh3 domain acting in parallel with the Neh4 and Neh5 domains has a transactivation-like activity to activate the transcription of NRF2 target genes [22,23]. The Neh6 domain includes two binding sites for β-transducin repeat-containing protein (β-TrCP), DSGIS, and DSAPGS, leading to glycogen synthase kinase 3 beta (GSK3β)-mediated NRF2 degradation in a KEAP1-independent manner [24,25]. Finally, Neh7 contains a domain that mediates a direct interaction between NRF2 and the DBD of retinoid X receptor α (RXRα), which suppresses the transcriptional activity of NRF2 by inhibiting the recruitment of coactivators to Neh4 and Neh5 domains [26].
Numerous studies indicate that NRF2 has a vital role in regulating redox and metabolic homeostasis by inducing corresponding target genes [27,28,29]. Due to the post-translational regulation by the ubiquitin-proteasome system (UPS), the cellular level of NRF2 is normally very low. However, when an organism is exposed to endogenous and environmental stresses, the action of the UPS is blocked, leading to the activation of NRF2 signaling. The newly translated NRF2 translocates to the nucleus, binds to sMAF proteins, and transcribes ARE-regulated genes. Multiple research data have confirmed that NRF2-deficient mice are more vulnerable to the toxicity and carcinogenesis of various xenobiotic stresses [30,31,32,33]. Many compounds, including natural products and others, have been found to activate NRF2 to protect cells from damage [34]. However, just as NRF2 can safeguard normal cells against insult, it can also protect tumor cells from damage, facilitating their transformation, growth, metastasis, and chemoresistance. Therefore, a growing number of researchers are committed to the discovery and development of NRF2 inhibitors.
Numerous studies have reported that STAT3 and NRF2 are hyperactive in tumors, with important and intricate regulatory functions. STAT3 and NRF2 interact to regulate tumor progression [35,36]. Depending on the situation, they can either prevent or promote cancer progression [35,36]. This review focuses on the crucial roles of two important transcription regulators (i.e., STAT3 and NRF2) and their interactions in the tumor microenvironment to identify potential antitumor drug targets and ultimately improve the health and survival of cancer patients.
2. Role of STAT3 Signaling
2.1. STAT3 Signal Transduction Cascade
Many factors including receptor tyrosine kinases (RTK), Janus kinases (JAKs), cytokines, and some non-receptor tyrosine kinases such as Src and Abl can induce the phosphorylation of tyrosine (705) and serine (727) residues of STAT3, which will activate the STAT3 signal transduction cascade [4,6,37,38,39,40]. Upon activation, STAT3 dissociates from the receptor/kinase complex to form homodimers or heterodimers via the SH2 domain. This is followed by nuclear translocation, DNA binding, and activation of target genes, including pro-proliferative/anti-apoptotic genes, angiogenic genes, metastatic genes, and the STAT3 gene itself [41]. Additionally, it is reported that unphosphorylated STAT3 (u-STAT3) can drive the expression of many genes, such as interleukin-6 (IL-6), IL-8, C-C motif chemokine ligand 5 (CCL5), regulated upon activation, normal T-cell expressed and secreted (RANTES), mesenchymal-epithelial transition (MET), and muscle RAS oncogene homolog (MRAS), via a non-canonical pathway independent of phosphorylation [42]. Therefore, phosphorylation is not necessary for STAT3 activation, and STAT3 regulates corresponding target genes through different methods.
STAT3 is tightly negatively regulated in unstimulated cells by a number of modulators, including the protein inhibitor of activated STAT3 (PIAS3), suppressor of cytokine signaling (SOCS) proteins, protein tyrosine phosphatases (PTPs), and ubiquitin enzymes, which can suppress the expression and nuclear translocation of STAT3 [43]. For example, PIAS3 expression negatively correlates with STAT3 signal transduction in cervical cancer (CC) cells, probably by repressing the DNA binding activity of STAT3 [44,45,46]. Baek et al. discovered that resveratrol can induce SOCS-1 expression, suppress STAT3 phosphorylation, and restrain proliferation, which thereby inhibits the STAT3 signaling pathway in squamous cell carcinoma of the head and neck (SCCHN) [47]. Numerous studies have shown that STAT3 signaling can be inhibited by various PTPs, such as protein tyrosine phosphatase non-receptor type 6 (PTPN6, also known as SHP1), protein tyrosine phosphatase non-receptor type 11 (PTPN11, also known as SHP2), CD45, protein tyrosine phosphatase non-receptor type 1 (PTPN1, also known as PTP1B), protein tyrosine phosphatase non-receptor type 2 (PTPN2, also known as TC-PTP), and phosphatase and tensin homologue deleted on chromosome 10 (PTEN) [48,49,50,51,52]. Furthermore, Nie et al. demonstrated that paeoniflorin inhibited proliferation and induced apoptosis in human glioma cells via ubiquitin–proteasome pathway (UPP)-mediated STAT3 degradation [53].
STAT3 activation and inactivation are highly regulated in normal cells, whereas in tumor cells, downregulation of the endogenous negative regulators of the STAT3 signaling pathway leads to enhanced proliferation and malignancy [54,55].
2.2. STAT3 in Tumor Cells
The activity of STAT3 is required for embryonic development, but can also lead to tumorigenesis and tumor progression, making it a double-edged sword (Figure 2).
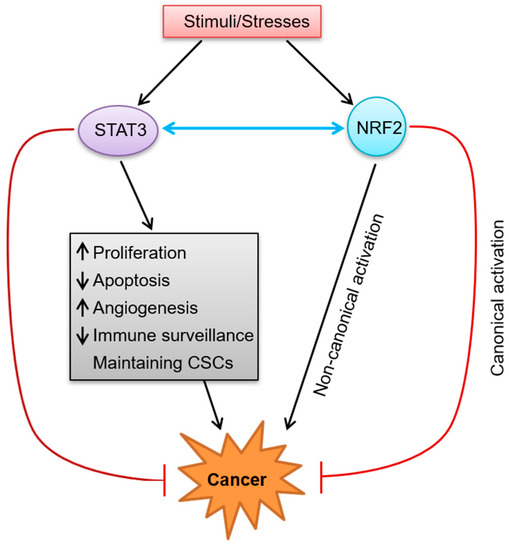
Figure 2.
The dual roles of STAT3 and NRF2 in cancer and their crosstalk. When cells are stimulated by various kinds of stimuli/stresses, STAT3 and NRF2 will be activated. Activated STAT3 promotes cancer development through multiple mechanisms including promoting cell proliferation, suppressing apoptosis, facilitating angiogenesis, escaping immune surveillance, and maintaining CSCs. On the other hand, activated STAT3 has anti-tumor function. Just as STAT3, NRF2 can not only promote carcinogenesis via a non-canonical activation mode, but also inhibit the development of cancer through a canonical mode. In addition, STAT3 and NRF2 can also interact with each other. (The black arrow represents activation, the red arrow represents inhibition, and the blue double-head arrow represents interaction/crosstalk.)
2.2.1. Functions of STAT3 in Cell Proliferation and Survival in Tumors
A growing body of research data indicates that sustained STAT3 activation is necessary for abnormal cell proliferation and survival during tumorigenesis, whereas blocking STAT3 signaling inhibits cell proliferation and promotes apoptosis in multiple cancers. You et al. found that IL-26 could promote proliferation and suppress apoptosis in human gastric cancer cells by increasing the expression of Bcl-2, Bcl-xL, and c-Myc, which are associated with STAT3 activation [56], whereas Kanai et al. found that differentiation-inducing factor-1 (DIF-1) suppressed gastric cancer cell proliferation by inhibiting STAT3 activity in a MEK/ERK-dependent manner [57]. LL1 can also induce apoptosis and inhibit metastasis in colorectal cancer cells by selectively blocking STAT3 activation [58]. In addition to the upregulation of STAT3 activity, the proliferation and survival of tumor cells also involve the downregulation of wild-type p53. Furthermore, STAT3 blockade in cancer cells can upregulate p53, leading to p53-dependent tumor cell apoptosis and UV-induced tumor cell growth arrest [59].
2.2.2. Contribution of STAT3 to Tumor Angiogenesis
Angiogenesis is considered to be a key step for tumor growth and metastasis. STAT3 can participate in angiogenesis by interacting with various growth factors including vascular endothelial growth factor (VEGF), basic fibroblast growth factor (bFGF), and platelet-derived growth factor (PDGF), leading to the degradation of vascular basement membrane, proliferation and migration of vascular epithelial cells, and reconstruction and dissolution of new blood vessels [60,61]. In addition, STAT3 can induce hypoxia-inducible factor-1α (HIF1α), another key regulator of angiogenesis, in the tumor microenvironment [62,63,64]. Notably, several studies have shown that matrix metalloproteinases (MMPs) such as MMP2 and MMP9 contribute to tumor angiogenesis, which can be suppressed by inhibiting STAT3 activity [64,65].
2.2.3. STAT3 in Immune System Evasion
The immune system exerts a pivotal function in cancer prevention by detecting and removing abnormal transformed cells that rely on some immune cells. First of all, Innate immune cells including macrophages, natural killer (NK) cells, and dendritic cells (DCs), as well as adaptive immune cells (T helper cell type 1 (Th1)), can destroy tumor cells through a variety of mechanisms [66]. Nevertheless, abnormal cells can escape immune surveillance and ultimately result in malignant tumors via tumor-associated macrophages (TAMs) and myeloid-derived suppressor cells (MDSCs) through complex mechanisms. These include decreased expression of cancer antigens and major histocompatibility complex (MHC)-I and MHC-II molecules on T cells, and increased angiogenetic, metastatic, and growth factors or immunosuppressive cytokines [4,67,68,69,70].
Increasing evidence suggests that STAT3 is involved in regulating tumor cell immune evasion. Wang et al. indicated that in tumors, activating STAT3 can negatively regulate the inflammatory cytokines’ expression and inhibit DC maturation, resulting in a decrease of MHC-II expression, antigen presentation, and T-cell immunity [71]. Numerous studies show that, within the tumor microenvironment, STAT3 signaling induces pro-carcinogenic cytokines (such as IL-6, IL-10, and IL-23) and inhibits anti-carcinogenic cytokines (e.g., IL-12), thereby promoting tumor immune evasion and cancer progression [72]. For instance, Kortylewski et al. found that STAT3 facilitated IL-23-mediated pro-carcinogenic immune responses and restrained IL-12-dependent antitumor immunity [73]. In addition, STAT3 inhibits the expression of CXC-chemokine ligand 10 (CXCL10), which can significantly enhance NK cell cytotoxicity against tumor cells [74]. STAT3 activation can induce cancer-promoting inflammation mediated by nuclear factor-kappa B (NF-κB) and IL-6/GP130/JAK pathways while suppressing NF-κB- and STAT1-mediated Th1 antitumor immune responses by decreasing the expression of antitumor cytokines such as IL-12 and IFN [73,75,76].
2.2.4. Function of STAT3 in Cancer Stem Cells
Many tumors contain a subpopulation of cells that possess the same properties as stem cells in normal tissue, known as cancer stem cells (CSCs) or cancer stem-like cells [77,78,79,80]. CSCs are capable of self-renewal and can generate a wide variety of tumor cells, thereby facilitating tumor heterogeneity. Moreover, CSCs cause tumor recurrence, metastasis, and drug resistance. Remarkably, STAT3 exerts a vital role in promoting cancer through regulating the activities of CSCs. STAT3 can maintain the population of CSCs and their “stem-like” characteristics through various intricate mechanisms. Firstly, STAT3 plays an essential role in maintaining the expression of CSC marker genes such as CD24, CD34, CD38, CD44, CD90, and CD133, which are essential for the stem cell phenotype. The evidence suggests that the function of STAT3 in CSCs is achieved via crosstalk between activated STAT3 and the marker genes of pluripotent embryonic stem (ES) cells, such as OCT3/4 and NANOG [81,82,83,84]. Secondly, STAT3 is involved in epithelial–mesenchymal transition (EMT)-related pathways; this is one of the chief accepted mechanisms for CSC formation [85,86]. Furthermore, STAT3 participates in the expression and protein stability of HIF-1α, and either regulates or is regulated by VEGF, which plays an essential role in maintaining CSCs’ self-renewal [87,88]. STAT3 can also protect CSCs from the innate immune system, as inhibiting activated STAT3 can reverse the suppression of phagocytosis and the secretion of IL-10 in glioma CSCs (gCSCs) [89]. Moreover, STAT3 feedback activation might perform an important role in mediating drug resistance to a wide range of targeted cancer therapies and chemotherapies [90].
Considering the prominent functions of STAT3 in maintaining the characteristics of CSCs, it is reasonable to speculate that STAT3 inhibition can markedly or permanently eliminate CSCs for achieving cancer prevention. One study found that STAT3 can be selectively inhibited by the chemical compound stattic or by siRNA, which can abolish CSC proliferation [91]. In another study, researchers found that BBI608, a small molecule STAT3 inhibitor known to inhibit cancer recurrence, progression, and metastasis, could suppress the expression of stemness-associated genes, deplete ALDH1-positive CSCs, and overcome cisplatin resistance in non-small cell lung cancer (NSCLC) [92].
2.3. Dual Roles of STAT3 in Cancer
In addition to the tumor-promoting effects described previously, plenty of evidence indicates that STAT3 can be used as a tumor suppressor in various tumors under certain conditions. For instance, in glial cells, STAT3 exerts a tumor-suppressive effect with complete PTEN function, whereas in epidermal growth factor receptor (EGFR)vIII-positive tumors, it plays an oncogenic role, both of which are mediated by different signaling pathways [93,94]. In a normal cell with intact PTEN function, the protein kinase B (Akt, also known as PKB) is inhibited, allowing forkhead box O3 (FOXO3) to activate transcription of the leukemia inhibitory factor receptor β (LIFRβ) gene. STAT3 is then activated by phosphorylation and represses transcription of the IL8 gene, which ultimately suppresses glioma cell proliferation and invasiveness [93,94]. However, in human glioblastoma cells, PTEN loss leads to the downregulation of LIFRβ expression via Akt inhibition of FOXO3, STAT3 is no longer active, and its inhibition of IL8 gene is removed, leading to upregulation of IL8, which drives malignant glial transformation [93,94]. In colorectal cancer, the tumor inhibitory effect of STAT3 is achieved by suppressing the expression of Snail-1 by promoting GSK3β activity [95]. The action of STAT3 in lung cancer is also ambivalent. In lung adenocarcinomas, STAT3 can be activated by mutant EGFR via driving the expression of the IL-6 cytokine, which activates the gp130/JAK signaling pathway, while blocking this pathway will repress cell-cycle progression, cell growth, and tumorigenesis [96]. However, in Kirsten rat sarcoma and viral oncogene (KRAS)-mutant lung adenocarcinoma, STAT3 exerts an unexpected tumor-suppressive effect by sequestering NF-κB in the cytoplasm, thus decreasing IL-8 expression induced by NF-κB [97]. Specifically, genetic ablation of Stat3 in murine as well as STAT3 in human cells leads to an increase of NF-κB-induced expression of CXCL1/IL-8, which contributes to infiltration of myeloid cells as well as vascularization; while inhibiting CXCL1’s cognate receptor, CXCR2 can normalize tumor vascularization and microenvironment and reduce tumor burden [97]. Considering the aforementioned roles of STAT3, we must consider the dual effects of the STAT3 signaling pathway when using it as a drug target. The therapeutic purpose can be achieved by seeking advantages and avoiding disadvantages.
3. Intricacies of NRF2 Regulation in the Tumor Microenvironment
3.1. NRF2 Signaling Pathway
Under normal conditions, NRF2 is negatively regulated by three E3 ubiquitin ligase complexes: the KEAP1-cullin 3 (CUL3)-ring box 1 (RBX1) complex, the β-TrCP-S-phase kinase-associated protein 1 (SKP1)-CUL1-RBX1 complex, and the Hmg-CoA reductase degradation protein 1 (HRD1) [29,98,99,100]. However, when the organism is exposed to endogenous and environmental stresses, NRF2 degradation is interrupted by the inhibition of the UPS, and newly translated NRF2 translocates to the nucleus, binds to sMAF proteins, and transcribes ARE-driven genes. The extensive cytoprotective genes regulated by NRF2 are crucial for suppressing the oxidative, proteotoxic, and metabolic stresses which facilitate malignant transformation. The transient activation of NRF2 during stress is beneficial to health, while a sustained activation of NRF2 has detrimental effects.
3.1.1. Canonical Activation of NRF2
A large body of evidence shows that KEAP1 has become a crucial regulator of the NRF2-mediated signaling pathway. It is generally believed that KEAP1 can function as a molecular switch to sense the imbalance in redox homeostasis and turn on or off the NRF2 signaling pathway [101,102]. Under general conditions, the activity of NRF2 is negatively regulated by KEAP1. During stress, NRF2 upregulation can be induced by oxidative or electrophilic modification of KEAP1 cysteines (i.e., Cys151), which suppresses the formation of the NRF2–KEAP1 complex and results in diminished NRF2 ubiquitination, thereby initiating the canonical NRF2 signaling pathway [103,104,105,106]. Thus, newly translated NRF2 translocates to the nucleus, binds to sMAF proteins, and transcribes ARE-regulated cytoprotective target genes to maintain redox homeostasis [20]. When redox homeostasis is restored, KEAP1 travels into the nucleus to dissociate NRF2 from the ARE and returns NRF2 to the cytosol for ubiquitination and degradation to inhibit the sustained activation of NRF2 [107]. This pattern of NRF2-activation regulation is an immediate consequence of oxidative or electrophilic stresses and is referred to as “canonical activation”. In terms of chemoprevention, NRF2 has been shown to be activated via the canonical mode by various dietary compounds or synthetic chemicals [108]. The treatment strategies for many diseases, including cancer, are based on utilizing the protective capacity of the NRF2 response, which is achieved by transient NRF2 activation via oxidative or electrophilic modification of KEAP1 [109,110,111,112]. Therefore, the canonical activation of NRF2 is crucial to switch on the detoxification of harmful carcinogens and relieve excessive stress to avoid malignant transformation (Figure 2).
3.1.2. Non-Canonical Activation of NRF2
Another important pathway during stress is the autophagy-lysosome pathway, a highly regulated cellular degradation pathway which is responsible for removing damaged, degenerative, and aging proteins and organelles, such as oxidatively damaged proteins and dysfunctional mitochondria. Autophagy dysfunction leads to the accumulation of pathogenic proteins and organelles, which is the root cause of many diseases, including metabolic disorders, neurodegenerative diseases, infectious diseases, cardiovascular diseases, and cancer [113,114]. To some extent, autophagy pathway dysfunction is associated with NRF2 activation. For instance, several studies have found that NRF2 can be activated through autophagy inhibition in a p62-dependent but Keap1-Cys151-independent manner, which is known as “non-canonical activation” [115,116,117,118]. Autophagy dysfunction has been shown to induce the accumulation of p62, a selective autophagy adaptor, which leads to the sequestration and loss of function of numerous binding partners, including KEAP1 [115,119,120]. p62 interacts directly with KEAP1 through its KEAP1-interacting region (KIR), which contains a DPSTGE motif similar to the ETGE motif in NRF2 for KEAP1 binding [117,121]. The KEAP1 sequestration by p62 stabilizes NRF2, which can initiate the transcription of target genes, including p62, creating a positive feedback loop and prolonging NRF2 activation [119]. However, the excessive accumulation of p62 induces sustained NRF2 activation, which facilitates the formation and development of tumors [122]. Deletion of p62 consistently inhibits NRF2 activation and arsenic-induced malignant transformation of human keratinocytes [123]. The relationship between the non-canonical activation of NRF2 and carcinogenesis must be thoroughly investigated to identify a potential target for cancer treatment or prevention.
3.2. Dual Roles of NRF2 in Tumor
NRF2 has traditionally been considered a tumor suppressor since the NRF2–KEAP1 signaling pathway is an essential cell protection mechanism that can defend against oxidative/electrophilic stresses and promote cell survival. The activation of the NRF2 pathway induced by natural compounds is an effective chemoprevention strategy [124]. Moreover, NRF2-deficient mice are more susceptible to develop cancer, and NRF2 deficiency is associated with cancer metastasis [125,126,127,128].
Transient activation of NRF2 during stress is beneficial to normal cells, whereas hyperactivation of NRF2 facilitates the survival of normal as well as malignant cells. Recent evidence suggests that the “dark” side of NRF2 may be mediated by excessive accumulation of p21 and p62 via disruption of NRF2–KEAP1 interactions [115,129]. In addition, NRF2 can exert a significant action of chemoresistance, inhibiting drug accumulation in cancer cells, and thereby contributing to survival of cancer cells. Considering the pro-tumorigenic effect of NRF2 in cancer cells, pharmacological suppression of the NRF2 pathway will emerge as a promising area of cancer research. Several groups have identified many NRF2 pharmacological inhibitors, such as brusatol, halofuginone, luteolin, and procyanidin [130,131,132,133]. However, there is currently no FDA-approved drug to suppress NRF2 activation. Therefore, extensive research is required to identify drugs that can prevent and treat cancer.
4. Crosstalk between the STAT3 and NRF2 Signaling Pathways in the Tumor Microenvironment
Interestingly, the STAT3 and NRF2 signaling pathways can interact with each other (Figure 2), which undoubtedly increases the complexity of their signal transduction and the diversity of drug treatment targets. There is increasing evidence that STAT3 and NRF2 have synergistic effects in cancer cells [134,135]. Wu et al. found that IL-6 secreted by pancreatic stellate cell (PSC)-induced EMT phenotypes and gene expression in Panc-1 cells by activating STAT3, which in turn induced the expression of NRF2 and its target genes to mediate EMT [36]. EMT is a process where epithelial cells lose their cell–cell adhesion and apical-basolateral polarity and obtain mesenchymal features [136,137]. Numerous studies indicate that several metastatic cancers are caused by IL-6-induced EMT events [136,137,138]. Results from Wu et al. show that PSC-secreted IL-6 binds to its receptor and activates JAK/STAT3 signaling, which then triggers intracellular NRF2 signaling and its downstream EMT-related transcription factors to drive the expression of EMT-related marker genes, thereby inducing EMT in Panc-1 cells [36]. This study showed that the IL-6/STAT3/NRF2 signaling pathway might play a role in the progression of pancreatic ductal adenocarcinoma (PDAC) [36]. Another study found that the expression levels of both STAT3 and NRF2 were increased in HT-29 colon cancer cells, but when treated with the combination of 5-fluorouracil (5-FU) and stattic, the level of NRF2 decreased after the reduction of STAT3 expression [35]. It may be assumed that the effect of 5-FU may inhibit STAT3 and NRF2 signal transduction by blocking IL-6. The specific mechanism still needs a great quantity of research.
Moreover, in osteosarcoma cells, overactivation of STAT3/NRF2 signaling can lead to cisplatin resistance by increasing glutathione peroxidase 4 (GPX4) activity, thereby suppressing ferroptosis [135]. However, when BP-1-102, a STAT3 inhibitor, was used, the expression levels of NRF2 and GPX4 were strikingly decreased, which reactivated ferroptosis and enhanced the sensitivity of osteosarcoma cells to cisplatin [135].
Furthermore, both Nrf2 and STAT3 are overexpressed in breast cancer, especially in basal-like breast cancer (BLBC). Kim et al. found that NRF2 can form a stable complex with Y705 phosphorylated dimeric form of STAT3, which may accelerate the progression of breast cancer by inducing IL-23A expression [139]. IL-23A is significantly overexpressed in almost half of BLBC patients. It is worth noting that the survival rate of breast cancer patients with high levels of IL-23A mRNA is worse than that of patients with no or low expression of IL-23A mRNA [139]. IL-23 is a common proinflammatory cytokine which mainly exists in activated macrophages, dendritic cells, and keratinocytes in healthy skin [140]. However, recent studies have shown that IL-23 is also involved in tumor growth and metastasis by directly binding to the IL-23 receptor, which is expressed in a variety of in inflammation-related malignant tumors, including breast cancer [141]. The STAT3–NRF2 complex located in the nucleus where it binds to the promoter region of the IL-23A gene and induces its transcription. The protein products of IL-23A can bind to their receptors in BLBC cells in an autocrine manner, which will amplify the intracellular signals for breast cancer cell proliferation, migration, metastasis, etc. [139]. In view of this, the STAT3/NRF2-IL-23A axis can emphasize the importance of subtype-specific molecular pathways, which can be a potential therapeutic target.
5. Conclusions
STAT3, as an essential transcription factor, regulates the expression of a great quantity of genes and participates in many physiological processes, including cell growth, apoptosis, differentiation, inflammation, immunity, and angiogenesis. In normal cells, activation and inactivation of STAT3 are highly regulated, whereas, in tumor cells, STAT3 is typically overactive. Persistent STAT3 signaling can directly promote tumorigenesis by facilitating cell proliferation and inhibiting apoptosis through the upregulation of genes encoding apoptosis inhibitors (Bcl-2, Bcl-xl, Mcl-1) and cell cycle regulators (cyclins D1/D2, c-Myc) [56,142]. Moreover, in the tumor microenvironment, the endogenous negative regulators of the STAT3 signaling pathway are downregulated, resulting in enhanced proliferation and malignancy of tumor cells [54,55]. Therefore, STAT3 signaling is a viable target for cancer therapy, and the use of its inhibitors may impede the progression of cancer. However, under certain circumstances, STAT3 can act as a tumor suppressor in various tumors. This means that treatments based on STAT3 modulators should take into account the dual roles of this transcription factor and be tailored to specific tumor types.
Similar to STAT3, NRF2 is a prominent transcription factor with dual roles. Stress-induced transient activation of NRF2 is beneficial to health, whereas sustained NRF2 activation has detrimental effects. NRF2 has been considered a tumor suppressor because the NRF2–KEAP1 signaling pathway is a major cytoprotective mechanism that can defend against oxidative/electrophilic stresses and promote cell survival. However, excessive activation of NRF2 facilitates the survival of both normal and malignant cells. Recent evidence has revealed the “dark” side of NRF2, which may be mediated by the activation of a non-canonical pathway [115,116,117,118].
In addition, there is increasing evidence that the STAT3 and NRF2 signaling pathways can interact, thereby enhancing the complexity of their signal transduction. Recent research indicates that the STAT3/NRF2 signaling pathway contributes to cancer progression [35,36,134,135]. Gao et al. found that remote limb ischemic postconditioning (RIPostC) attenuated apoptosis and protected mice from myocardial ischemia/reperfusion (IR) injury, possibly by activating the JAK/STAT3-mediated NRF2-antioxidant signaling pathway [143]. This raises the question of whether the STAT3/NRF2 signaling pathway has an antioxidant effect on tumor cells by protecting them from oxidative stress damage and promoting their survival. Another study indicates that NRF2 promotes carcinogenesis in nickel-transformed cells by suppressing apoptosis and promoting autophagy via STAT3 signaling [144]. The specific regulatory mechanisms underlying STAT3 and NRF2 signaling need to be studied further under different pathological conditions.
To conclude, the signal transduction and functions of STAT3 and NRF2 are extremely intricate under physiological conditions as well as pathological conditions, particularly in tumors. Because crosstalk between STAT3 and NRF2 signaling can occur in various tumors, the specific mechanisms and functions must be determined to better guide clinical medication and new drug development.
Author Contributions
Y.T., H.L., M.W., R.W., G.Y. and M.Z. wrote the manuscript. R.C. formulated and revised the manuscript. Y.T., H.L. and R.C. obtained funding, reviewed, and edited the draft. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by National Nature Science Foundation of China (21807041, 81901341), Lin He’s Academician Workstation of New Medicine and Clinical Translation in Jining Medical University (JYHL2022ZD04), the Key Research and Development Program of Shandong Province (2019GSF107047), Shandong Medical and Health Science and Technology Development Project (202006020902), the Key Research and Development Project of Jining (2020YXNS004), Academic Promotion Program of Shandong First Medical University (2019QL013), and Innovation training program for University Students (cx2022129z, cx2022007z, cx2022198).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Lim, C.P.; Cao, X. Structure, function, and regulation of STAT proteins. Mol. Biosyst. 2006, 2, 536–550. [Google Scholar] [CrossRef] [PubMed]
- Darnell, J.E.; Kerr, I.M.; Stark, G.R. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science 1994, 264, 1415–1421. [Google Scholar] [CrossRef] [PubMed]
- Forbes, L.R.; Milner, J.; Haddad, E. Signal transducer and activator of transcription 3: A year in review. Curr. Opin. Hematol. 2016, 23, 23–27. [Google Scholar] [CrossRef] [PubMed]
- Xiong, A.; Yang, Z.; Shen, Y.; Zhou, J.; Shen, Q. Transcription Factor STAT3 as a Novel Molecular Target for Cancer Prevention. Cancers 2014, 6, 926–957. [Google Scholar] [CrossRef]
- Takeda, K.; Noguchi, K.; Shi, W.; Tanaka, T.; Matsumoto, M.; Yoshida, N.; Kishimoto, T.; Akira, S. Targeted disruption of the mouse Stat3 gene leads to early embryonic lethality. Proc. Natl. Acad. Sci. USA 1997, 94, 3801–3804. [Google Scholar] [CrossRef]
- Mohan, C.D.; Bharathkumar, H.; Bulusu, K.C.; Pandey, V.; Rangappa, S.; Fuchs, J.E.; Shanmugam, M.K.; Dai, X.; Li, F.; Deivasigamani, A.; et al. Development of a novel azaspirane that targets the Janus kinase-signal transducer and activator of transcription (STAT) pathway in hepatocellular carcinoma in vitro and in vivo. J. Biol. Chem. 2014, 289, 34296–34307. [Google Scholar] [CrossRef]
- Sonnenblick, A.; Agbor-Tarh, D.; de Azambuja, E.; Hultsch, S.; Izquierdo, M.; Liu, M.; Pruneri, G.; Harbeck, N.; Piccart, M.; Moreno-Aspita, A.; et al. STAT3 activation in HER2-positive breast cancers: Analysis of data from a large prospective trial. Int. J. Cancer 2021, 148, 1529–1535. [Google Scholar] [CrossRef]
- Yuan, K.; Ye, J.; Liu, Z.; Ren, Y.; He, W.; Xu, J.; He, Y.; Yuan, Y. Complement C3 overexpression activates JAK2/STAT3 pathway and correlates with gastric cancer progression. J. Exp. Clin. Cancer Res. 2020, 39, 9. [Google Scholar] [CrossRef]
- Liang, Z.; Wu, G.; Fan, C.; Xu, J.; Jiang, S.; Yan, X.; Di, S.; Ma, Z.; Hu, W.; Yang, Y. The emerging role of signal transducer and activator of transcription 3 in cerebral ischemic and hemorrhagic stroke. Prog. Neurobiol. 2016, 137, 1–16. [Google Scholar] [CrossRef]
- Xia, Y.; Ling, X.; Hu, G.; Zhu, Q.; Zhang, J.; Li, Q.; Zhao, B.; Wang, Y.; Deng, Z. Small extracellular vesicles secreted by human iPSC-derived MSC enhance angiogenesis through inhibiting STAT3-dependent autophagy in ischemic stroke. Stem. Cell Res. Ther. 2020, 11, 313. [Google Scholar] [CrossRef]
- Cao, X.; Li, B.; Han, X.; Zhang, X.; Dang, M.; Wang, H.; Du, F.; Zeng, X.; Guo, C. Soluble receptor for advanced glycation end-products promotes angiogenesis through activation of STAT3 in myocardial ischemia/reperfusion injury. Apoptosis 2020, 25, 341–353. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Zhang, W.; Lee, L.; Hong, M.; Lee, M.; Chou, G.; Yu, L.; Sui, Y.; Chou, B. Down-regulated microRNA-195-5p and up-regulated CXCR4 attenuates the heart function injury of heart failure mice via inactivating JAK/STAT pathway. Int. Immunopharmacol. 2020, 82, 106225. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Lv, J.; Han, M.; Yang, Z.; Li, T.; Jiang, S.; Yang, Y. STAT3: The art of multi-tasking of metabolic and immune functions in obesity. Prog. Lipid. Res. 2018, 70, 17–28. [Google Scholar] [CrossRef] [PubMed]
- Akira, S.; Nishio, Y.; Inoue, M.; Wang, X.J.; Wei, S.; Matsusaka, T.; Yoshida, K.; Sudo, T.; Naruto, M.; Kishimoto, T. Molecular cloning of APRF, a novel IFN-stimulated gene factor 3 p91-related transcription factor involved in the gp130-mediated signaling pathway. Cell 1994, 77, 63–71. [Google Scholar] [CrossRef]
- Choi, J.Y.; Li, W.L.; Kouri, R.E.; Yu, J.; Kao, F.T.; Ruano, G. Assignment of the acute phase response factor (APRF) gene to 17q21 by microdissection clone sequencing and fluorescence in situ hybridization of a P1 clone. Genomics 1996, 37, 264–265. [Google Scholar] [CrossRef]
- Hu, T.; Yeh, J.E.; Pinello, L.; Jacob, J.; Chakravarthy, S.; Yuan, G.C.; Chopra, R.; Frank, D.A. Impact of the N-Terminal Domain of STAT3 in STAT3-Dependent Transcriptional Activity. Mol. Cell. Biol. 2015, 35, 3284–3300. [Google Scholar] [CrossRef]
- Chai, E.Z.; Shanmugam, M.K.; Arfuso, F.; Dharmarajan, A.; Wang, C.; Kumar, A.P.; Samy, R.P.; Lim, L.H.; Wang, L.; Goh, B.C.; et al. Targeting transcription factor STAT3 for cancer prevention and therapy. Pharmacol. Ther. 2016, 162, 86–97. [Google Scholar] [CrossRef]
- Zouein, F.A.; Altara, R.; Chen, Q.; Lesnefsky, E.J.; Kurdi, M.; Booz, G.W. Pivotal Importance of STAT3 in Protecting the Heart from Acute and Chronic Stress: New Advancement and Unresolved Issues. Front. Cardiovasc. Med. 2015, 2, 36. [Google Scholar] [CrossRef]
- Timofeeva, O.A.; Tarasova, N.I.; Zhang, X.; Chasovskikh, S.; Cheema, A.K.; Wang, H.; Brown, M.L.; Dritschilo, A. STAT3 suppresses transcription of proapoptotic genes in cancer cells with the involvement of its N-terminal domain. Proc. Natl. Acad. Sci. USA 2013, 110, 1267–1272. [Google Scholar] [CrossRef]
- Yamamoto, M.; Kensler, T.W.; Motohashi, H. The KEAP1-NRF2 System: A Thiol-Based Sensor-Effector Apparatus for Maintaining Redox Homeostasis. Physiol. Rev. 2018, 98, 1169–1203. [Google Scholar] [CrossRef]
- Itoh, K.; Wakabayashi, N.; Katoh, Y.; Ishii, T.; Igarashi, K.; Engel, J.D.; Yamamoto, M. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 1999, 13, 76–86. [Google Scholar] [CrossRef] [PubMed]
- Katoh, Y.; Itoh, K.; Yoshida, E.; Miyagishi, M.; Fukamizu, A.; Yamamoto, M. Two domains of Nrf2 cooperatively bind CBP, a CREB binding protein, and synergistically activate transcription. Genes Cells 2001, 6, 857–868. [Google Scholar] [CrossRef] [PubMed]
- Nioi, P.; Nguyen, T.; Sherratt, P.J.; Pickett, C.B. The carboxy-terminal Neh3 domain of Nrf2 is required for transcriptional activation. Mol. Cell. Biol. 2005, 25, 10895–10906. [Google Scholar] [CrossRef] [PubMed]
- McMahon, M.; Thomas, N.; Itoh, K.; Yamamoto, M.; Hayes, J.D. Redox-regulated turnover of Nrf2 is determined by at least two separate protein domains, the redox-sensitive Neh2 degron and the redox-insensitive Neh6 degron. J. Biol. Chem. 2004, 279, 31556–31567. [Google Scholar] [CrossRef] [PubMed]
- Rada, P.; Rojo, A.I.; Evrard-Todeschi, N.; Innamorato, N.G.; Cotte, A.; Jaworski, T.; Tobón-Velasco, J.C.; Devijver, H.; García-Mayoral, M.F.; Van Leuven, F.; et al. Structural and functional characterization of Nrf2 degradation by the glycogen synthase kinase 3/beta-TrCP axis. Mol. Cell. Biol. 2012, 32, 3486–3499. [Google Scholar] [CrossRef]
- Wang, H.; Liu, K.; Geng, M.; Gao, P.; Wu, X.; Hai, Y.; Li, Y.; Li, Y.; Luo, L.; Hayes, J.D.; et al. RXRalpha inhibits the NRF2-ARE signaling pathway through a direct interaction with the Neh7 domain of NRF2. Cancer Res. 2013, 73, 3097–3108. [Google Scholar] [CrossRef]
- Itoh, K.; Igarashi, K.; Hayashi, N.; Nishizawa, M.; Yamamoto, M. Cloning and characterization of a novel erythroid cell-derived CNC family transcription factor heterodimerizing with the small Maf family proteins. Mol. Cell. Biol. 1995, 15, 4184–4193. [Google Scholar] [CrossRef]
- Itoh, K.; Tong, K.I.; Yamamoto, M. Molecular mechanism activating Nrf2-Keap1 pathway in regulation of adaptive response to electrophiles. Free Radic. Biol. Med. 2004, 36, 1208–1213. [Google Scholar] [CrossRef]
- Dodson, M.; de la Vega, M.R.; Cholanians, A.B.; Schmidlin, C.J.; Chapman, E.; Zhang, D.D. Modulating NRF2 in Disease: Timing Is Everything. Annu. Rev. Pharmacol. Toxicol. 2019, 59, 555–575. [Google Scholar] [CrossRef]
- Reisman, S.A.; Csanaky, I.L.; Aleksunes, L.M.; Klaassen, C.D. Altered disposition of acetaminophen in Nrf2-null and Keap1-knockdown mice. Toxicol. Sci. 2009, 109, 31–40. [Google Scholar] [CrossRef]
- Iizuka, T.; Ishii, Y.; Itoh, K.; Kiwamoto, T.; Kimura, T.; Matsuno, Y.; Morishima, Y.; Hegab, A.E.; Homma, S.; Nomura, A.; et al. Nrf2-deficient mice are highly susceptible to cigarette smoke-induced emphysema. Genes Cells 2005, 10, 1113–1125. [Google Scholar] [CrossRef] [PubMed]
- Ramos-Gomez, M.; Kwak, M.K.; Dolan, P.M.; Itoh, K.; Yamamoto, M.; Talalay, P.; Kensler, T.W. Sensitivity to carcinogenesis is increased and chemoprotective efficacy of enzyme inducers is lost in nrf2 transcription factor-deficient mice. Proc. Natl. Acad. Sci. USA 2001, 98, 3410–3415. [Google Scholar] [CrossRef] [PubMed]
- Itoh, K.; Chiba, T.; Takahashi, S.; Ishii, T.; Igarashi, K.; Katoh, Y.; Oyake, T.; Hayashi, N.; Satoh, K.; Hatayama, I.; et al. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem. Biophys. Res. Commun. 1997, 236, 313–322. [Google Scholar] [CrossRef] [PubMed]
- Sivinski, J.; Zhang, D.D.; Chapman, E. Targeting NRF2 to treat cancer. Semin. Cancer Biol. 2021, 76, 61–73. [Google Scholar] [CrossRef] [PubMed]
- Tajmohammadi, I.; Mohammadian, J.; Sabzichi, M.; Mahmuodi, S.; Ramezani, M.; Aghajani, M.; Ramezani, F. Identification of Nrf2/STAT3 axis in induction of apoptosis through sub-G 1 cell cycle arrest mechanism in HT-29 colon cancer cells. J. Cell. Biochem. 2019, 120, 14035–14043. [Google Scholar] [CrossRef]
- Wu, Y.S.; Chung, I.; Wong, W.F.; Masamune, A.; Sim, M.S.; Looi, C.Y. Paracrine IL-6 signaling mediates the effects of pancreatic stellate cells on epithelial-mesenchymal transition via Stat3/Nrf2 pathway in pancreatic cancer cells. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 296–306. [Google Scholar] [CrossRef]
- Dai, X.; Ahn, K.S.; Kim, C.; Siveen, K.S.; Ong, T.H.; Shanmugam, M.K.; Li, F.; Shi, J.; Kumar, A.P.; Wang, L.Z.; et al. Ascochlorin, an isoprenoid antibiotic inhibits growth and invasion of hepatocellular carcinoma by targeting STAT3 signaling cascade through the induction of PIAS3. Mol. Oncol. 2015, 9, 818–833. [Google Scholar] [CrossRef]
- Lee, J.H.; Kim, C.; Sethi, G.; Ahn, K.S. Brassinin inhibits STAT3 signaling pathway through modulation of PIAS-3 and SOCS-3 expression and sensitizes human lung cancer xenograft in nude mice to paclitaxel. Oncotarget 2015, 6, 6386–6405. [Google Scholar] [CrossRef]
- Mali, S.B. Review of STAT3 (Signal Transducers and Activators of Transcription) in head and neck cancer. Oral Oncol. 2015, 51, 565–569. [Google Scholar] [CrossRef]
- Siveen, K.S.; Sikka, S.; Surana, R.; Dai, X.; Zhang, J.; Kumar, A.P.; Tan, B.K.; Sethi, G.; Bishayee, A. Targeting the STAT3 signaling pathway in cancer: Role of synthetic and natural inhibitors. Biochim. Biophys. Acta 2014, 1845, 136–154. [Google Scholar] [CrossRef]
- Sgrignani, J.; Garofalo, M.; Matkovic, M.; Merulla, J.; Catapano, C.V.; Cavalli, A. Structural Biology of STAT3 and Its Implications for Anticancer Therapies Development. Int. J. Mol. Sci. 2018, 19, 1591. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Liao, X.; Agarwal, M.K.; Barnes, L.; Auron, P.E.; Stark, G.R. Unphosphorylated STAT3 accumulates in response to IL-6 and activates transcription by binding to NFkappaB. Genes Dev. 2007, 21, 1396–1408. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.E.; O’Keefe, R.A.; Grandis, J.R. Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat. Rev. Clin. Oncol. 2018, 15, 234–248. [Google Scholar] [CrossRef] [PubMed]
- Chung, C.D.; Liao, J.; Liu, B.; Rao, X.; Jay, P.; Berta, P.; Shuai, K. Specific inhibition of Stat3 signal transduction by PIAS3. Science 1997, 278, 1803–1805. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Yang, B.; Yao, Y.Y.; Zhong, L.X.; Chen, X.Y.; Kong, Q.Y.; Wu, M.L.; Li, C.; Li, H.; Liu, J. PIAS3, SHP2 and SOCS3 Expression patterns in Cervical Cancers: Relevance with activation and resveratrol-caused inactivation of STAT3 signaling. Gynecol. Oncol. 2015, 139, 529–535. [Google Scholar] [CrossRef] [PubMed]
- Shuai, K.; Liu, B. Regulation of gene-activation pathways by PIAS proteins in the immune system. Nat. Rev. Immunol. 2005, 5, 593–605. [Google Scholar] [CrossRef] [PubMed]
- Baek, S.H.; Ko, J.H.; Lee, H.; Jung, J.; Kong, M.; Lee, J.W.; Lee, J.; Chinnathambi, A.; Zayed, M.E.; Alharbi, S.A.; et al. Resveratrol inhibits STAT3 signaling pathway through the induction of SOCS-1: Role in apoptosis induction and radiosensitization in head and neck tumor cells. Phytomedicine 2016, 23, 566–577. [Google Scholar] [CrossRef]
- Baek, S.H.; Lee, J.H.; Ko, J.H.; Lee, H.; Nam, D.; Lee, S.G.; Yang, W.M.; Um, J.Y.; Lee, J.; Kim, S.H.; et al. Ginkgetin Blocks Constitutive STAT3 Activation and Induces Apoptosis through Induction of SHP-1 and PTEN Tyrosine Phosphatases. Phytother. Res. 2016, 30, 567–576. [Google Scholar] [CrossRef]
- Xu, D.; Qu, C.K. Protein tyrosine phosphatases in the JAK/STAT pathway. Front. Biosci. 2008, 13, 4925–4932. [Google Scholar] [CrossRef]
- Sun, S.; Steinberg, B.M. PTEN is a negative regulator of STAT3 activation in human papillomavirus-infected cells. J. Gen. Virol. 2002, 83, 1651–1658. [Google Scholar] [CrossRef]
- Tartaglia, M.; Niemeyer, C.M.; Fragale, A.; Song, X.; Buechner, J.; Jung, A.; Hählen, K.; Hasle, H.; Licht, J.D.; Gelb, B.D. Somatic mutations in PTPN11 in juvenile myelomonocytic leukemia; myelodysplastic syndromes and acute myeloid leukemia. Nat. Genet. 2003, 34, 148–150. [Google Scholar] [CrossRef] [PubMed]
- Buchert, M.; Burns, C.J.; Ernst, M. Targeting JAK kinase in solid tumors: Emerging opportunities and challenges. Oncogene 2016, 35, 939–951. [Google Scholar] [CrossRef] [PubMed]
- Nie, X.H.; Ou-yang, J.; Xing, Y.; Li, D.Y.; Dong, X.Y.; Liu, R.E.; Xu, R.X. Paeoniflorin inhibits human glioma cells via STAT3 degradation by the ubiquitin-proteasome pathway. Drug Des. Dev. Ther. 2015, 9, 5611–5622. [Google Scholar] [PubMed]
- Hu, F.; Li, G.; Huang, C.; Hou, Z.; Yang, X.; Luo, X.; Feng, Y.; Wang, G.; Hu, J.; Cao, Z. The autophagy-independent role of BECN1 in colorectal cancer metastasis through regulating STAT3 signaling pathway activation. Cell Death Dis. 2020, 11, 304. [Google Scholar] [CrossRef] [PubMed]
- Kadye, R.; Stoffels, M.; Fanucci, S.; Mbanxa, S.; Prinsloo, E. A STAT3 of Addiction: Adipose Tissue, Adipocytokine Signalling and STAT3 as Mediators of Metabolic Remodelling in the Tumour Microenvironment. Cells 2020, 9, 1043. [Google Scholar] [CrossRef] [PubMed]
- You, W.; Tang, Q.; Zhang, C.; Wu, J.; Gu, C.; Wu, Z.; Li, X. IL-26 promotes the proliferation and survival of human gastric cancer cells by regulating the balance of STAT1 and STAT3 activation. PLoS ONE 2013, 8, e63588. [Google Scholar] [CrossRef]
- Kanai, M.; Konda, Y.; Nakajima, T.; Izumi, Y.; Kanda, N.; Nanakin, A.; Kubohara, Y.; Chiba, T. Differentiation-inducing factor-1 (DIF-1) inhibits STAT3 activity involved in gastric cancer cell proliferation via MEK-ERK-dependent pathway. Oncogene 2003, 22, 548–554. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Wang, H.; Guan, L.; Lai, C.; Yu, W.; Lai, M. LL1, a novel and highly selective STAT3 inhibitor, displays anti-colorectal cancer activities in vitro and in vivo. Br. J. Pharmacol. 2020, 177, 298–313. [Google Scholar] [CrossRef]
- Niu, G.; Wright, K.L.; Ma, Y.; Wright, G.M.; Huang, M.; Irby, R.; Briggs, J.; Karras, J.; Cress, W.D.; Pardoll, D.; et al. Role of Stat3 in regulating p53 expression and function. Mol. Cell. Biol. 2005, 25, 7432–7440. [Google Scholar] [CrossRef]
- Grunstein, J.; Roberts, W.G.; Mathieu-Costello, O.; Hanahan, D.; Johnson, R.S. Tumor-derived expression of vascular endothelial growth factor is a critical factor in tumor expansion and vascular function. Cancer Res. 1999, 59, 1592–1598. [Google Scholar]
- Kalluri, R. Basement membranes: Structure, assembly and role in tumour angiogenesis. Nat. Rev. Cancer 2003, 3, 422–433. [Google Scholar] [CrossRef] [PubMed]
- Semenza, G.L. Targeting HIF-1 for cancer therapy. Nat. Rev. Cancer 2003, 3, 721–732. [Google Scholar] [CrossRef] [PubMed]
- Oh, M.K.; Park, H.J.; Kim, N.H.; Park, S.J.; Park, I.Y.; Kim, I.S. Hypoxia-inducible factor-1alpha enhances haptoglobin gene expression by improving binding of STAT3 to the promoter. J. Biol. Chem. 2011, 286, 8857–8865. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.H.; Yu, M.O.; Park, K.J.; Chi, S.G.; Park, D.H.; Chung, Y.G. Activated STAT3 regulates hypoxia-induced angiogenesis and cell migration in human glioblastoma. Neurosurgery 2010, 67, 1386–1395, discussion 1395. [Google Scholar] [CrossRef]
- Bergers, G.; Brekken, R.; McMahon, G.; Vu, T.H.; Itoh, T.; Tamaki, K.; Tanzawa, K.; Thorpe, P.; Itohara, S.; Werb, Z.; et al. Matrix metalloproteinase-9 triggers the angiogenic switch during carcinogenesis. Nat. Cell Biol. 2000, 2, 737–744. [Google Scholar] [CrossRef]
- Vesely, M.D.; Kershaw, M.H.; Schreiber, R.D.; Smyth, M.J. Natural innate and adaptive immunity to cancer. Annu. Rev. Immunol. 2011, 29, 235–271. [Google Scholar] [CrossRef]
- Cheng, P.; Corzo, C.A.; Luetteke, N.; Yu, B.; Nagaraj, S.; Bui, M.M.; Ortiz, M.; Nacken, W.; Sorg, C.; Vogl, T.; et al. Inhibition of dendritic cell differentiation and accumulation of myeloid-derived suppressor cells in cancer is regulated by S100A9 protein. J. Exp. Med. 2008, 205, 2235–2249. [Google Scholar] [CrossRef]
- Gabrilovich, D.I.; Nagaraj, S. Myeloid-derived suppressor cells as regulators of the immune system. Nat. Rev. Immunol. 2009, 9, 162–174. [Google Scholar] [CrossRef]
- Pollard, J.W. Tumour-educated macrophages promote tumour progression and metastasis. Nat. Rev. Cancer 2004, 4, 71–78. [Google Scholar] [CrossRef]
- Matsukawa, A.; Kudo, S.; Maeda, T.; Numata, K.; Watanabe, H.; Takeda, K.; Akira, S.; Ito, T. Stat3 in resident macrophages as a repressor protein of inflammatory response. J. Immunol. 2005, 175, 3354–3359. [Google Scholar] [CrossRef]
- Wang, T.; Niu, G.; Kortylewski, M.; Burdelya, L.; Shain, K.; Zhang, S.; Bhattacharya, R.; Gabrilovich, D.; Heller, R.; Coppola, D.; et al. Regulation of the innate and adaptive immune responses by Stat-3 signaling in tumor cells. Nat. Med. 2004, 10, 48–54. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Kortylewski, M.; Pardoll, D. Crosstalk between cancer and immune cells: Role of STAT3 in the tumour microenvironment. Nat. Rev. Immunol. 2007, 7, 41–51. [Google Scholar] [CrossRef] [PubMed]
- Kortylewski, M.; Xin, H.; Kujawski, M.; Lee, H.; Liu, Y.; Harris, T.; Drake, C.; Pardoll, D.; Yu, H. Regulation of the IL-23 and IL-12 balance by Stat3 signaling in the tumor microenvironment. Cancer Cell 2009, 15, 114–123. [Google Scholar] [CrossRef] [PubMed]
- Saudemont, A.; Jouy, N.; Hetuin, D.; Quesnel, B. NK cells that are activated by CXCL10 can kill dormant tumor cells that resist CTL-mediated lysis and can express B7-H1 that stimulates T cells. Blood 2005, 105, 2428–2435. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Pardoll, D.; Jove, R. STATs in cancer inflammation and immunity: A leading role for STAT3. Nat. Rev. Cancer 2009, 9, 798–809. [Google Scholar] [CrossRef] [PubMed]
- Ho, H.H.; Ivashkiv, L.B. Role of STAT3 in type I interferon responses. Negative regulation of STAT1-dependent inflammatory gene activation. J. Biol. Chem. 2006, 281, 14111–14118. [Google Scholar] [CrossRef]
- Nassar, D.; Blanpain, C. Cancer Stem Cells: Basic Concepts and Therapeutic Implications. Annu. Rev. Pathol. 2016, 11, 47–76. [Google Scholar] [CrossRef]
- Pattabiraman, D.R.; Weinberg, R.A. Tackling the cancer stem cells—What challenges do they pose? Nat. Rev. Drug Discov. 2014, 13, 497–512. [Google Scholar] [CrossRef]
- Korkaya, H.; Liu, S.; Wicha, M.S. Regulation of cancer stem cells by cytokine networks: Attacking cancer’s inflammatory roots. Clin. Cancer Res. 2011, 17, 6125–6129. [Google Scholar] [CrossRef]
- Rosen, J.M.; Jordan, C.T. The increasing complexity of the cancer stem cell paradigm. Science 2009, 324, 1670–1673. [Google Scholar] [CrossRef]
- Mitsui, K.; Tokuzawa, Y.; Itoh, H.; Segawa, K.; Murakami, M.; Takahashi, K.; Maruyama, M.; Maeda, M.; Yamanaka, S. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell 2003, 113, 631–642. [Google Scholar] [CrossRef] [PubMed]
- Chambers, I.; Colby, D.; Robertson, M.; Nichols, J.; Lee, S.; Tweedie, S.; Smith, A. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell 2003, 113, 643–655. [Google Scholar] [CrossRef] [PubMed]
- Gibbs, C.P.; Kukekov, V.G.; Reith, J.D.; Tchigrinova, O.; Suslov, O.N.; Scott, E.W.; Ghivizzani, S.C.; Ignatova, T.N.; Steindler, D.A. Stem-like cells in bone sarcomas: Implications for tumorigenesis. Neoplasia 2005, 7, 967–976. [Google Scholar] [CrossRef] [PubMed]
- Chambers, I. The molecular basis of pluripotency in mouse embryonic stem cells. Cloning Stem Cells 2004, 6, 386–391. [Google Scholar] [CrossRef]
- Vazquez-Santillan, K.; Melendez-Zajgla, J.; Jimenez-Hernandez, L.; Martínez-Ruiz, G.; Maldonado, V. NF-kappaB signaling in cancer stem cells: A promising therapeutic target? Cell. Oncol. 2015, 38, 327–339. [Google Scholar] [CrossRef]
- Wang, S.S.; Jiang, J.; Liang, X.H.; Tang, Y.L. Links between cancer stem cells and epithelial-mesenchymal transition. Onco Targets Ther. 2015, 8, 2973–2980. [Google Scholar]
- Zhao, D.; Pan, C.; Sun, J.; Gilbert, C.; Drews-Elger, K.; Azzam, D.J.; Picon-Ruiz, M.; Kim, M.; Ullmer, W.; El-Ashry, D.; et al. VEGF drives cancer-initiating stem cells through VEGFR-2/Stat3 signaling to upregulate Myc and Sox2. Oncogene 2015, 34, 3107–3119. [Google Scholar] [CrossRef]
- Won, C.; Kim, B.H.; Yi, E.H.; Choi, K.J.; Kim, E.K.; Jeong, J.M.; Lee, J.H.; Jang, J.J.; Yoon, J.H.; Jeong, W.I.; et al. Signal transducer and activator of transcription 3-mediated CD133 up-regulation contributes to promotion of hepatocellular carcinoma. Hepatology 2015, 62, 1160–1173. [Google Scholar] [CrossRef]
- Wu, A.; Wei, J.; Kong, L.Y.; Wang, Y.; Priebe, W.; Qiao, W.; Sawaya, R.; Heimberger, A.B. Glioma cancer stem cells induce immunosuppressive macrophages/microglia. Neuro Oncol. 2010, 12, 1113–1125. [Google Scholar] [CrossRef]
- Zhao, C.; Li, H.; Lin, H.J.; Yang, S.; Lin, J.; Liang, G. Feedback Activation of STAT3 as a Cancer Drug-Resistance Mechanism. Trends Pharmacol. Sci. 2016, 37, 47–61. [Google Scholar] [CrossRef]
- Villalva, C.; Martin-Lannerée, S.; Cortes, U.; Dkhissi, F.; Wager, M.; Le Corf, A.; Tourani, J.M.; Dusanter-Fourt, I.; Turhan, A.G.; Karayan-Tapon, L. STAT3 is essential for the maintenance of neurosphere-initiating tumor cells in patients with glioblastomas: A potential for targeted therapy? Int. J. Cancer 2011, 128, 826–838. [Google Scholar] [CrossRef] [PubMed]
- MacDonagh, L.; Gray, S.G.; Breen, E.; Cuffe, S.; Finn, S.P.; O’Byrne, K.J.; Barr, M.P. BBI608 inhibits cancer stemness and reverses cisplatin resistance in NSCLC. Cancer Lett. 2018, 428, 117–126. [Google Scholar] [CrossRef] [PubMed]
- de la Iglesia, N.; Puram, S.V.; Bonni, A. STAT3 regulation of glioblastoma pathogenesis. Curr. Mol. Med. 2009, 9, 580–590. [Google Scholar] [CrossRef] [PubMed]
- de la Iglesia, N.; Konopka, G.; Puram, S.V.; Chan, J.A.; Bachoo, R.M.; You, M.J.; Levy, D.E.; Depinho, R.A.; Bonni, A. Identification of a PTEN-regulated STAT3 brain tumor suppressor pathway. Genes Dev. 2008, 22, 449–462. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Kim, J.C.; Lee, S.E.; Quinley, C.; Kim, H.; Herdman, S.; Corr, M.; Raz, E. Signal transducer and activator of transcription 3 (STAT3) protein suppresses adenoma-to-carcinoma transition in Apcmin/+ mice via regulation of Snail-1 (SNAI) protein stability. J. Biol. Chem. 2012, 287, 18182–18189. [Google Scholar] [CrossRef]
- Gao, S.P.; Mark, K.G.; Leslie, K.; Pao, W.; Motoi, N.; Gerald, W.L.; Travis, W.D.; Bornmann, W.; Veach, D.; Clarkson, B.; et al. Mutations in the EGFR kinase domain mediate STAT3 activation via IL-6 production in human lung adenocarcinomas. J. Clin. Investig. 2007, 117, 3846–3856. [Google Scholar] [CrossRef]
- Grabner, B.; Schramek, D.; Mueller, K.M.; Moll, H.P.; Svinka, J.; Hoffmann, T.; Bauer, E.; Blaas, L.; Hruschka, N.; Zboray, K.; et al. Disruption of STAT3 signalling promotes KRAS-induced lung tumorigenesis. Nat. Commun. 2015, 6, 6285. [Google Scholar] [CrossRef]
- Tebay, L.E.; Robertson, H.; Durant, S.T.; Vitale, S.R.; Penning, T.M.; Dinkova-Kostova, A.T.; Hayes, J.D. Mechanisms of activation of the transcription factor Nrf2 by redox stressors, nutrient cues, and energy status and the pathways through which it attenuates degenerative disease. Free Radic. Biol. Med. 2015, 88, 108–146. [Google Scholar] [CrossRef]
- Wu, T.; Zhao, F.; Gao, B.; Tan, C.; Yagishita, N.; Nakajima, T.; Wong, P.K.; Chapman, E.; Fang, D.; Zhang, D.D. Hrd1 suppresses Nrf2-mediated cellular protection during liver cirrhosis. Genes Dev. 2014, 28, 708–722. [Google Scholar] [CrossRef]
- Cuadrado, A. Structural and functional characterization of Nrf2 degradation by glycogen synthase kinase 3/beta-TrCP. Free Radic. Biol. Med. 2015, 88, 147–157. [Google Scholar] [CrossRef]
- Lau, A.; Villeneuve, N.F.; Sun, Z.; Wong, P.K.; Zhang, D.D. Dual roles of Nrf2 in cancer. Pharmacol. Res. 2008, 58, 262–270. [Google Scholar] [CrossRef] [PubMed]
- Baird, L.; Dinkova-Kostova, A.T. The cytoprotective role of the Keap1-Nrf2 pathway. Arch. Toxicol. 2011, 85, 241–272. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.D.; Hannink, M. Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress. Mol. Cell. Biol. 2003, 23, 8137–8151. [Google Scholar] [CrossRef] [PubMed]
- Eggler, A.L.; Liu, G.; Pezzuto, J.M.; van Breemen, R.B.; Mesecar, A.D. Modifying specific cysteines of the electrophile-sensing human Keap1 protein is insufficient to disrupt binding to the Nrf2 domain Neh2. Proc. Natl. Acad. Sci. USA 2005, 102, 10070–10075. [Google Scholar] [CrossRef]
- Eggler, A.L.; Luo, Y.; van Breemen, R.B.; Mesecar, A.D. Identification of the highly reactive cysteine 151 in the chemopreventive agent-sensor Keap1 protein is method-dependent. Chem. Res. Toxicol. 2007, 20, 1878–1884. [Google Scholar] [CrossRef]
- Luo, Y.; Eggler, A.L.; Liu, D.; Liu, G.; Mesecar, A.D.; van Breemen, R.B. Sites of alkylation of human Keap1 by natural chemoprevention agents. J. Am. Soc. Mass Spectrom. 2007, 18, 2226–2232. [Google Scholar] [CrossRef]
- Sun, Z.; Zhang, S.; Chan, J.Y.; Zhang, D.D. Keap1 controls postinduction repression of the Nrf2-mediated antioxidant response by escorting nuclear export of Nrf2. Mol. Cell. Biol. 2007, 27, 6334–6349. [Google Scholar] [CrossRef]
- Thimmulappa, R.K.; Mai, K.H.; Srisuma, S.; Kensler, T.W.; Yamamoto, M.; Biswal, S. Identification of Nrf2-regulated genes induced by the chemopreventive agent sulforaphane by oligonucleotide microarray. Cancer Res. 2002, 62, 5196–5203. [Google Scholar]
- Su, Z.Y.; Shu, L.; Khor, T.O.; Lee, J.H.; Fuentes, F.; Kong, A.N. A perspective on dietary phytochemicals and cancer chemoprevention: Oxidative stress, nrf2, and epigenomics. Top. Curr. Chem. 2013, 329, 133–162. [Google Scholar]
- Zheng, H.; Whitman, S.A.; Wu, W.; Wondrak, G.T.; Wong, P.K.; Fang, D.; Zhang, D.D. Therapeutic potential of Nrf2 activators in streptozotocin-induced diabetic nephropathy. Diabetes 2011, 60, 3055–3066. [Google Scholar] [CrossRef]
- Jaramillo, M.C.; Zhang, D.D. The emerging role of the Nrf2-Keap1 signaling pathway in cancer. Genes Dev. 2013, 27, 2179–2191. [Google Scholar] [CrossRef] [PubMed]
- Slocum, S.L.; Kensler, T.W. Nrf2: Control of sensitivity to carcinogens. Arch. Toxicol. 2011, 85, 273–284. [Google Scholar] [CrossRef] [PubMed]
- Murrow, L.; Debnath, J. Autophagy as a stress-response and quality-control mechanism: Implications for cell injury and human disease. Annu. Rev. Pathol. 2013, 8, 105–137. [Google Scholar] [CrossRef]
- Choi, A.M.; Ryter, S.W.; Levine, B. Autophagy in human health and disease. N. Engl. J. Med. 2013, 368, 1845–1846. [Google Scholar] [CrossRef] [PubMed]
- Lau, A.; Zheng, Y.; Tao, S.; Wang, H.; Whitman, S.A.; White, E.; Zhang, D.D. Arsenic inhibits autophagic flux, activating the Nrf2-Keap1 pathway in a p62-dependent manner. Mol. Cell. Biol. 2013, 33, 2436–2446. [Google Scholar] [CrossRef] [PubMed]
- Riley, B.E.; Kaiser, S.E.; Kopito, R.R. Autophagy inhibition engages Nrf2-p62 Ub-associated signaling. Autophagy 2011, 7, 338–340. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lau, A.; Wang, X.J.; Zhao, F.; Villeneuve, N.F.; Wu, T.; Jiang, T.; Sun, Z.; White, E.; Zhang, D.D. A noncanonical mechanism of Nrf2 activation by autophagy deficiency: Direct interaction between Keap1 and p62. Mol. Cell. Biol. 2010, 30, 3275–3285. [Google Scholar] [CrossRef]
- Wang, X.J.; Sun, Z.; Chen, W.; Li, Y.; Villeneuve, N.F.; Zhang, D.D. Activation of Nrf2 by arsenite and monomethylarsonous acid is independent of Keap1-C151: Enhanced Keap1-Cul3 interaction. Toxicol. Appl. Pharmacol. 2008, 230, 383–389. [Google Scholar] [CrossRef]
- Komatsu, M.; Kurokawa, H.; Waguri, S.; Taguchi, K.; Kobayashi, A.; Ichimura, Y.; Sou, Y.S.; Ueno, I.; Sakamoto, A.; Tong, K.I.; et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat. Cell Biol. 2010, 12, 213–223. [Google Scholar] [CrossRef]
- Katsuragi, Y.; Ichimura, Y.; Komatsu, M. p62/SQSTM1 functions as a signaling hub and an autophagy adaptor. FEBS J. 2015, 282, 4672–4678. [Google Scholar] [CrossRef]
- Jain, A.; Lamark, T.; Sjøttem, E.; Larsen, K.B.; Awuh, J.A.; Øvervatn, A.; McMahon, M.; Hayes, J.D.; Johansen, T. p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J. Biol. Chem. 2010, 285, 22576–22591. [Google Scholar] [CrossRef] [PubMed]
- Inami, Y.; Waguri, S.; Sakamoto, A.; Kouno, T.; Nakada, K.; Hino, O.; Watanabe, S.; Ando, J.; Iwadate, M.; Yamamoto, M.; et al. Persistent activation of Nrf2 through p62 in hepatocellular carcinoma cells. J. Cell Biol. 2011, 193, 275–284. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Sun, R.; Wang, H.; Yang, B.; Wang, F.; Xu, H.; Chen, S.; Zhao, R.; Pi, J.; Xu, Y. Enhanced p62-NRF2 Feedback Loop due to Impaired Autophagic Flux Contributes to Arsenic-Induced Malignant Transformation of Human Keratinocytes. Oxid. Med. Cell. Longev. 2019, 2019, 1038932. [Google Scholar] [CrossRef] [PubMed]
- Schmidlin, C.J.; Shakya, A.; Dodson, M.; Chapman, E.; Zhang, D.D. The intricacies of NRF2 regulation in cancer. Semin. Cancer Biol. 2021, 76, 110–119. [Google Scholar] [CrossRef]
- Iida, K.; Itoh, K.; Kumagai, Y.; Oyasu, R.; Hattori, K.; Kawai, K.; Shimazui, T.; Akaza, H.; Yamamoto, M. Nrf2 is essential for the chemopreventive efficacy of oltipraz against urinary bladder carcinogenesis. Cancer Res. 2004, 64, 6424–6431. [Google Scholar] [CrossRef]
- Ramos-Gomez, M.; Dolan, P.M.; Itoh, K.; Yamamoto, M.; Kensler, T.W. Interactive effects of nrf2 genotype and oltipraz on benzo[a]pyrene-DNA adducts and tumor yield in mice. Carcinogenesis 2003, 24, 461–467. [Google Scholar] [CrossRef]
- Rachakonda, G.; Sekhar, K.R.; Jowhar, D.; Samson, P.C.; Wikswo, J.P.; Beauchamp, R.D.; Datta, P.K.; Freeman, M.L. Increased cell migration and plasticity in Nrf2-deficient cancer cell lines. Oncogene 2010, 29, 3703–3714. [Google Scholar] [CrossRef]
- Satoh, H.; Moriguchi, T.; Taguchi, K.; Takai, J.; Maher, J.M.; Suzuki, T.; Winnard, P.T.; Jr Raman, V.; Ebina, M.; Nukiwa, T.; et al. Nrf2-deficiency creates a responsive microenvironment for metastasis to the lung. Carcinogenesis 2010, 31, 1833–1843. [Google Scholar] [CrossRef]
- Chen, W.; Sun, Z.; Wang, X.J.; Jiang, T.; Huang, Z.; Fang, D.; Zhang, D.D. Direct interaction between Nrf2 and p21(Cip1/WAF1) upregulates the Nrf2-mediated antioxidant response. Mol. Cell 2009, 34, 663–673. [Google Scholar] [CrossRef]
- Ren, D.; Villeneuve, N.F.; Jiang, T.; Wu, T.; Lau, A.; Toppin, H.A.; Zhang, D.D. Brusatol enhances the efficacy of chemotherapy by inhibiting the Nrf2-mediated defense mechanism. Proc. Natl. Acad. Sci. USA 2011, 108, 1433–1438. [Google Scholar] [CrossRef]
- Tsuchida, K.; Tsujita, T.; Hayashi, M.; Ojima, A.; Keleku-Lukwete, N.; Katsuoka, F.; Otsuki, A.; Kikuchi, H.; Oshima, Y.; Suzuki, M.; et al. Halofuginone enhances the chemo-sensitivity of cancer cells by suppressing NRF2 accumulation. Free Radic. Biol. Med. 2017, 103, 236–247. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Wang, H.; Fan, L.; Wu, X.; Xin, A.; Ren, H.; Wang, X.J. Luteolin inhibits Nrf2 leading to negative regulation of the Nrf2/ARE pathway and sensitization of human lung carcinoma A549 cells to therapeutic drugs. Free Radic. Biol. Med. 2011, 50, 1599–1609. [Google Scholar] [CrossRef] [PubMed]
- Ohnuma, T.; Matsumoto, T.; Itoi, A.; Kawana, A.; Nishiyama, T.; Ogura, K.; Hiratsuka, A. Enhanced sensitivity of A549 cells to the cytotoxic action of anticancer drugs via suppression of Nrf2 by procyanidins from Cinnamomi Cortex extract. Biochem. Biophys. Res. Commun. 2011, 413, 623–629. [Google Scholar] [CrossRef] [PubMed]
- Yu, B.; Liang, J.; Li, X.; Liu, L.; Yao, J.; Chen, X.; Chen, R. Renieramycin T Inhibits Melanoma B16F10 Cell Metastasis and Invasion via Regulating Nrf2 and STAT3 Signaling Pathways. Molecules 2022, 27, 5337. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Wang, K. The induction of ferroptosis by impairing STAT3/Nrf2/GPx4 signaling enhances the sensitivity of osteosarcoma cells to cisplatin. Cell Biol. Int. 2019, 43, 1245–1256. [Google Scholar] [CrossRef]
- Liu, H.; Ren, G.; Wang, T.; Chen, Y.; Gong, C.; Bai, Y.; Wang, B.; Qi, H.; Shen, J.; Zhu, L.; et al. Aberrantly expressed Fra-1 by IL-6/STAT3 transactivation promotes colorectal cancer aggressiveness through epithelial-mesenchymal transition. Carcinogenesis 2015, 36, 459–468. [Google Scholar] [CrossRef]
- Sullivan, N.J.; Sasser, A.K.; Axel, A.E.; Vesuna, F.; Raman, V.; Ramirez, N.; Oberyszyn, T.M.; Hall, B.M. Interleukin-6 induces an epithelial-mesenchymal transition phenotype in human breast cancer cells. Oncogene 2009, 28, 2940–2947. [Google Scholar] [CrossRef]
- Yadav, A.; Kumar, B.; Datta, J.; Teknos, T.N.; Kumar, P. IL-6 promotes head and neck tumor metastasis by inducing epithelial-mesenchymal transition via the JAK-STAT3-SNAIL signaling pathway. Mol. Cancer Res. 2011, 9, 1658–1667. [Google Scholar] [CrossRef]
- Kim, S.J.; Saeidi, S.; Cho, N.C.; Kim, S.H.; Lee, H.B.; Han, W.; Noh, D.Y.; Surh, Y.J. Interaction of Nrf2 with dimeric STAT3 induces IL-23 expression: Implications for breast cancer progression. Cancer Lett. 2021, 500, 147–160. [Google Scholar] [CrossRef]
- Piskin, G.; Sylva-Steenland, R.M.; Bos, J.D.; Teunissen, M.B. In vitro and in situ expression of IL-23 by keratinocytes in healthy skin and psoriasis lesions: Enhanced expression in psoriatic skin. J. Immunol. 2006, 176, 1908–1915. [Google Scholar] [CrossRef]
- Sheng, S.; Zhang, J.; Ai, J.; Hao, X.; Luan, R. Aberrant expression of IL-23/IL-23R in patients with breast cancer and its clinical significance. Mol. Med. Rep. 2018, 17, 4639–4644. [Google Scholar] [CrossRef]
- Buettner, R.; Mora, L.B.; Jove, R. Activated STAT signaling in human tumors provides novel molecular targets for therapeutic intervention. Clin. Cancer Res. 2002, 8, 945–954. [Google Scholar] [PubMed]
- Gao, S.; Zhan, L.; Yang, Z.; Shi, R.; Li, H.; Xia, Z.; Yuan, S.; Wu, Q.P.; Wang, T.; Yao, S. Remote Limb Ischaemic Postconditioning Protects Against Myocardial Ischaemia/Reperfusion Injury in Mice: Activation of JAK/STAT3-Mediated Nrf2-Antioxidant Signalling. Cell Physiol. Biochem. 2017, 43, 1140–1151. [Google Scholar] [CrossRef] [PubMed]
- Son, Y.O.; Pratheeshkumar, P.; Divya, S.P.; Zhang, Z.; Shi, X. Nuclear factor erythroid 2-related factor 2 enhances carcinogenesis by suppressing apoptosis and promoting autophagy in nickel-transformed cells. J. Biol. Chem. 2017, 292, 8315–8330. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).