Abstract
Coexisting salt and alkaline stresses seriously threaten plant survival. Most studies have focused on halophytes; however, knowledge on how plants defend against saline–alkali stress is limited. This study investigated the role of Taraxacum mongolicum in a Puccinellia tenuiflora community under environmental saline–alkali stress to analyse the response of elements and metabolites in T. mongolicum, using P. tenuiflora as a control. The results show that the macroelements Ca and Mg are significantly accumulated in the aboveground parts (particularly in the stem) of T. mongolicum. Microelements B and Mo are also accumulated in T. mongolicum. Microelement B can adjust the transformation of sugars, and Mo contributes to the improvement in nitrogen metabolism. Furthermore, the metabolomic results demonstrate that T. mongolicum leads to decreased sugar accumulation and increased amounts of amino acids and organic acids to help plants resist saline–alkali stress. The resource allocation of carbon (sugar) and nitrogen (amino acids) results in the accumulation of only a few phenolic metabolites (i.e., petunidin, chlorogenic acid, and quercetin-3-O-rhamnoside) in T. mongolicum. These phenolic metabolites help to scavenge excess reactive oxygen species. Our study primarily helps in understanding the contribution of T. mongolicum in P. tenuiflora communities on coping with saline–alkali stress.
1. Introduction
Soil salinisation is a serious ecological problem that exerts complex adverse effects on plant metabolism [1]. With the aggravation of inappropriate anthropogenic activities, the natural environment has deteriorated, and soil saline-alkalisation has become increasingly serious [2]. According to statistics, approximately 7% of land worldwide (i.e., more than 900 million ha) is threatened by saline-alkalisation [3]. Plants grown on saline–alkali soil respond to both salt and alkali stress. Salt stress primarily results from NaCl, Na2SO4, and other neutral salts, producing a range of adverse effects. The excess accumulation of Cl− and Na+ induces specific ionic toxicities and disrupts ion homeostasis [4,5,6]. High salinity decreases the osmotic potential of the soil solution, reduces the uptake of water, diminishes the photosynthetic ability of natural plants, and ultimately inhibits plant growth [4,7]. Salt stress also increases the generation of reactive oxygen species (ROS) and induces damage to intracellular components [4]. Alkali stress is primarily caused by high amounts of NaHCO3 and Na2CO3. High pH induces serious changes in the morphology and physiological functions of most plants [5,8]. Elevated pH levels lead to a deficiency in external protons and an increase in plant root-growth resistance, which further disturbs the ion balance in plants [8,9,10]. High pH levels also severely disturb intracellular pH stability, destroy cell membrane integrity, and impair photosynthetic function [11,12]. Soil salinisation and alkalisation are typically observed together in many cases. The coexistence of soil salinisation and alkalisation further aggravate their adverse effects [1,13].
Prior research has demonstrated that in order to adapt to salt stress, plants have developed a series of morphological, physiological, biochemical, and molecular defence mechanisms, such as maintaining ion homeostasis, regulating osmotic balance, scavenging ROS, inducing signalling transduction, modulating antioxidative enzyme activities, mediating excretion, and performing the intracellular compartmentalisation of salts [9,14,15,16,17]. Some metabolites, such as betaine, proline, polyamine, and polyhydric alcohol, are considered to be beneficial for salt-stress tolerance in plants [1]. Fructose and sucrose, signalling molecules which can stimulate related enzymes, are widely recognised as regulators of salt responses. Unsaturated free fatty acids were found to enhance the activity of plasma membrane H+-ATPase in Arabidopsis under salt stress [18,19]. Moreover, glucose was reported to improve salt tolerance by regulating the expression of the glucose sensor hexokinase1 in apple (Malus domestica Borkh) [20].
Plant responses to alkali stress have also been investigated. Under mild alkali stress, bermudagrass reduces carbohydrate degradation and suppresses nitrogen metabolic processes to maintain basic growth. However, under moderate and severe alkali stress, bermudagrass utilises different response strategies. Higher amounts of carbohydrates, as well as significantly elevated ROS and malondialdehyde contents, are observed [21]. After arbuscular mycorrhizal fungi inoculation, Puccinellia tenuiflora (P. tenuiflora) exhibit higher amounts of amino acids, organic acids, flavonoids, and hormones. These elevated metabolites adjust and benefit the maintenance of cell membrane stability under alkali stress [22]. The responses to salt and alkali stresses have been thoroughly investigated. In contrast, little attention has been given to responses to saline–alkali stress, although soil salinisation and alkalisation are commonly observed together [3,22].
Considering that plants collectively compose community responses to saline–alkali stress in the natural environment, it is crucial to decipher the roles of plants in their community. In this study, Taraxacum mongolicum (T. mongolicum) was selected as the experimental subject and P. tenuiflora was selected as a control to investigate the role of T. mongolicum on elements and metabolites in the P. tenuiflora community under saline–alkali stress. The findings may provide novel insights into saline–alkali tolerance mechanisms from the perspective of plant communities.
2. Results
2.1. Accumulation and Distribution of Elements in T. mongolicum
The rhizosphere soil around T. mongolicum is defined as saline–alkali soil, based on our previous research [23]. The parameters of the rhizosphere soil were as follows: pH of 8.76, exchangeable sodium percentage of 40.58%, soluble salt content of 0.57%, electrical conductivity of 895 mS/cm, (CO32− HCO3−)/(Cl− SO42−) ratio of 0.81, and Cl−/SO42− ratio of 0.37.
The results from the PCA plot and the OPLS-DA plot identified the diverse expression patterns of elements in P. tenuiflora and T. mongolicum (VIP > 1, p-value < 0.05) (Figure 1). The elements K, Ca, Na, Mg, B, and Mo were differentially expressed between P. tenuiflora and T. mongolicum, and significantly accumulated in T. mongolicum (Figure 2). The measurements of elements in different tissues (leaf, stem, and root) showed that Na contents in the leaf, stem, and root of T. mongolicum increased 2.8-fold, 5.4-fold, and 2.3-fold, respectively (Figure 2a), compared with P. tenuiflora. Calculated factors demonstrated that Na was absorbed from soil by T. mongolicum (Figure 2b). In addition to Na, macroelements K, Ca, and Mg were evidently accumulated in the aboveground parts of T. mongolicum, particularly in the stem (Figure 2c–h). This viewpoint was further supported by the ratio of K/Na in T. mongolicum being evidently higher than that in P. tenuiflora (Figure 2i). Ca and Mg play important roles in resisting the toxicity of Na in T. mongolicum. The relative contents of Ca and Mg were even larger than that of K in T. mongolicum. In T. mongolicum, Ca contents increased 3.9-fold and 4.7-fold in the leaf and stem, respectively (Figure 2f). Mg contents increased 5.1-fold and 8.1-fold in the leaf and stem, respectively, in T. mongolicum compared with those in P. tenuiflora (Figure 2g). These significantly accumulated macroelements in T. mongolicum were primarily actively absorbed from the soil. The microelements B and Mo were also significantly accumulated in T. mongolicum (Figure 2j–m). The calculated relative contents of B indicated that B was primarily distributed in the stem and root. BF and TF values revealed that B was absorbed from the soil and accumulated in the root. Mo was transported from the root to the leaf (Figure 2k,m).
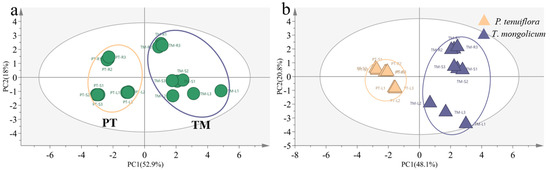
Figure 1.
Comparison of elements between T. mongolicum and P. tenuiflora. (a) PCA score plot. Purple circle: T. mongolicum, orange circle: P. tenuiflora. TM: T. mongolicum group, PT: P. tenuiflora group. (b) OPLS−DA score plot. Purple triangle: P. tenuiflora, orange triangle: T. mongolicum.
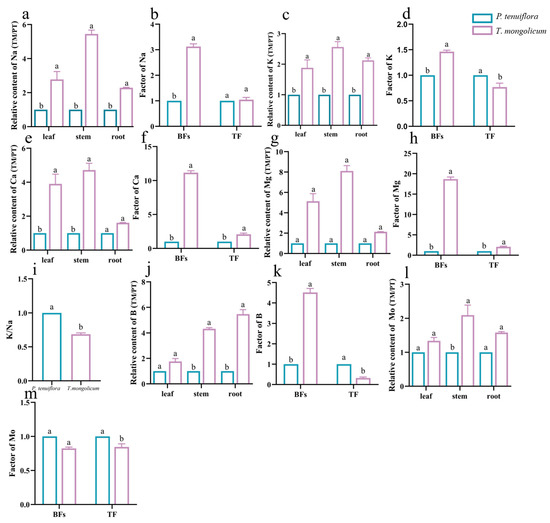
Figure 2.
Relative abundances of significantly different elements in T. mongolicum. (a,c,e,g,j,l) Relative contents of (a) Na; (c) K; (e) Ca; (g) Mg; (j) B; and (l) Mo in T. mongolicum compared to their contents in P. tenuiflora. The contents of elements in P. tenuiflora are normalized to 1. (b,d,f,h,k,m) Factors of (b) Na; (d) K; (f) Ca; (h) Mg; (k) B; and (m) Mo in T. mongolicum compared to factors in P. tenuiflora. The factors of elements in P. tenuiflora are normalized to 1. (i) The ratio of Na and K. Element contents and factors are summarized from 3 biological replicates and presented as mean ± SEM. Different letters indicate significant differences between groups (p-value < 0.05).
2.2. Responses of Primary Metabolites to Saline–Alkali Stress
In total, 226 effective compounds were detected in the leaf, stem, and root of P. tenuiflora and T. mongolicum via GC-MS. Similar to the PCA score plot of elements in P. tenuiflora and T. mongolicum, the PCA score plot of primary metabolites exhibited robust differences between P. tenuiflora and T. mongolicum (Figure 3a). The OPLS-DA score plot showed a PC1 of 11.2% and a PC2 of 26.4%, revealing a clear distinction between P. tenuiflora and T. mongolicum (Figure 3b). Among these 226 compounds, 88 primary metabolites were significantly differentially expressed between P. tenuiflora and T. mongolicum, screened by a threshold of VIP > 1 and p-value < 0.05. These significantly different metabolites could be divided into 13 sugars, 10 alcohols, 7 amino acids, 13 esters, 28 acids, and 17 other compounds (not shown) (Table 1). Calculated Q-values indicated that sugars were significantly accumulated in P. tenuiflora (Figure 4a); acids and esters were significantly accumulated in the aboveground parts of T. mongolicum (Figure 4b,c); and alcohols and amino acids were markedly accumulated in all parts of T. mongolicum (Figure 4d,e). These findings indicated that there might be variable distributions of energy and materials between P. tenuiflora and T. mongolicum.
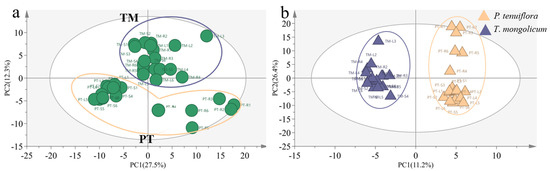
Figure 3.
Comparison of primary metabolites between T. mongolicum and P. tenuiflora. (a) PCA score plot. Purple circle: T. mongolicum, orange circle: P. tenuiflora. TM: T. mongolicum group, PT: P. tenuiflora group. (b) OPLS−DA score plot. Purple triangle: P. tenuiflora, orange triangle: T. mongolicum.

Table 1.
List of significantly different primary metabolites between T. mongolicum and P. tenuiflora.
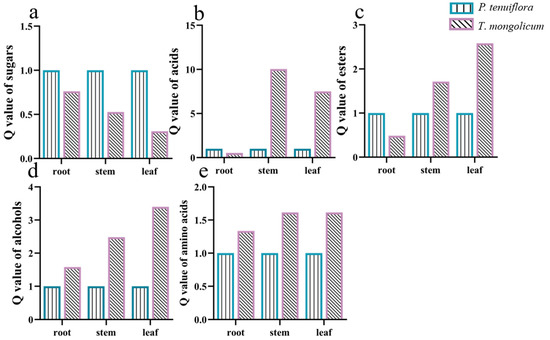
Figure 4.
Q-values of significantly different metabolites in T. mongolicum. Relative Q-values of (a) sugars; (b) acids; (c) esters; (d) alcohols; and (e) amino acids in T. mongolicum compared to Q-values in P. tenuiflora. Q-values of metabolites in P. tenuiflora are normalised to 1.
To explore the metabolic mechanisms underlying variable distributions of energy and materials, metabolic pathways of different tissues under saline–alkali stress were visualised (Figure 5). Amino acid metabolism was more activated in T. mongolicum. Valine exhibited a 1.7-fold increase in the leaf, a 29.8-fold increase in the stem, and a 5.2-fold increase in the root of T. mongolicum, compared with those of P. tenuiflora. Nicotinoylglycine and hydroxynorvaline markedly accumulated in the leaf of T. mongolicum, exhibiting a 3.0-fold increase and a 3.7-fold increase, respectively. Nicotinoylglycine and hydroxynorvaline also exhibited a 1.7-fold increase and a 2.9-fold increase in the stem of T. mongolicum, respectively. Carbamylglutamate exhibited a 4.5-fold increase in the leaf, 5.6-fold increase in the stem, and 5.2-fold increase in the root of T. mongolicum, compared with those of P. tenuiflora. Tyrosine only accumulated in T. mongolicum, exhibiting a relative content of 0.5 in the leaf, 0.5 in the stem, and 1.0 in the root (Figure 5).
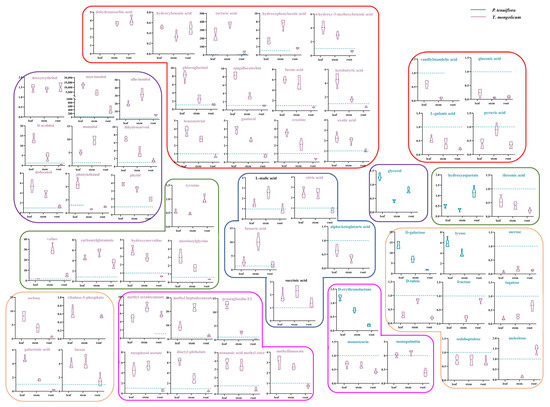
Figure 5.
The metabolic networks of significantly different primary metabolites. The contents of primary metabolites in P. tenuiflora are normalised to 1 and represented as dashed lines in blue. Purple frame represents alcohols, orange frame represents sugars, green frame represents amino acids, rose pink frame represents esters, red frame represents acids, and dark blue frame represents components in the TCA cycle. Pink title indicates that the contents of primary metabolites in T. mongolicum are higher than in P. tenuiflora while blue title represents that the contents of primary metabolites in P. tenuiflora are higher than in T. mongolicum. Black title indicates that the primary metabolite was not a significantly different metabolite.
Unlike amino acid metabolism, sugar metabolism was more involved in P. tenuiflora, indicating a choice between carbon metabolism (represented by sugar) and nitrogen metabolism (represented by amino acid) in plants via the tricarboxylic acid (TCA) cycle in response to saline–alkali stress. Among them, fumaric acid and citric acid were significantly different metabolites. Fumaric acid increased by 2.7-fold (leaf), 10.1-fold (stem), and 1.8-fold (root) in T. mongolicum, whereas citric acid increased by 2.5-fold (leaf) and 2.4-fold (stem) in T. mongolicum. For sugar metabolism, in T. mongolicum, galactonic acid exhibited a 4.6-fold increase in the leaf and a 1.7-fold increase in the stem, whereas fucose exhibited a 3.9-fold increase in the leaf and a 4.1-fold increase in the stem. Sorbose and ribulose-5-phosphate were only accumulated in T. mongolicum (Figure 5).
The metabolites of alcohols, esters, and acids are involved in carbon–TCA–amino acid metabolism as supplements. More alcohols were enriched in T. mongolicum, increasing 12,102-fold (leaf), 4314-fold (stem), and 388-fold (root) for myo-inositol; 19.4-fold (leaf), 32.1-fold (stem), and 5.5-fold (root) for allo-inositol; 13.3-fold (leaf) and 4.4-fold (stem) or D-arabitol; 5.1-fold (leaf) and 9.7-fold (stem) for mannitol; 7.0-fold (leaf) and 3.5-fold (stem) for dihydrocarveol; 5.4-fold (leaf) and 3.9-fold (stem) for dodecanol; 4.3-fold (leaf) for phenylethanol; and 3.9-fold (leaf) and 3.3-fold (stem) for phytol, compared with those in P. tenuiflora. Moreover, deoxyerythritol was only enriched in T. mongolicum. Esters were more accumulated in the aboveground parts of T. mongolicum. Specifically, methyl heptadecanoate exhibited 9.1-fold (leaf), 23.0-fold (stem), and 2.4-fold (root) increases in T. mongolicum; prostaglandin E2 exhibited 12.7-fold (leaf) and 2.8-fold (stem) increases; tocopherol acetate exhibited 4.6-fold (leaf) and 5.4-fold (stem) increases; dioctyl phthalate exhibited 6.3-fold (leaf) and 2.9-fold (stem) increases; nonanoic acid methyl ester exhibited 3.7-fold (leaf) and 3.5-fold (stem) increases; and methylfumarate exhibited 3.1-fold (leaf) and 2.6-fold (stem) increases. Methyl octadecenoate only accumulated in the aboveground parts of T. mongolicum (Figure 5).
Acids involved in sugar–TCA–amino acid metabolism were also massively accumulated in the aboveground parts of T. mongolicum, increasing 243.6-fold (leaf), 367.5-fold (stem), and 30.4-fold (root) for tartaric acid; 3.4-fold (leaf) and 7.5-fold (stem) for hydroxyphenylacetic acid; 4.1-fold (leaf) and 3.0-fold (stem) for 4-hydroxy-3-methoxybenzoic acid; 8.1-fold (leaf) and 2.6-fold (stem) for phloroglucinol; 8.4-fold (leaf) and 3.3-fold (stem) for epigallocatechin; 6.0-fold (leaf) and 5.3-fold (stem) for furoic acid; 4.5-fold (leaf) and 1.6-fold (stem) for ketobutyric acid; 3.7-fold (leaf) and 2.8-fold (stem) for benzenetriol; 3.9-fold (leaf) and 2.6-fold (stem) for guaiacol; 3.5-fold (leaf) and 2.1-fold (stem) for creatine; and 2.3-fold (leaf) and 2.0-fold (stem) for oxalic acid, compared with those in P. tenuiflora. Hydroxybenzoic acid and dehydroascorbic acid were only accumulated in T. mongolicum.
2.3. Responses of Phenolic Metabolites to Saline–Alkali Stress
To further explore the response mechanisms of T. mongolicum against saline–alkali stress, the relative abundances of phenolic metabolites were summarised. In total, 20 effective phenolic metabolites were detected from 34 phenolic metabolites. Among these 20 effective phenolic metabolites, 9 significantly different phenolic metabolites were screened, based on the PCA plot and OPLS-DA score plot between P. tenuiflora and T. mongolicum (Figure 6, Table 2). These phenolic metabolites were accumulated less in T. mongolicum than in P. tenuiflora. Among these phenolic metabolites, chlorogenic acid was significantly accumulated in T. mongolicum, increasing 7.0-fold in the leaf, 72.0-fold in the stem, and 2.4-fold in the root compared with P. tenuiflora (Figure 7a). In the aboveground parts of T. mongolicum, petunidin increased 15.0-fold in the leaf and 286.6-fold in the stem compared with P. tenuiflora (Figure 7b). Another prominent compound, quercetin-3-O-rhamnoside, was only detected in T. mongolicum (Figure 7c). The other PCs were more markedly accumulated in P. tenuiflora than T. mongolicum (Figure 7d–i).
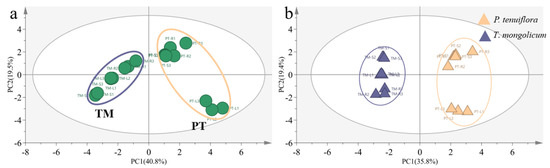
Figure 6.
Comparison of phenolic metabolites between T. mongolicum and P. tenuiflora. (a) PCA score plot. Purple circle: T. mongolicum, orange circle: P. tenuiflora. TM: T. mongolicum group, PT: P. tenuiflora group. (b) OPLS−DA score plot. Purple triangle: P. tenuiflora, orange triangle: T. mongolicum.

Table 2.
List of significantly different phenolic metabolites between T. mongolicum and P. tenuiflora.
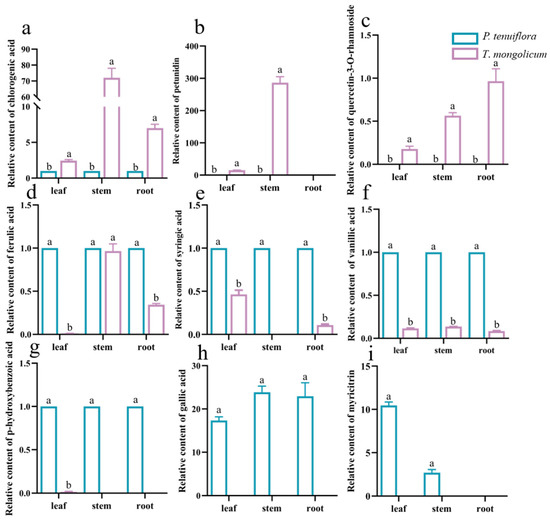
Figure 7.
Relative contents of significantly different phenolic metabolites in T. mongolicum. Relative contents of (a) chlorogenic acid; (b) petunidin; (c) quercetin-3-O-rhamnoside; (d) ferulic acid; (e) syringic acid; (f) vanillic acid; (g) p-hydroxybenzoic acid; (h) gallic acid; and (i) myricitrin in T. mongolicum compared to their contents in P. tenuiflora. The contents of phenolic metabolites in P. tenuiflora are normalised to 1. Phenolic metabolite contents are summarised from 3 biological replicates and presented as mean ± SEM. Different letters indicate significant differences between groups (p-value < 0.05).
3. Materials and Methods
3.1. Plant Materials and Growth Conditions
T. mongolicum and P. tenuiflora were gathered from saline–alkaline land in the Hulun Buir Grassland, Inner Mongolia Autonomous Region, China. Chen Qi identified T. mongolicum and P. tenuiflora. T. mongolicum belongs to the P. tenuiflora community, which is one of the dominant community types in the Hulun Buir Grassland. These two plants were collected from three different sample plots and stored at low temperature (<4 °C) for further study. P. tenuiflora was considered as the control group.
Measurement of Elemental Contents
Tissue samples (leaf, stem, and root) were dried at 60 °C and digested in concentrated HNO3 (95%) using a graphite plate (EH45A plus, Shanghai, China) at 130 °C. The elemental contents (Na, K, Ca, Mg, B, Fe, Mn, Ni, Mo, Cu, and Zn) were determined using an inductively-coupled plasma-emission spectrometer (ICP-OES Optima 8000, Perkin Elmer, Boston, MA, USA) and calculated from the standard curve of each element. The factors of elements, including the bioaccumulation factor (BF) and transfer factor (TF), were calculated according to a previously described method [24], where:
- BF = Mplant/Mmedium;
- TF = Mleaf/Mroot; and
Mplant: mass of element in plant (mg); Mmedium: mass of element in medium (mg); Mleaf: mass of element in leave (mg); Mroot: mass of element in root (mg).
3.2. Measurement of Primary Metabolites
The extraction and determination of primary metabolites were performed as described by Chen et al. with a few improvements [24]. Briefly, 90 mg of the sample was weighed, and 600 μL extract solution (including 540 μL cold methanol and 60 μL internal standard) was added. After 30 min of ultra-sonication, 300 μL chloroform and 600 μL water were added to the mixture. After 2 min of vortexing, the mixture was ultra-sonicated for 30 min and centrifuged at 13,000× g rpm at 4 °C for 10 min. Afterwards, 700 μL supernatant was transferred to a glass vial and dried in a vacuum concentrator. Dried samples were reconstituted by adding 400 μL methoxyamine (15 mg/mL in pyridine). The obtained solution was derived through sequential reaction with 400 μL BSTFA (15 mg/mL in 1% trimethylcholorosilane) and 80 μL n-hexane for GC-MS analysis.
Extracted samples were injected into the GC-MS system (Agilent 7890A-5975C, Agilent Technologies, Inc., Santa Clara, CA, USA). The constant flow rate was 1.0 mL/min, and the model of the non-polar DB-5 capillary column was 30 m × 250 μm I.D. (J&W Scientific, Folsom, CA, USA). The scanning range was set from 50 to 500 m/z, and the electron impact (EI) ion source was maintained at 70 Ev. The quality control sample was prepared by mixing the aliquots of tissues samples together to produce a pooled sample.
3.3. Measurement of Phenolic Metabolites
The extraction and determination of phenolic metabolites were performed as described by Chen et al. [24]. Briefly, 1.0 g of the sample was homogenised with 10 mL 70% methanol, ultra-sonicated for 40 min, and centrifuged at 8000× g rpm for 10 min. The supernatant was retained, the residual residue was re-extracted, and the supernatant was mixed and dried under vacuum twice. The dried sample was re-dissolved in 1 mL 70% methanol and filtered with 0.22 μm nylon membrane for ultra-high-performance liquid chromatography quadrupole time-of-flight mass spectrometry (UPLC-qTOF-MS) analysis. The UPLC (Waters, Tokyo, Japan) conditions were as follows: A%: 0.05% formic acid-water, B%: 0.05% formic acid–acetonitrile, and ACQUIT UPLC-BEH C18 Column (1.7 mm, 2.1 mm × 50 mm, Waters, Milford, MA, USA). The volume injected was 2 μL, and the column temperature was maintained at 30 °C. Mass spectrometry (Waters® Xevo G2 QTOF mass spectrometer, Waters) conditions were as follows: positive ion mode, capillary voltage of 3.0 kV, cone voltage of 45 V, source temperature of 400 °C, and desolvation temperature of 500 °C. The scanning range was set between 50 and −1000 m/z, with an ion-acquisition rate of 0.2/s. Extracted samples were injected into the mass spectrometer every 10 s at a flow rate of 5 μL/min. Leu-enkephalin was applied as an internal standard.
3.4. Statistical Analysis
The GC-MS data were analysed using Chroma TOF software (LECO, San Joes, CA, USA). The UPLC-qTOF-MS data were analysed and normalised using MassLynxTM (Waters). Unsupervised principal component analysis (PCA) and orthogonal partial least squares discriminant analysis (OPLS-DA) were performed to visualise the differences between groups. Significantly different compounds were screened by Student’s t-test and the multivariate statistical method (p-value < 0.05 and VIP > 1.0). The metabolic pathway was analysed by Kyoto Enrichment of Genes and Genomes (KEGG). The data underwent log2 transformation and min–max normalisation for improving data normality. Data were presented as the mean ± standard error (SEM). The score of principal component “Q” was calculated using SPSS (IBM, New York, NY, USA). Boxplot and histograms were generated using GraphPad Prism8 (Harvey, GraphPad Software, San Diego, CA, USA). Pathway maps were drawn with Adobe illustrator (Adobe, San Diego, CA, USA).
4. Discussion
Soil alkalisation is a major abiotic stress that affects plant growth, vegetation distribution, and crop yield [19,25]. Saline–alkali stress damages the physiological condition of plants, interrupts biochemical metabolism, and may even lead to the plant’s death [3,26,27]. Plant communities are naturally distributed in the saline–alkali soil and normally function as a team to resist saline–alkali stress. However, the roles of plants in the community under saline–alkali stress are typically neglected. In this study, the accumulation of elements, carbon–TCA–nitrogen metabolites, and phenolic metabolites were analysed to explore the role of T. mongolicum in a P. tenuiflora community under saline–alkali stress.
Compared with P. tenuiflora, Na was significantly accumulated in the aboveground parts of T. mongolicum, indicating that saline–alkali stress disrupts the ionic balance and induces Na toxicity. Maintaining high K and low Na concentrations is the main mechanism of plants to maintain osmotic regulation and ensure the proper functioning of many enzymes [1]. Several studies have demonstrated that competitive relationships exist between K and Na during their uptake under the conditions of high salt and alkali stress. The amount of Na increased, whereas the total K content decreased [1]. In this study, K was significantly accumulated in T. mongolicum, indicating a potential lack of competitive inhibition between the absorption of Na and K, which may be due to the recretohalophyte attributes of T. mongolicum. These kinds of plants may have a unique pathway of Na absorption, independent of K. Our findings suggest that in T. mongolicum, K cannot fight Na toxicity independently.
The macroelements Ca and Mg were enriched in the aboveground parts of T. mongolicum, indicating that they play a key role in resisting Na toxicity. Ca is a key signalling component of the salt-overly sensitive (SOS) pathway, which contributes to the maintenance of K/Na balance by extruding Na [28]. Our results showed that Ca is massively absorbed from soil and is accumulated in the aboveground parts of T. mongolicum, particularly in the stem. The distribution of Ca was consistent with that of Na, indicating that saline–alkali stress may activate the SOS–Na system to exclude Na and diminish the damage caused by Na toxicity.
Superfluous Na can destroy the structures and suppress the functions of chloroplasts, negatively affecting the photosynthetic capacity and chlorophyll content [1,29]. Mg is a central element of Mg-protoporphyrin and is thus involved in the biosynthesis of chlorophyll. In this study, Mg was significantly accumulated in the aboveground parts of T. mongolicum, indicating that the photosynthesis of T. mongolicum is not affected by saline–alkali stress. Mg was absorbed from the soil and transported to the aboveground parts of T. mongolicum to regulate chlorophyll biosynthesis and the photosynthetic rate. This suggests that T. mongolicum has relatively strong Mg-uptake capability. Mg content was considered as a signal arising from pools of adenylates, could reflecting lower energy status (less ATP) of a tissue in response to salt stress [30,31]. In our studies, leaves and stem have a lower-energy status. Saline–alkali stress may activate Mg-uptake channels, reduce the accumulation of Na, and improve osmotic adjustment. Remarkably, Na, Ca, and Mg were significantly accumulated in the stem of T. mongolicum, compared with P. tenuiflora, implying that the stem is the defensive battlefield of T. mongolicum in response to Na toxicity.
Microelement B can promote sugar transformation and transportation. Additionally, it forms peroxides to improve oxygen supply to plant roots [32]. It also plays an important role in regulating water distribution in plants [32,33]. In this study, T. mongolicum accumulated more B than P. tenuiflora under saline–alkali stress, implying that sugar metabolites and water distribution are regulated to respond to saline–alkali stress in T. mongolicum. In addition, B can limit vegetative growth by regulating the contents of phenolic metabolites under stress. Compared with P. tenuiflora, in T. mongolicum, B accumulated in the root. Accumulated B may improve the supply of oxygen and H2O2 and regulate phenolic metabolites in response to stress in the leaf. The microelement Mo is an important component of nitrogenase and nitrate reductase that affects plant nitrogen metabolism [34]. Our results show that a higher amount of Mo was accumulated in T. mongolicum than in P. tenuiflora, indicating that nitrogen metabolism plays an important role in the response of T. mongolicum to saline–alkali stress.
Sugar metabolism and nitrogen metabolism were highlighted in this study using the microelements B and Mo. To investigate the relationship between sugar metabolism and nitrogen metabolism under saline–alkali stress, primary metabolites were detected via GC-MS. The metabolic networks of some significantly different primary metabolites, including sugars, amino acids, esters, and acids, were also established. In this study, amino acids and esters accumulated more in the aboveground parts of T. mongolicum, whereas sugars accumulated more in P. tenuiflora. This demonstrates that a different response strategy is present. Under saline–alkali stress, plants can alter the contents of metabolites associated with sugar metabolism and change amino acid synthesis as well as the TCA cycle to counteract stress [35]. The concentrations of sugars, such as glucose, fructose, and sucrose, were increased in response to neutral salt stress [35]. These sugars are enhanced under neutral salt stress, implying that the degradation of polysaccharides as a carbon source likely promotes the maintenance of osmotic balance. Nevertheless, the concentrations of sugars were significantly reduced in response to saline–alkali stress; this was caused by the inhibition of reducing forces and the limitation of nitrogen metabolism [1,36]. This is similar to our findings in the P. tenuiflora community. Our results suggest that P. tenuiflora improves saline–alkali stress via the selection of metabolic strategies for other plants within the community. We speculate that nitrogen metabolism and ester metabolism are used by T. mongolicum to further respond to saline–alkali stress.
Previous studies have indicated that amino acids serve as key precursors for the synthesis of some secondary metabolites involved in plant defence responses [37]. At high pH levels, alkaline salt stress leads to the reduced production of amino acids and limited nitrogen intake/transport and metabolism [1,38,39,40]. In this study, amino acids were significantly accumulated in T. mongolicum, indicating that nitrogen metabolism still plays an important role in the defence against saline–alkali stress. The accumulation of amino acids can improve plant tolerance to saline–alkali stress by mediating the removal of ROS [7,41]. The accumulation of amino acids is a critical mechanism to compensate cellular osmolarity [42]. A choice exists between carbon and nitrogen metabolism, and T. mongolicum is more inclined to undergo nitrogen metabolism compared with P. tenuiflora.
Fatty acids are also significantly accumulated in the aboveground parts of T. mongolicum. Fatty acids reserve energy to participate in sugar–TCA–nitrogen metabolism. Fatty acids and their derivatives are also involved in plant resistance to abiotic stress [43,44]. As important components of the cytomembrane, polyunsaturated fatty acids play an important role in maintaining and regulating the normal biological functions of cells [17,45].
The accumulation of organic acids may be important for plants to adapt to saline–alkali stress [5,46]. In this study, acids were significantly accumulated in the aboveground parts of T. mongolicum. These organic acids play a key role in regulating pH. The accumulation of organic acids can reduce cell water potential and maintain the balance of ions and pH in cells [22,40,43,47]. In P. tenuiflora, organic acids are primarily accumulated in the root. Increased organic acid secretion may increase the acidity of the rhizosphere, thereby neutralising alkalinity surrounding the plant roots and promoting root growth [48]. In addition, alcohols, considered as osmotic regulators, were significantly accumulated in T. mongolicum. These alcohols can maintain osmotic pressure balance and scavenge ROS. Phenolic metabolites are essential secondary metabolites. In plants, they play a vital role against abiotic stress [49,50,51]. Phenolic metabolites and their derivatives reportedly improve soybean salt tolerance [52]. Similar results have also been reported in other species. In Arabidopsis, increasing the levels of flavonoids by transgenes can increase the tolerance to salt stress [53,54,55]. To investigate the role of phenolic metabolites in T. mongolicum under saline–alkali stress, the levels of phenolic metabolites were detected. The condition of surplus carbon in plants is a beneficial signal for phenolic metabolism. Therefore, the accumulation of phenolic metabolites is increased more when plants have higher carbohydrate reserves [56,57]. Consistent with the observed accumulation of sugars in P. tenuiflora, we found that phenolic metabolites accumulated more in P. tenuiflora. This observation further reflects the distribution trade-off between carbon and nitrogen in plants under saline–alkali stress [58,59]. Sugar accumulation also provides more carbon resources for specialised metabolites in plants. From a total of 20 phenolic metabolites, only petunidin, chlorogenic acid, and quercetin-3-O-rhamnoside were significantly accumulated in T. mongolicum. These phenolic metabolites were highlighted in conditions of limited resource distribution, demonstrating that they are crucial components in dealing with saline–alkali stress. Petunidin, quercetin-3-O-rhamnoside, and chlorogenic acid are established phenylpropanoid compounds which play a positive role in scavenging excess ROS. Therefore, accumulated petunidin, quercetin-3-O-rhamnoside, and chlorogenic acid may benefit T. mongolicum in the regulation of osmotic balance [15,52]. Phenolic metabolites can enhance plant tolerance to abiotic and biotic stresses as they can remove dangerous stress-response substances, including free radicals, singlet oxygen molecules, and peroxides, from the cell [22,60]. Some phenolic metabolites are found to have a strong binding affinity to superoxide dismutase 1 (SOD1). These phenolic metabolites stabilise the SOD1 dimer and inhibit the aggregation of SOD1 [60,61]. Phenolic metabolites and their derivatives play critical roles as antioxidants in response to saline–alkali stress [55,62]. Therefore, in T. mongolicum, petunidin, chlorogenic acid, and quercetin-3-O-rhamnoside may play an important role in ameliorating saline–alkali stress [63].
5. Conclusions
T. mongolicum plays an important role in the P. tenuiflora community in balancing the elements Ca, Mg, B, and Mo; the metabolism of nitrogen and fatty acids; as well as the accumulation of organic acids, metabolic solutes, and some phenolic metabolites. The macroelements Ca and Mg are accumulated in the aboveground parts, particularly in the stem of T. mongolicum, to defend against Na toxicity. B accumulated in T. mongolicum regulates sugar transformation and transportation, whereas Mo affects nitrogen metabolism. Compared with P. tenuiflora, T. mongolicum decreases the accumulation of carbohydrates, rather than synthesising more amino acids and organic acids, to help resist saline–alkali stressed environments. Enriched petunidin, chlorogenic acid, and quercetin-3-O-rhamnoside in T. mongolicum play positive roles in scavenging excess ROS.
Author Contributions
X.L. and Q.C. performed the experiments, analysed and interpreted the data, and prepared the figures. Q.C. and Y.J. wrote the manuscript. Z.T. revised the manuscript. M.X. and X.G. performed part of the data analysis. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Research Initiation Funds for the Natural Science Foundation of the Jiangsu Higher Education Institutions (21KJB180009), the Nantong Science and Technology Foundation of China (JC2021058).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data related to this study is available from the corresponding author upon reasonable request.
Conflicts of Interest
The authors declare no conflict of interest.
Sample Availability
Samples of the compounds significantly different TIAs or PCs are available from the authors.
References
- Guo, R.; Shi, L.X.; Yan, C.; Zhong, X.; Gu, F.X.; Liu, Q.; Xia, X.; Li, H. Ionomic and metabolic responses to neutral salt or alkaline salt stresses in Maize (Zea mays L.) seedlings. BMC Plant Biol. 2017, 17, 41–54. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Takano, T.; Liu, S. Screening and evaluation of saline-alkaline tolerant germplasm of rice (Oryza sativa L.) in soda saline-alkali Soil. Agronomy 2018, 8, 205–221. [Google Scholar] [CrossRef]
- Fang, S.; Hou, X.; Liang, X. Response mechanisms of plants under saline-alkali stress. Front. Plant Sci. 2021, 12, 667458. [Google Scholar] [CrossRef] [PubMed]
- Youssef, R.; Giampaolo, R.; Luigi, L.; Petronia, C.; Kyriacou, M.C.; Giuseppe, C.; Valerio, C.; Antonio, P.; Christophe, E.N.; Stefania, D.P. Physiological and metabolic responses triggered by omeprazole improve tomato plant tolerance to NaCl stress. Front. Plant Sci. 2018, 9, 249–267. [Google Scholar]
- Jia, X.M.; Wang, H.; Svetla, S.; Zhu, Y.F.; Hu, Y.; Cheng, L.; Zhao, T.; Wang, Y. Comparative physiological responses and adaptive strategies of apple Malus halliana to salt, alkali and saline-alkali stress. Sci. Hortic. 2019, 245, 154–162. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, Y.; Yan, X.; Guo, J. Physiological and transcriptomic analyses of yellow horn (Xanthoceras sorbifolia) provide important insights into salt and saline-alkali stress tolerance. PLoS ONE 2020, 15, e0244365. [Google Scholar] [CrossRef]
- Zhang, K.; Tang, J.; Wang, Y.; Kang, H.; Zeng, J. The tolerance to saline-alkaline stress was dependent on the roots in wheat. Physiol. Mol. Biol. Plants 2020, 26, 947–954. [Google Scholar]
- Chuamnakthong, S.; Mampei, M.; Ueda, A. Characterization of Na+ exclusion mechanism in rice under saline-alkaline stress conditions. Plant Sci. 2019, 287, 110171. [Google Scholar] [CrossRef]
- Liu, A.; Xiao, Z.; Li, M.W.; Wong, F.L.; Yung, W.S.; Ku, Y.S.; Wang, Q.; Wang, X.; Xie, M.; Yim, A.K.Y.; et al. Transcriptomic reprogramming in soybean seedlings under salt stress. Plant Cell Environ. 2019, 42, 98–114. [Google Scholar] [CrossRef]
- Zhang, H.; Li, X.; Guan, Y.A.; Li, M.A.; Wang, Y.B.; An, M.C.; Zhang, Y.C.; Liu, G.D.; Xu, N.; Sun, G. Physiological and proteomic responses of reactive oxygen species metabolism and antioxidant machinery in mulberry (Morus alba L.) seedling leaves to NaCl and NaHCO3 stress-sciencedirect. Ecotoxicol. Environ. Saf. 2020, 193, 110259–110270. [Google Scholar]
- Guo, k.; Xu, Z.; Huo, Y.; Sun, Q.; Wang, Y.; Che, Y.; Wang, J.; Li, W.; Zhang, H. Effects of salt concentration, pH, and their interaction on plant growth, nutrient uptake, and photochemistry of alfalfa (Medicago sativa) leaves. Plant Signal. Behav. 2020, 12, 1832373. [Google Scholar] [CrossRef] [PubMed]
- Hui, Z.; Liu, X.L.; Zhang, R.X.; Yuan, H.Y.; Wang, M.M.; Yang, H.; Ma, H.Y.; Duo, L.; Jiang, C.J.; Liang, Z.-W. Root damage under alkaline stress is associated with reactive oxygen species accumulation in rice (Oryza sativa L.). Front. Plant Sci. 2017, 8, 1580. [Google Scholar]
- Latef, A.A.A.H.; Tran, L.S. Impacts of priming with silicon on the growth and tolerance of maize plants to alkaline stress. Front. Plant Sci. 2016, 7, 243–253. [Google Scholar]
- Hannachi, S.; Labeke, M.V. Salt Stress Affects Germination, Seedling growth and physiological responses differentially in eggplant cultivars (Solanum melongena L.). Sci. Hortic. 2018, 228, 56–65. [Google Scholar] [CrossRef]
- Jia, X.M.; Zhu, Y.; Zhang, R.; Zhu, Z.; Wang, Y. Ionomic and metabolomic analyses reveal the resistance response mechanism to saline-alkali stress in Malus halliana seedlings. Plant Physiol. Biochem. 2019, 147, 77–90. [Google Scholar] [CrossRef]
- Arif, Y.; Singh, P.; Siddiqui, H.; Bajguz, A.; Hayat, S. Salinity induced physiological and biochemical changes in plants: An omic approach towards salt stress tolerance. Plant Physiol. Biochem. 2020, 156, 64–77. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Pang, J.; Zhang, F.; Sun, L.; Yang, L.; Zhao, Y.; Yang, Y.; Wang, Y.; Siddique, K. Integrated transcriptomics and metabolomics analysis to characterize alkali stress responses in Canola (Brassica napus L.). Plant Physiol. Biochem. 2021, 166, 605–620. [Google Scholar] [CrossRef]
- Han, X.; Yang, Y.; Wu, Y.; Liu, X.; Lei, X.; Guo, Y. A bioassay-guided fractionation system to identify endogenous small molecules that activate plasma membrane H+-atpase activity in Arabidopsis. J. Exp. Bot. 2017, 11, 2951–2962. [Google Scholar] [CrossRef]
- Jia, X.M.; Zhu, Y.F.; Hu, Y.; Zhang, R.; Wang, Y.X. Integrated physiologic, proteomic, and metabolomic analyses of Malus halliana adaptation to saline-alkali stress. Hortic. Res. 2019, 6, 91–110. [Google Scholar] [CrossRef]
- Sun, M.H.; Ma, Q.J.; Hu, D.G.; Zhu, X.P.; Hao, Y.J. The glucose sensor mdhxk1 phosphorylates a tonoplast Na+/H+ exchanger to improve salt tolerance. Plant Physiol. 2018, 176, 2977–2990. [Google Scholar] [CrossRef]
- Ye, T.; Wang, Y.; Feng, Y.Q.; Chan, Z. Physiological and metabolomic responses of bermudagrass (Cynodon dactylon) to alkali stress. Physiol. Plant. 2020, 171, 22–33. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Zhao, W.; Wang, Y.; Zhang, L.; Lin, J. Metabolomics analysis reveals the alkali tolerance mechanism in Puccinellia tenuiflora plants inoculated with Arbuscular mycorrhizal Fungi. Microorganisms 2020, 8, 327–347. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Jin, Y.; Zhang, Z.H.; Cao, M.; Wei, G.Y.; Guo, X.; Zhang, J.; Lu, X.; Tang, Z. Ionomic and metabolomic analyses reveal different response mechanisms to saline-alkali stress between Suaeda salsa Community and Puccinellia tenuiflora Community. Front. Plant Sci. 2021, 12, 2500. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Lu, X.; Guo, X.; Pan, Y.; Yu, B.; Tang, Z.; Guo, Q. Differential responses to Cd Stress induced by exogenous application of Cu, Zn or Ca in the medicinal plant Catharanthus roseus. Ecotoxicol. Environ. Saf. 2018, 157, 266–275. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Zhang, K.; Sun, Y.; Cui, H.; Cao, S.; Xu, M. Growth, physiology, and transcriptional analysis of two contrasting Carex rigescens genotypes under salt stress reveals salt-tolerance mechanisms. J. Plant Physiol. 2018, 229, 77–88. [Google Scholar]
- Qian, L.; Yang, A.; Zhang, W.H. Efficient acquisition of iron confers greater tolerance to saline-alkaline stress in rice (Oryza sativa L.). J. Exp. Bot. 2016, 67, 6431–6444. [Google Scholar]
- Wang, X.S.; Ren, H.L.; Wei, Z.W.; Wang, Y.W.; Ren, W.B. Effects of neutral salt and alkali on ion distributions in the roots, shoots, and leaves of two Alfalfa cultivars with differing degrees of salt tolerance. J. Integr. Agric. 2017, 16, 1800–1807. [Google Scholar] [CrossRef]
- Yang, Y.; Guo, Y. Unraveling salt stress signaling in plants. J. Integr. Plant Biol. 2018, 60, 58–66. [Google Scholar] [CrossRef]
- Zhang, Y.; Fang, J.; Wu, X.; Dong, L. Na+/K+ balance and transport regulatory mechanisms in weedy and cultivated rice (Oryza sativa L.) under salt stress. BMC Plant Biol. 2018, 18, 375. [Google Scholar] [CrossRef]
- Kleczkowski, L.A.; Igamberdiev, A.U. Magnesium signaling in plants. Int. J. Mol. Sci. 2021, 22, 1159. [Google Scholar] [CrossRef]
- Raven, J.A. Interactions between above and below ground plant structures: Mechanisms and ecosystem services. Front. Agric. Sci. Eng. 2022, 9, 17. [Google Scholar]
- Alpaslan, M.; Gunes, A. Interactive effects of boron and salinity stress on the growth, membrane permeability and mineral composition of tomato and cucumber plants. Plant Soil 2001, 236, 123–128. [Google Scholar] [CrossRef]
- Bellaloui, N. Effect of water stress and foliar boron application on seed protein, oil, fatty acids, and nitrogen metabolism in soybean. Am. J. Plant Sci. 2011, 2, 692–701. [Google Scholar] [CrossRef]
- Sun, X.; Hu, C.; Tan, Q.; Liu, J.; Liu, H. Endogenous hormone in response to molybdenum in winter wheat roots under low temperature stress. J. Food Agric. Environ. 2010, 8, 597–601. [Google Scholar]
- Mizuno, H.; Kasuga, S.; Kawahigashi, H. Root lodging is a physical stress that changes gene expression from sucrose accumulation to degradation in sorghum. BMC Plant Biol. 2018, 18, 2–14. [Google Scholar] [CrossRef]
- Forieri, I.; Hildebrandt, U.; Rostás, M. Salinity stress effects on direct and indirect defence metabolites in Maize. Environ. Exp. Bot. 2016, 122, 68–77. [Google Scholar] [CrossRef]
- Abdallah, S.B.; Aung, B.; Amyot, L.; Lalin, I.; Lachaal, M.; Karray-Bouraoui, N.; Hannoufa, A. Salt Stress (NaCl) affects plant growth and branch pathways of carotenoid and flavonoid biosyntheses in Solanum nigrum. Acta Physiol. Plant. 2016, 38, 72. [Google Scholar] [CrossRef]
- Lin, J.; Yu, D.; Shi, Y.; Sheng, H.; Li, C.; Wang, Y.; Mu, C.; Li, X. Salt-alkali tolerance during germination and establishment of Leymus chinensis in the Songnen Grassland of China. Ecol. Eng. 2016, 95, 763–769. [Google Scholar] [CrossRef]
- Song, T.; Xu, H.; Na, S.; Liu, J.; Tian, P.; Yong, Y.; Yang, W.; Cai, H.; Cui, G. Metabolomic analysis of Alfalfa (Medicago sativa L.) root-symbiotic rhizobia responses under alkali stress. Front. Plant Sci. 2017, 8, 1208–1223. [Google Scholar] [CrossRef]
- Zhao, Z.; Liu, J.; Jia, R.; Bao, S.; Haixia; Chen, X. Physiological and Tmt-based proteomic analysis of oat early seedlings in response to alkali stress. J. Proteom. 2019, 193, 10–26. [Google Scholar] [CrossRef]
- Fan, W.; Ge, G.; Liu, Y.; Wang, W.; Liu, L.; Jia, Y. Proteomics integrated with metabolomics: Analysis of the internal causes of nutrient changes in Alfalfa at different growth stages. BMC Plant Biol. 2018, 18, 78–93. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Wang, Y.; He, Q.; Li, H.; Zhang, X.; Zhang, F. Comparative proteomics illustrates the complexity of drought resistance mechanisms in two wheat (Triticum aestivum L.) cultivars under dehydration and rehydration. BMC Plant Biol. 2016, 16, 188–211. [Google Scholar] [CrossRef] [PubMed]
- Carvalhais, L.C.; Schenk, P.M.; Dennis, P.G. Jasmonic acid signalling and the plant holobiont. Curr. Opin. Microbiol. 2017, 37, 42–47. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.S.; Zhang, J.; Li, M.X.; Shi, L.X. Metabolomics analysis reveals the salt-tolerant mechanism in Glycine soja. J. Plant Growth Regul. 2017, 36, 460–471. [Google Scholar] [CrossRef]
- Salama, K.H.A.; Mansour, M.M.F. Choline priming-induced plasma membrane lipid alterations contributed to improved wheat salt tolerance. Acta Physiol. Plant. 2015, 37, 170–177. [Google Scholar] [CrossRef]
- Khan, M.I.R.; Per, F.; Anjum, T.S.; Khan, N.A. Salicylic acid-induced abiotic stress tolerance and underlying mechanisms in plants. Front. Plant Sci. 2015, 6, 462–479. [Google Scholar] [CrossRef]
- Pang, Q.Y.; Zhang, Q.; Zang, W.; Wei, L.; Yan, X. Integrated proteomics and metabolomics for dissecting the mechanism of global responses to salt and alkali stress in Suaeda corniculata. Plant Soil 2016, 402, 379–394. [Google Scholar] [CrossRef]
- Dünser, K.; Kleine-Vehn, J. Differential growth regulation in plants-the acid growth balloon theory. Curr. Opin. Plant Biol. 2015, 28, 55–59. [Google Scholar] [CrossRef]
- Petropoulos, S.A.; Levizou, E.; Ntatsi, G.; Fernandes, A.; Pertrotos, K.; Akoumianakis, K.; Barros, L.; Ferreira, I. Salinity effect on nutritional value, chemical composition and bioactive compounds content of Cichorium spinosum L. Food Chem. 2017, 214, 129–136. [Google Scholar] [CrossRef]
- Sarker, U.; Oba, S. Augmentation of leaf color parameters, pigments, vitamins, phenolic acids, flavonoids and antioxidant activity in selected Amaranthus tricolor under salinity stress. Entific. Rep. 2018, 8, 12349. [Google Scholar] [CrossRef]
- Kumar, S.; Li, G.; Yang, J.; Huang, X.F.; Ji, Q.; Zhou, K.; Khan, S.; Ke, W.D.; Hou, H.W. Investigation of an antioxidative system for salinity tolerance in Oenanthe javanica. Antioxidants 2020, 9, 940. [Google Scholar] [CrossRef] [PubMed]
- Pi, E.; Zhu, C.; Wei, F.; Huang, Y.; Qu, L.; Li, Y.; Zhao, Q.; Ding, F.; Qiu, L.; Wang, H.; et al. Quantitative phosphoproteomic and metabolomic analyses reveal Gmmyb173 optimizes flavonoid metabolism in soybean under salt stress. Mol. Cell. Proteom. 2018, 17, 1209–1224. [Google Scholar] [CrossRef] [PubMed]
- Ma, D.; Sun, D.; Wang, C.; Li, Y.; Guo, T. Expression of flavonoid biosynthesis genes and accumulation of flavonoid in wheat leaves in response to drought stress. Plant Physiol. Biochem. 2014, 2014, 80. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.; Wang, S.; Huang, Z.; Zhang, S.; Liao, Q.; Zhang, C.; Tao, L.; Mao, Q.; Meng, P.; Yang, C.; et al. Rewiring of the fruit metabolome in tomato breeding. Cell 2018, 172, 249. [Google Scholar] [CrossRef]
- Ma, S.; Lv, L.; Meng, C.; Zhang, C.; Li, Y. Integrative analysis of the metabolome and transcriptome of sorghum bicolor reveals dynamic changes in flavonoids accumulation under saline-alkali stress. J. Agric. Food Chem. 2020, 68, 14781–14789. [Google Scholar] [CrossRef]
- Deng, B.; Li, Y.; Lei, G.; Liu, G. Effects of nitrogen availability on mineral nutrient balance and flavonoid accumulation in Cyclocarya paliurus. Plant Physiol. Biochem. 2019, 135, 111–118. [Google Scholar] [CrossRef]
- Li, Z.; Jiang, H.; Yan, H.; Jiang, X.; Ma, Y.; Qin, Y. Carbon and nitrogen metabolism under nitrogen variation affects flavonoid accumulation in the leaves of Coreopsis tinctoria. Peer J. 2021, 9, e12152. [Google Scholar] [CrossRef]
- Kennington, S. Signal transduction in the light regulation of the flavonoid biosynthetic pathway in Arabidopsis. Atherosclerosis 2006, 174, 343–347. [Google Scholar]
- Li, Z.Y.; Jiang, H.; Qin, Y.N.; Yan, H.Z.; Jiang, X.M.; Qin, Y. Nitrogen deficiency maintains the yield and improves the antioxidant activity of Coreopsis tinctoria Nutt. Biosci. Biotechnol. Biochem. 2021, 85, 1492–1505. [Google Scholar] [CrossRef]
- Wang, F.; Kong, W.; Wong, G.; Fu, L.; Peng, R.; Li, Z.; Yao, Q. Atmyb12 regulates flavonoids accumulation and abiotic stress tolerance in transgenic Arabidopsis thaliana. Mol. Genet. Genom. 2016, 291, 1545–1559. [Google Scholar] [CrossRef]
- Zhuang, X.Y.; Zhao, B.; Liu, S.; Song, F.R.; Cui, F.C.; Liu, Z.Q.; Li, Y.Q. Noncovalent interactions between superoxide dismutase and flavonoids studied by native mass spectrometry combined with molecular simulations. Anal. Chem. 2016, 88, 11720–11726. [Google Scholar] [CrossRef] [PubMed]
- Kovinich, N.; Kayanja, G.; Chanoca, A.; Otegui, M.S.; Grotewold, E. Abiotic stresses induce different localizations of anthocyanins in Arabidopsis. Plant Signal. Behav. 2015, 10, e1027850. [Google Scholar] [CrossRef] [PubMed]
- Rietjens, I.M.; Huseiny, W.A.; Boersma, M.G. Flavonoids and alkenylbenzenes: New concepts in bioactivation studies. Chem. Biol. Interact. 2011, 192, 87–95. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).