CD70-Targeted Micelles Enhance HIF2α siRNA Delivery and Inhibit Oncogenic Functions in Patient-Derived Clear Cell Renal Carcinoma Cells
Abstract
1. Introduction
2. Results and Discussion
2.1. Synthesis of HIF2α-CD27 PAMs and Characterization of siRNA Loading and Release
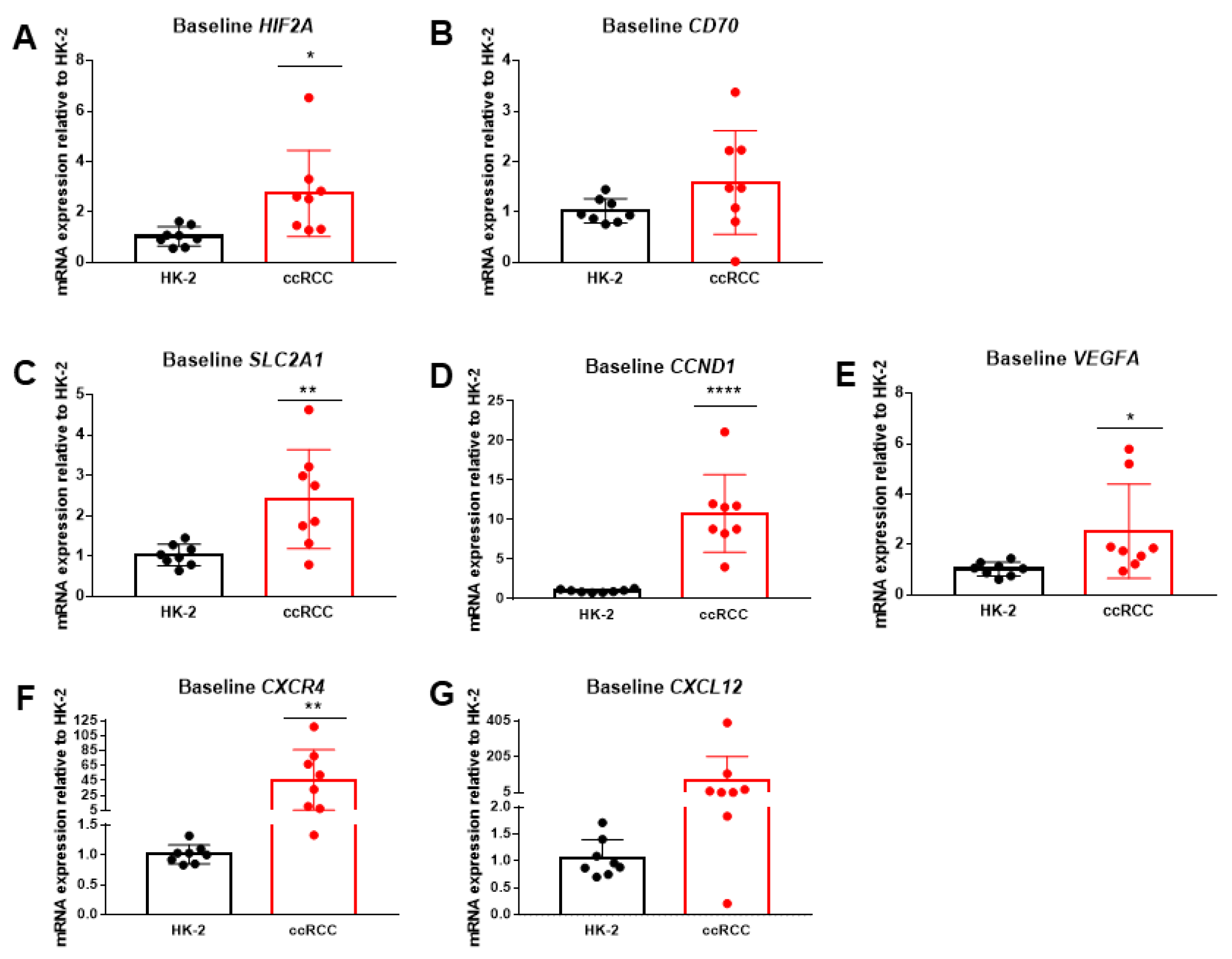
2.2. HIF2α and Downstream Genes Are Upregulated in Patient-Derived ccRCC Cells
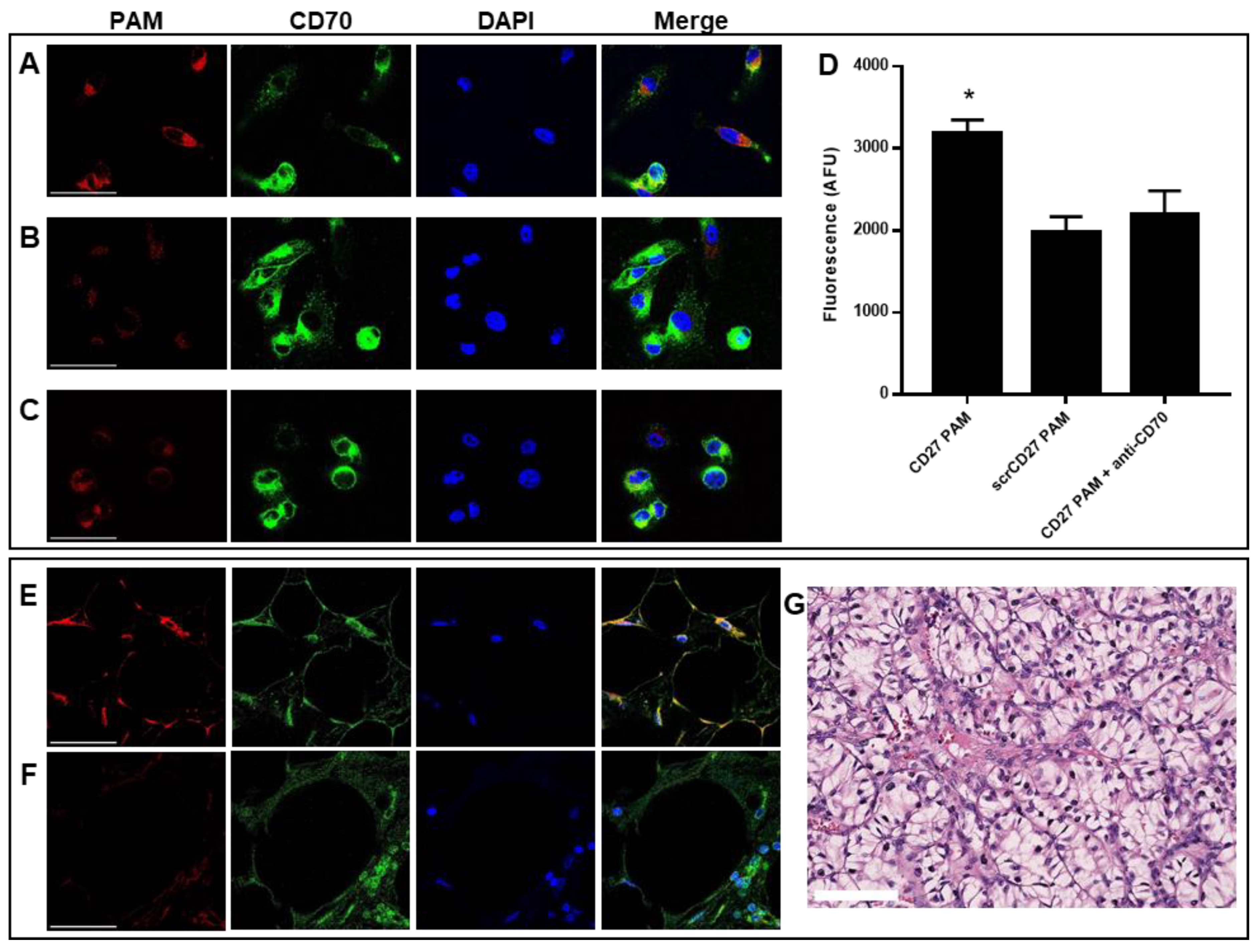
2.3. CD70-Targeting Micelles Bind to Patient-Derived ccRCC Cells In Vitro
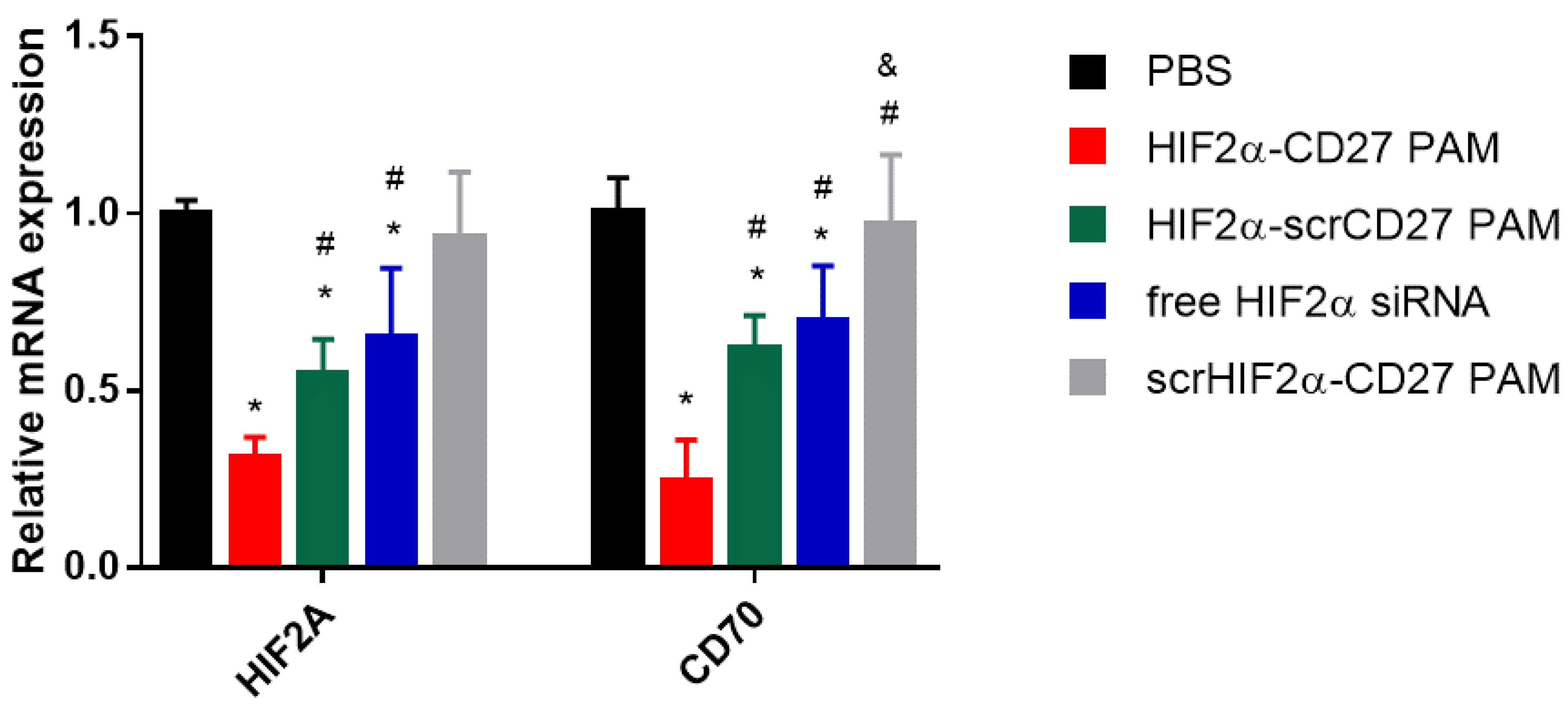
2.4. HIF2α-CD27 PAM Treatment Reduces HIF2α mRNA Expression In Vitro
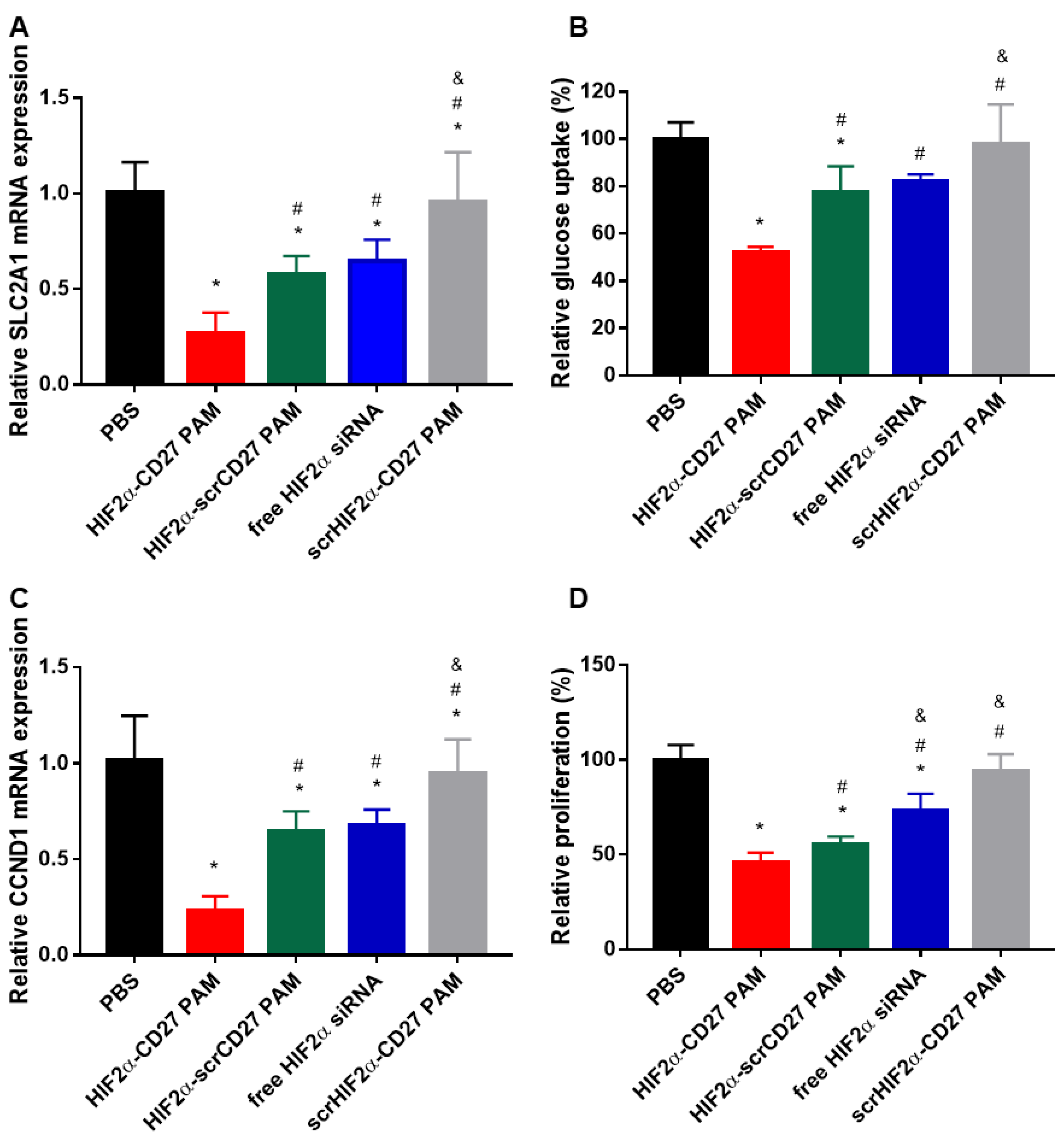
2.5. HIF2α-CD27 PAMs Inhibit In Vitro Glucose Transport and Proliferation by Reducing SLC2A1 and CCND1 Expression
2.6. Culture Medium Collected from HIF2α-CD27 PAM-Treated ccRCC Cells Shows Anti-Angiogenic Properties
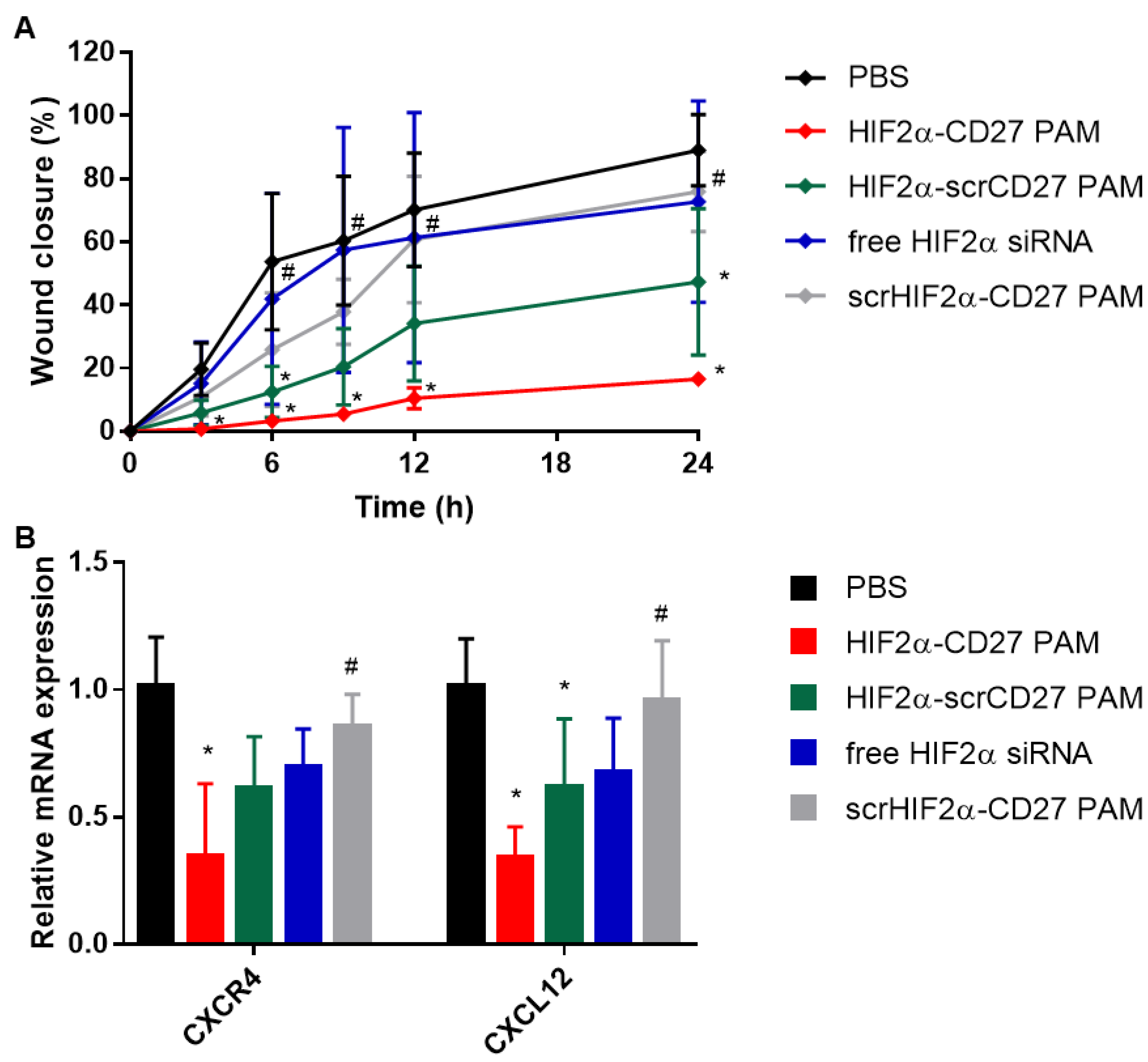
2.7. HIF2α-CD27 PAM Treatment Reduces Patient-Derived ccRCC Cell Migration and Wound Closure
3. Conclusions
4. Materials and Methods
4.1. Materials and Cells
4.2. Synthesis of HIF2α-CD27 Peptide Amphiphile Micelles
4.3. Dynamic Light Scattering (DLS) and Zeta Potential
4.4. Transmission Electron Microscopy (TEM)
4.5. Gel Electrophoresis Assay
4.6. Characterization of siRNA Release
4.7. Isolation and Culture of Patient-Derived ccRCC Cells
4.8. In Vitro Micelle Binding to Patient-Derived ccRCC Cells
4.9. Immunohistochemical (IHC) Staining and PAM Binding to ccRCC Patient Tissue Sections
4.10. In Vitro Transfection and mRNA Expression of Patient-Derived ccRCC Cells
4.11. MTS Assay
4.12. Glucose Uptake Assay
4.13. Collection of ccRCC-Conditioned Cell Culture Medium and In Vitro Culture and Growth of Endothelial Cells
4.14. Wound Healing Assay
4.15. Statistics
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Padala, S.A.; Barsouk, A.; Thandra, K.C.; Saginala, K.; Mohammed, A.; Vakiti, A.; Rawla, P.; Barsouk, A. Epidemiology of Renal Cell Carcinoma. World J. Oncol. 2020, 11, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Wierzbicki, P.M.; Klacz, J.; Kotulak-Chrzaszcz, A.; Wronska, A.; Stanislawowski, M.; Rybarczyk, A.; Ludziejewska, A.; Kmiec, Z.; Matuszewski, M. Prognostic significance of VHL, HIF1A, HIF2A, VEGFA and p53 expression in patients with clearcell renal cell carcinoma treated with sunitinib as firstline treatment. Int. J. Oncol. 2019, 55, 371–390. [Google Scholar] [CrossRef] [PubMed]
- Dagher, J.; Kammerer-Jacquet, S.F.; Dugay, F.; Beaumont, M.; Lespagnol, A.; Cornevin, L.; Verhoest, G.; Bensalah, K.; Rioux-Leclercq, N.; Belaud-Rotureau, M.A. Clear cell renal cell carcinoma: A comparative study of histological and chromosomal characteristics between primary tumors and their corresponding metastases. Virchows Arch. 2017, 471, 107–115. [Google Scholar] [CrossRef] [PubMed]
- Serzan, M.T.; Atkins, M.B. Current and emerging therapies for first line treatment of metastatic clear cell renal cell carcinoma. J. Cancer Metastasis. Treat. 2021, 7, 39. [Google Scholar] [CrossRef]
- Motzer, R.; Alekseev, B.; Rha, S.Y.; Porta, C.; Eto, M.; Powles, T.; Grunwald, V.; Hutson, T.E.; Kopyltsov, E.; Mendez-Vidal, M.J.; et al. Lenvatinib plus Pembrolizumab or Everolimus for Advanced Renal Cell Carcinoma. N. Engl. J. Med. 2021, 384, 1289–1300. [Google Scholar] [CrossRef]
- Motzer, R.J.; Penkov, K.; Haanen, J.; Rini, B.; Albiges, L.; Campbell, M.T.; Venugopal, B.; Kollmannsberger, C.; Negrier, S.; Uemura, M.; et al. Avelumab plus Axitinib versus Sunitinib for Advanced Renal-Cell Carcinoma. N. Engl. J. Med. 2019, 380, 1103–1115. [Google Scholar] [CrossRef]
- Rini, B.I.; Plimack, E.R.; Stus, V.; Gafanov, R.; Hawkins, R.; Nosov, D.; Pouliot, F.; Alekseev, B.; Soulieres, D.; Melichar, B.; et al. Pembrolizumab plus Axitinib versus Sunitinib for Advanced Renal-Cell Carcinoma. N. Engl. J. Med. 2019, 380, 1116–1127. [Google Scholar] [CrossRef]
- Razafinjatovo, C.; Bihr, S.; Mischo, A.; Vogl, U.; Schmidinger, M.; Moch, H.; Schraml, P. Characterization of VHL missense mutations in sporadic clear cell renal cell carcinoma: Hotspots, affected binding domains, functional impact on pVHL and therapeutic relevance. BMC Cancer 2016, 16, 638. [Google Scholar] [CrossRef]
- Nickerson, M.L.; Jaeger, E.; Shi, Y.; Durocher, J.A.; Mahurkar, S.; Zaridze, D.; Matveev, V.; Janout, V.; Kollarova, H.; Bencko, V.; et al. Improved identification of von Hippel-Lindau gene alterations in clear cell renal tumors. Clin. Cancer Res. 2008, 14, 4726–4734. [Google Scholar] [CrossRef]
- Young, A.C.; Craven, R.A.; Cohen, D.; Taylor, C.; Booth, C.; Harnden, P.; Cairns, D.A.; Astuti, D.; Gregory, W.; Maher, E.R.; et al. Analysis of VHL Gene Alterations and their Relationship to Clinical Parameters in Sporadic Conventional Renal Cell Carcinoma. Clin. Cancer Res. 2009, 15, 7582–7592. [Google Scholar] [CrossRef]
- Baldewijns, M.M.; van Vlodrop, I.J.; Vermeulen, P.B.; Soetekouw, P.M.; van Engeland, M.; de Bruine, A.P. VHL and HIF signalling in renal cell carcinogenesis. J. Pathol. 2010, 221, 125–138. [Google Scholar] [CrossRef] [PubMed]
- Ruf, M.; Moch, H.; Schraml, P. PD-L1 expression is regulated by hypoxia inducible factor in clear cell renal cell carcinoma. Int. J. Cancer 2016, 139, 396–403. [Google Scholar] [CrossRef]
- Martinez-Saez, O.; Gajate Borau, P.; Alonso-Gordoa, T.; Molina-Cerrillo, J.; Grande, E. Targeting HIF-2 alpha in clear cell renal cell carcinoma: A promising therapeutic strategy. Crit. Rev. Oncol. Hematol. 2017, 111, 117–123. [Google Scholar] [CrossRef] [PubMed]
- Arnaiz, E.; Miar, A.; Bridges, E.; Prasad, N.; Hatch, S.B.; Ebner, D.; Lawrie, C.H.; Harris, A.L. Differential effects of HIF2alpha antagonist and HIF2alpha silencing in renal cancer and sensitivity to repurposed drugs. BMC Cancer 2021, 21, 896. [Google Scholar] [CrossRef] [PubMed]
- Micucci, C.; Matacchione, G.; Valli, D.; Orciari, S.; Catalano, A. HIF2alpha is involved in the expansion of CXCR4-positive cancer stem-like cells in renal cell carcinoma. Br. J. Cancer 2015, 113, 1178–1185. [Google Scholar] [CrossRef]
- Kondo, K.; Kim, W.Y.; Lechpammer, M.; Kaelin, W.G., Jr. Inhibition of HIF2alpha is sufficient to suppress pVHL-defective tumor growth. PLoS Biol. 2003, 1, E83. [Google Scholar] [CrossRef]
- Jonasch, E.; Donskov, F.; Iliopoulos, O.; Rathmell, W.K.; Narayan, V.K.; Maughan, B.L.; Oudard, S.; Else, T.; Maranchie, J.K.; Welsh, S.J.; et al. Belzutifan for Renal Cell Carcinoma in von Hippel-Lindau Disease. N. Engl. J. Med. 2021, 385, 2036–2046. [Google Scholar] [CrossRef]
- McCabe, E.M.; Lee, S.; Rasmussen, T.P. Belzutifan (Welireg) for von Hippel Lindau disease. Trends Pharmacol. Sci. 2022, 43, 882–883. [Google Scholar] [CrossRef]
- Wong, S.C.; Cheng, W.; Hamilton, H.; Nicholas, A.L.; Wakefield, D.H.; Almeida, A.; Blokhin, A.V.; Carlson, J.; Neal, Z.C.; Subbotin, V.; et al. HIF2alpha-Targeted RNAi Therapeutic Inhibits Clear Cell Renal Cell Carcinoma. Mol. Cancer Ther. 2018, 17, 140–149. [Google Scholar] [CrossRef]
- Rankin, E.B.; Biju, M.P.; Liu, Q.; Unger, T.L.; Rha, J.; Johnson, R.S.; Simon, M.C.; Keith, B.; Haase, V.H. Hypoxia-inducible factor-2 (HIF-2) regulates hepatic erythropoietin in vivo. J. Clin. Investig. 2007, 117, 1068–1077. [Google Scholar] [CrossRef]
- Huang, Y.; Wang, J.; Jiang, K.; Chung, E.J. Improving kidney targeting: The influence of nanoparticle physicochemical properties on kidney interactions. J. Control. Release 2021, 334, 127–137. [Google Scholar] [CrossRef] [PubMed]
- Trac, N.T.; Chung, E.J. Peptide-based targeting of immunosuppressive cells in cancer. Bioact. Mater. 2020, 5, 92–101. [Google Scholar] [CrossRef] [PubMed]
- Chenthamara, D.; Subramaniam, S.; Ramakrishnan, S.G.; Krishnaswamy, S.; Essa, M.M.; Lin, F.H.; Qoronfleh, M.W. Therapeutic efficacy of nanoparticles and routes of administration. Biomater. Res. 2019, 23, 20. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Parveen, R.; Chatterji, B.P. Toxicology of Nanoparticles in Drug Delivery. Curr. Pathobiol. Rep. 2021, 9, 133–144. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Li, Q.; Chen, D.; Wu, B.; Wu, Y.; Tong, W.; Huang, P. Kidney-targeted rhein-loaded liponanoparticles for diabetic nephropathy therapy via size control and enhancement of renal cellular uptake. Theranostics 2019, 9, 6191–6208. [Google Scholar] [CrossRef]
- Kamaly, N.; He, J.C.; Ausiello, D.A.; Farokhzad, O.C. Nanomedicines for renal disease: Current status and future applications. Nat. Rev. Nephrol. 2016, 12, 738–753. [Google Scholar] [CrossRef]
- Wang, J.; Poon, C.; Chin, D.; Milkowski, S.; Lu, V.; Hallows, K.R.; Chung, E.J. Design and in vivo characterization of kidney-targeting multimodal micelles for renal drug delivery. Nano Res. 2018, 11, 5584–5595. [Google Scholar] [CrossRef]
- Tripathy, N.; Wang, J.; Tung, M.; Conway, C.; Chung, E.J. Transdermal Delivery of Kidney-Targeting Nanoparticles Using Dissolvable Microneedles. Cell Mol. Bioeng. 2020, 13, 475–486. [Google Scholar] [CrossRef]
- Pal, S.K.; Forero-Torres, A.; Thompson, J.A.; Morris, J.C.; Chhabra, S.; Hoimes, C.J.; Vogelzang, N.J.; Boyd, T.; Bergerot, P.G.; Adashek, J.J.; et al. A phase 1 trial of SGN-CD70A in patients with CD70-positive, metastatic renal cell carcinoma. Cancer 2019, 125, 1124–1132. [Google Scholar] [CrossRef]
- Jilaveanu, L.B.; Sznol, J.; Aziz, S.A.; Duchen, D.; Kluger, H.M.; Camp, R.L. CD70 expression patterns in renal cell carcinoma. Hum. Pathol. 2012, 43, 1394–1399. [Google Scholar] [CrossRef]
- Teplyakov, A.; Obmolova, G.; Malia, T.J.; Gilliland, G.L. Crystal structure of CD27 in complex with a neutralizing noncompeting antibody. Acta Cryst. F Struct. Biol. Commun. 2017, 73, 294–299. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Jiang, K.; Zhang, X.; Chung, E.J. The effect of size, charge, and peptide ligand length on kidney targeting by small, organic nanoparticles. Bioeng. Transl. Med. 2020, 5, e10173. [Google Scholar] [CrossRef] [PubMed]
- Poon, C.; Chowdhuri, S.; Kuo, C.H.; Fang, Y.; Alenghat, F.J.; Hyatt, D.; Kani, K.; Gross, M.E.; Chung, E.J. Protein Mimetic and Anticancer Properties of Monocyte-Targeting Peptide Amphiphile Micelles. ACS Biomater. Sci. Eng. 2017, 3, 3273–3282. [Google Scholar] [CrossRef] [PubMed]
- Sato, Y.; Matsui, H.; Sato, R.; Harashima, H. Neutralization of negative charges of siRNA results in improved safety and efficient gene silencing activity of lipid nanoparticles loaded with high levels of siRNA. J. Control. Release 2018, 284, 179–187. [Google Scholar] [CrossRef] [PubMed]
- Kasai, H.; Inoue, K.; Imamura, K.; Yuvienco, C.; Montclare, J.K.; Yamano, S. Efficient siRNA delivery and gene silencing using a lipopolypeptide hybrid vector mediated by a caveolae-mediated and temperature-dependent endocytic pathway. J. Nanobiotechnol. 2019, 17, 11. [Google Scholar] [CrossRef]
- Jeong, E.J.; Lee, J.; Kim, H.S.; Lee, K.Y. In Vitro Cellular Uptake and Transfection of Oligoarginine-Conjugated Glycol Chitosan/siRNA Nanoparticles. Polymers 2021, 13, 4219. [Google Scholar] [CrossRef]
- Houseley, J.; Tollervey, D. The many pathways of RNA degradation. Cell 2009, 136, 763–776. [Google Scholar] [CrossRef]
- Aranda, P.S.; LaJoie, D.M.; Jorcyk, C.L. Bleach gel: A simple agarose gel for analyzing RNA quality. Electrophoresis 2012, 33, 366–369. [Google Scholar] [CrossRef]
- Lushchak, V.I. Glutathione homeostasis and functions: Potential targets for medical interventions. J. Amino Acids 2012, 2012, 736837. [Google Scholar] [CrossRef]
- Forman, H.J.; Zhang, H.; Rinna, A. Glutathione: Overview of its protective roles, measurement, and biosynthesis. Mol. Asp. Med. 2009, 30, 1–12. [Google Scholar] [CrossRef]
- Lv, H.; Zhen, C.; Liu, J.; Yang, P.; Hu, L.; Shang, P. Unraveling the Potential Role of Glutathione in Multiple Forms of Cell Death in Cancer Therapy. Oxid. Med. Cell Longev. 2019, 2019, 3150145. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Meierhofer, D. Glutathione Metabolism in Renal Cell Carcinoma Progression and Implications for Therapies. Int. J. Mol. Sci. 2019, 20, 3672. [Google Scholar] [CrossRef] [PubMed]
- Kurozumi, R.; Kojima, S. Increase of intracellular glutathione by low-level NO mediated by transcription factor NF-kappaB in RAW 264.7 cells. Biochim. Biophys. Acta 2005, 1744, 58–67. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Deponte, M. Glutathione catalysis and the reaction mechanisms of glutathione-dependent enzymes. Biochim. Biophys. Acta 2013, 1830, 3217–3266. [Google Scholar] [CrossRef] [PubMed]
- Melendez-Rodriguez, F.; Roche, O.; Sanchez-Prieto, R.; Aragones, J. Hypoxia-Inducible Factor 2-Dependent Pathways Driving Von Hippel-Lindau-Deficient Renal Cancer. Front. Oncol. 2018, 8, 214. [Google Scholar] [CrossRef]
- Chen, W.; Hill, H.; Christie, A.; Kim, M.S.; Holloman, E.; Pavia-Jimenez, A.; Homayoun, F.; Ma, Y.; Patel, N.; Yell, P.; et al. Targeting renal cell carcinoma with a HIF-2 antagonist. Nature 2016, 539, 112–117. [Google Scholar] [CrossRef]
- Adam, P.J.; Terrett, J.A.; Steers, G.; Stockwin, L.; Loader, J.A.; Fletcher, G.C.; Lu, L.S.; Leach, B.I.; Mason, S.; Stamps, A.C.; et al. CD70 (TNFSF7) is expressed at high prevalence in renal cell carcinomas and is rapidly internalised on antibody binding. Br. J. Cancer 2006, 95, 298–306. [Google Scholar] [CrossRef]
- Raval, R.R.; Lau, K.W.; Tran, M.G.; Sowter, H.M.; Mandriota, S.J.; Li, J.L.; Pugh, C.W.; Maxwell, P.H.; Harris, A.L.; Ratcliffe, P.J. Contrasting properties of hypoxia-inducible factor 1 (HIF-1) and HIF-2 in von Hippel-Lindau-associated renal cell carcinoma. Mol. Cell Biol. 2005, 25, 5675–5686. [Google Scholar] [CrossRef]
- Shinojima, T.; Oya, M.; Takayanagi, A.; Mizuno, R.; Shimizu, N.; Murai, M. Renal cancer cells lacking hypoxia inducible factor (HIF)-1alpha expression maintain vascular endothelial growth factor expression through HIF-2alpha. Carcinogenesis 2007, 28, 529–536. [Google Scholar] [CrossRef]
- Choi, W.S.W.; Boland, J.; Lin, J. Hypoxia-Inducible Factor-2alpha as a Novel Target in Renal Cell Carcinoma. J. Kidney Cancer VHL 2021, 8, 1–7. [Google Scholar] [CrossRef]
- Delahunt, B. Advances and controversies in grading and staging of renal cell carcinoma. Mod. Pathol. 2009, 22 (Suppl. 2), S24–S36. [Google Scholar] [CrossRef] [PubMed]
- Warren, A.Y.; Harrison, D. WHO/ISUP classification, grading and pathological staging of renal cell carcinoma: Standards and controversies. World J. Urol. 2018, 36, 1913–1926. [Google Scholar] [CrossRef]
- Samaratunga, H.; Gianduzzo, T.; Delahunt, B. The ISUP system of staging, grading and classification of renal cell neoplasia. J. Kidney Cancer VHL 2014, 1, 26–39. [Google Scholar] [CrossRef] [PubMed]
- Ruf, M.; Mittmann, C.; Nowicka, A.M.; Hartmann, A.; Hermanns, T.; Poyet, C.; van den Broek, M.; Sulser, T.; Moch, H.; Schraml, P. pVHL/HIF-regulated CD70 expression is associated with infiltration of CD27+ lymphocytes and increased serum levels of soluble CD27 in clear cell renal cell carcinoma. Clin. Cancer Res. 2015, 21, 889–898. [Google Scholar] [CrossRef]
- Kitajima, S.; Lee, K.L.; Fujioka, M.; Sun, W.; You, J.; Chia, G.S.; Wanibuchi, H.; Tomita, S.; Araki, M.; Kato, H.; et al. Hypoxia-inducible factor-2 alpha up-regulates CD70 under hypoxia and enhances anchorage-independent growth and aggressiveness in cancer cells. Oncotarget 2018, 9, 19123–19135. [Google Scholar] [CrossRef] [PubMed]
- Pragallapati, S.; Manyam, R. Glucose transporter 1 in health and disease. J. Oral Maxillofac. Pathol. 2019, 23, 443–449. [Google Scholar] [CrossRef]
- Chan, D.A.; Sutphin, P.D.; Nguyen, P.; Turcotte, S.; Lai, E.W.; Banh, A.; Reynolds, G.E.; Chi, J.T.; Wu, J.; Solow-Cordero, D.E.; et al. Targeting GLUT1 and the Warburg effect in renal cell carcinoma by chemical synthetic lethality. Sci. Transl. Med. 2011, 3, 94ra70. [Google Scholar] [CrossRef]
- Adekola, K.; Rosen, S.T.; Shanmugam, M. Glucose transporters in cancer metabolism. Curr. Opin. Oncol. 2012, 24, 650–654. [Google Scholar] [CrossRef]
- Fu, M.; Wang, C.; Li, Z.; Sakamaki, T.; Pestell, R.G. Minireview: Cyclin D1: Normal and abnormal functions. Endocrinology 2004, 145, 5439–5447. [Google Scholar] [CrossRef]
- Alao, J.P. The regulation of cyclin D1 degradation: Roles in cancer development and the potential for therapeutic invention. Mol. Cancer 2007, 6, 24. [Google Scholar] [CrossRef]
- Alhawarat, F.M.; Hammad, H.M.; Hijjawi, M.S.; Sharab, A.S.; Abuarqoub, D.A.; Al Shhab, M.A.; Zihlif, M.A. The effect of cycling hypoxia on MCF-7 cancer stem cells and the impact of their microenvironment on angiogenesis using human umbilical vein endothelial cells (HUVECs) as a model. PeerJ. 2019, 7, e5990. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.; Yang, C.; Huang, J.; Liu, L.; Zhang, Y.; Li, S.; Zhang, L.; Sun, Q.; Yang, P. Conditioned medium derived from FGF-2-modified GMSCs enhances migration and angiogenesis of human umbilical vein endothelial cells. Stem Cell Res. Ther. 2020, 11, 68. [Google Scholar] [CrossRef] [PubMed]
- Barrett, J.C.; Ulery, B.D.; Trent, A.; Liang, S.; David, N.A.; Tirrell, M.V. Modular Peptide Amphiphile Micelles Improving an Antibody-Mediated Immune Response to Group A Streptococcus. ACS Biomater. Sci. Eng. 2017, 3, 144–152. [Google Scholar] [CrossRef] [PubMed]
- Veber, D.F.; Milkowski, J.D.; Varga, S.L.; Denkewalter, R.G.; Hirschmann, R. Acetamidomethyl. A novel thiol protecting group for cysteine. J. Am. Chem. Soc. 1972, 94, 5456–5461. [Google Scholar] [CrossRef]
- Trac, N.; Chen, L.Y.; Zhang, A.; Liao, C.P.; Poon, C.; Wang, J.; Ando, Y.; Joo, J.; Garri, C.; Shen, K.; et al. CCR2-targeted micelles for anti-cancer peptide delivery and immune stimulation. J. Control. Release 2021, 329, 614–623. [Google Scholar] [CrossRef]
- Chin, D.D.; Poon, C.; Wang, J.; Joo, J.; Ong, V.; Jiang, Z.; Cheng, K.; Plotkin, A.; Magee, G.A.; Chung, E.J. miR-145 micelles mitigate atherosclerosis by modulating vascular smooth muscle cell phenotype. Biomaterials 2021, 273, 120810. [Google Scholar] [CrossRef]
- He, Y.T.; Wan, J.; Tokunaga, T. Kinetic stability of hematite nanoparticles: The effect of particle sizes. J. Nanoparticle Res. 2008, 10, 321–332. [Google Scholar] [CrossRef]
- Christie, R.J.; Miyata, K.; Matsumoto, Y.; Nomoto, T.; Menasco, D.; Lai, T.C.; Pennisi, M.; Osada, K.; Fukushima, S.; Nishiyama, N.; et al. Effect of polymer structure on micelles formed between siRNA and cationic block copolymer comprising thiols and amidines. Biomacromolecules 2011, 12, 3174–3185. [Google Scholar] [CrossRef]
- Wu, Y.; Wang, W.; Chen, Y.; Huang, K.; Shuai, X.; Chen, Q.; Li, X.; Lian, G. The investigation of polymer-siRNA nanoparticle for gene therapy of gastric cancer in vitro. Int. J. Nanomed. 2010, 5, 129–136. [Google Scholar] [CrossRef]
- Valente, M.J.; Henrique, R.; Costa, V.L.; Jeronimo, C.; Carvalho, F.; Bastos, M.L.; de Pinho, P.G.; Carvalho, M. A rapid and simple procedure for the establishment of human normal and cancer renal primary cell cultures from surgical specimens. PLoS ONE 2011, 6, e19337. [Google Scholar] [CrossRef]
- Dowling, P.; Clynes, M. Conditioned media from cell lines: A complementary model to clinical specimens for the discovery of disease-specific biomarkers. Proteomics 2011, 11, 794–804. [Google Scholar] [CrossRef] [PubMed]
- Staton, C.A.; Stribbling, S.M.; Tazzyman, S.; Hughes, R.; Brown, N.J.; Lewis, C.E. Current methods for assaying angiogenesis in vitro and in vivo. Int. J. Exp. Pathol. 2004, 85, 233–248. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.C.; Park, A.Y.; Guan, J.L. In vitro scratch assay: A convenient and inexpensive method for analysis of cell migration in vitro. Nat. Protoc. 2007, 2, 329–333. [Google Scholar] [CrossRef] [PubMed]


| Sample | Grade | Tumor Size (cm) | Stage |
|---|---|---|---|
| 1 | 3 | 2.5 | I |
| 2 | 2 | 6.3 | III |
| 3 | 3 | 4.7 | III |
| 4 | 3 | 11 | III |
| 5 | 2 | 3.6 | IV * |
| 6 | 2 | 4.1 | I |
| 7 | 3 | 15.6 | III |
| 8 | 4 | 13.5 | III |
| Gene | N (%) |
|---|---|
| HIF2A | 5/8 (63%) |
| CD70 | 5/8 (63%) |
| SLC2A1 | 5/8 (63%) |
| CCND1 | 8/8 (100%) |
| VEGFA | 7/8 (88%) |
| CXCR4 | 7/8 (88%) |
| CXCL12 | 7/8 (88%) |
| Sample | ||||||||
|---|---|---|---|---|---|---|---|---|
| Gene | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 |
| HIF2A | 6.54 * ± 1.99 | 3.31 * ± 0.39 | 1.28 ± 0.39 | 1.47 ± 0.18 | 2.84 * ± 0.72 | 1.32 ± 0.19 | 2.61 * ± 0.50 | 2.52 * ± 0.67 |
| CD70 | 0.81 ± 0.25 | 1.08 ± 0.14 | 2.23 * ± 0.36 | 2.22 * ± 0.45 | 3.38 * ± 0.24 | 1.48 * ± 0.22 | 0.02 * ± 0.00 | 1.48 * ± 0.18 |
| SLC2A1 | 1.32 ± 0.08 | 4.63 * ± 1.10 | 1.86 * ± 0.77 | 1.75 * ± 0.17 | 3.22 * ± 0.68 | 2.75 * ± 0.47 | 0.79 ± 0.11 | 2.99 * ± 0.02 |
| CCND1 | 8.83 * ± 1.23 | 8.82 * ± 1.02 | 11.75 * ± 1.55 | 11.58 * ± 0.28 | 8.27 * ± 1.22 | 4.04 * ± 0.80 | 21.09 * ± 3.35 | 12.00 * ± 2.54 |
| VEGFA | 1.91 * ± 0.18 | 5.20 * ± 0.95 | 0.95 ± 0.19 | 1.75 * ± 0.14 | 5.79 * ± 1.40 | 1.86 * ± 0.21 | 1.23 * ± 0.16 | 1.55 * ± 0.14 |
| CXCR4 | 116.90 * ± 75.61 | 66.95 * ± 25.37 | 33.25 * ± 8.98 | 7.40 * ± 3.89 | 10.45 * ± 4.19 | 52.47 * ± 18.75 | 1.33 ± 0.55 | 77.82 * ± 14.67 |
| CXCL12 | 393.77 * ± 219.76 | 0.21 * ± 0.13 | 1.83 * ± 0.44 | 11.59 * ± 3.00 | 4.06 * ± 1.85 | 20.73 * ± 7.77 | 109.59 * ± 18.41 | 4.38 * ± 2.76 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Trac, N.; Oh, H.S.; Jones, L.I.; Caliliw, R.; Ohtake, S.; Shuch, B.; Chung, E.J. CD70-Targeted Micelles Enhance HIF2α siRNA Delivery and Inhibit Oncogenic Functions in Patient-Derived Clear Cell Renal Carcinoma Cells. Molecules 2022, 27, 8457. https://doi.org/10.3390/molecules27238457
Trac N, Oh HS, Jones LI, Caliliw R, Ohtake S, Shuch B, Chung EJ. CD70-Targeted Micelles Enhance HIF2α siRNA Delivery and Inhibit Oncogenic Functions in Patient-Derived Clear Cell Renal Carcinoma Cells. Molecules. 2022; 27(23):8457. https://doi.org/10.3390/molecules27238457
Chicago/Turabian StyleTrac, Noah, Hyun Seok Oh, Leila Izzy Jones, Randy Caliliw, Shinji Ohtake, Brian Shuch, and Eun Ji Chung. 2022. "CD70-Targeted Micelles Enhance HIF2α siRNA Delivery and Inhibit Oncogenic Functions in Patient-Derived Clear Cell Renal Carcinoma Cells" Molecules 27, no. 23: 8457. https://doi.org/10.3390/molecules27238457
APA StyleTrac, N., Oh, H. S., Jones, L. I., Caliliw, R., Ohtake, S., Shuch, B., & Chung, E. J. (2022). CD70-Targeted Micelles Enhance HIF2α siRNA Delivery and Inhibit Oncogenic Functions in Patient-Derived Clear Cell Renal Carcinoma Cells. Molecules, 27(23), 8457. https://doi.org/10.3390/molecules27238457








