Can Crude Oil Exploration Influence the Phytochemicals and Bioactivity of Medicinal Plants? A Case of Nigerian Vernonia amygdalina and Ocimum gratissimum
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials and Reagents
2.2. Sample Preparation and Extraction
2.3. Mass Spectral Data Acquisition
2.4. Phytochemical Analysis of Metabolites of VAL and OGL Using UNIFI Platform
2.5. Metabolomic Analysis
2.6. Blood Sample Collection and Ethical Approval
2.7. In Vitro Sickling Reversal Assay
2.8. Isolation of Secondary Metabolites
2.9. Molecular Docking
2.10. 3D Pharmacophore Model Generation
2.11. Statistical Analysis
3. Results and Discussion
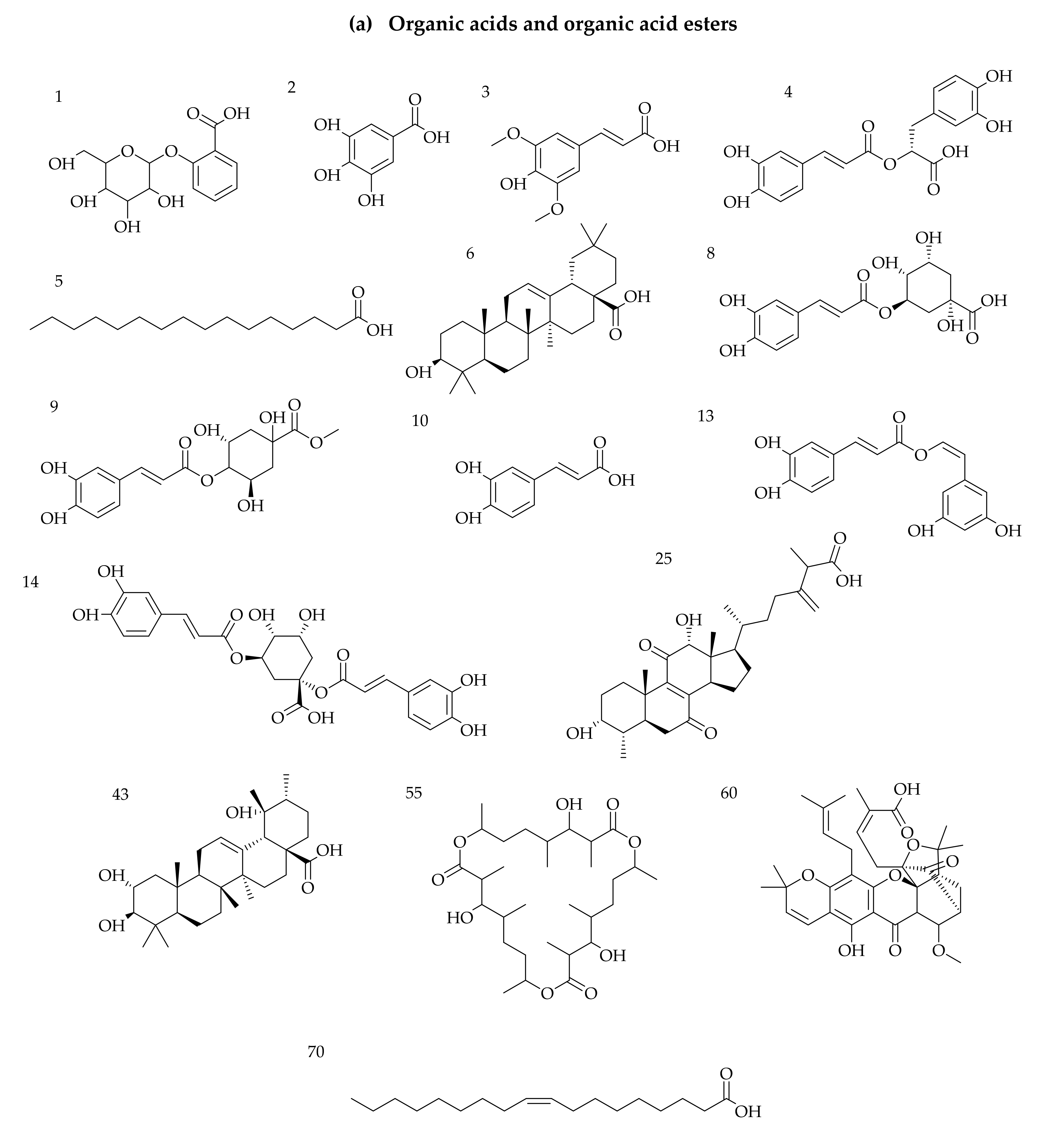
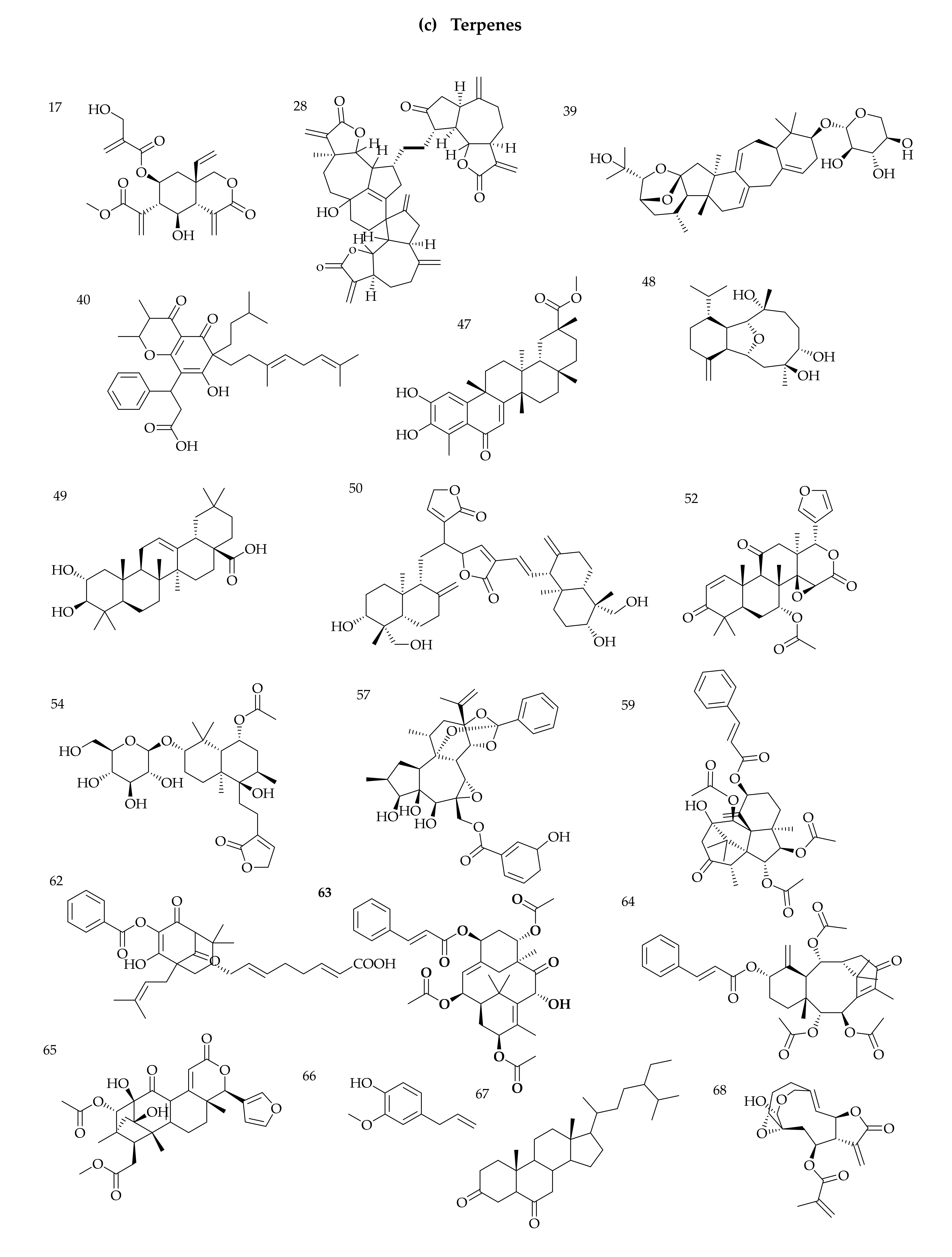
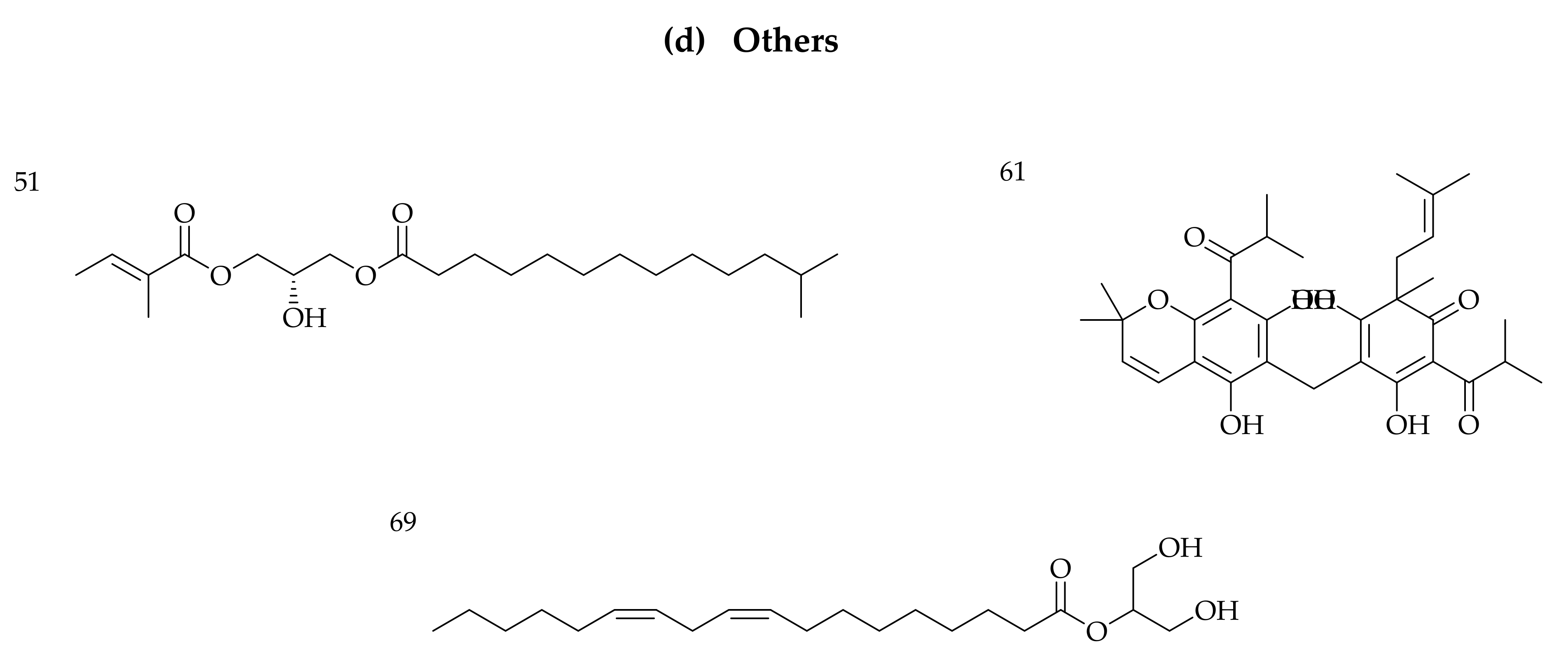
3.1. Identification of Phytochemicals from VAL and OGL Based on the UNIFI Platform
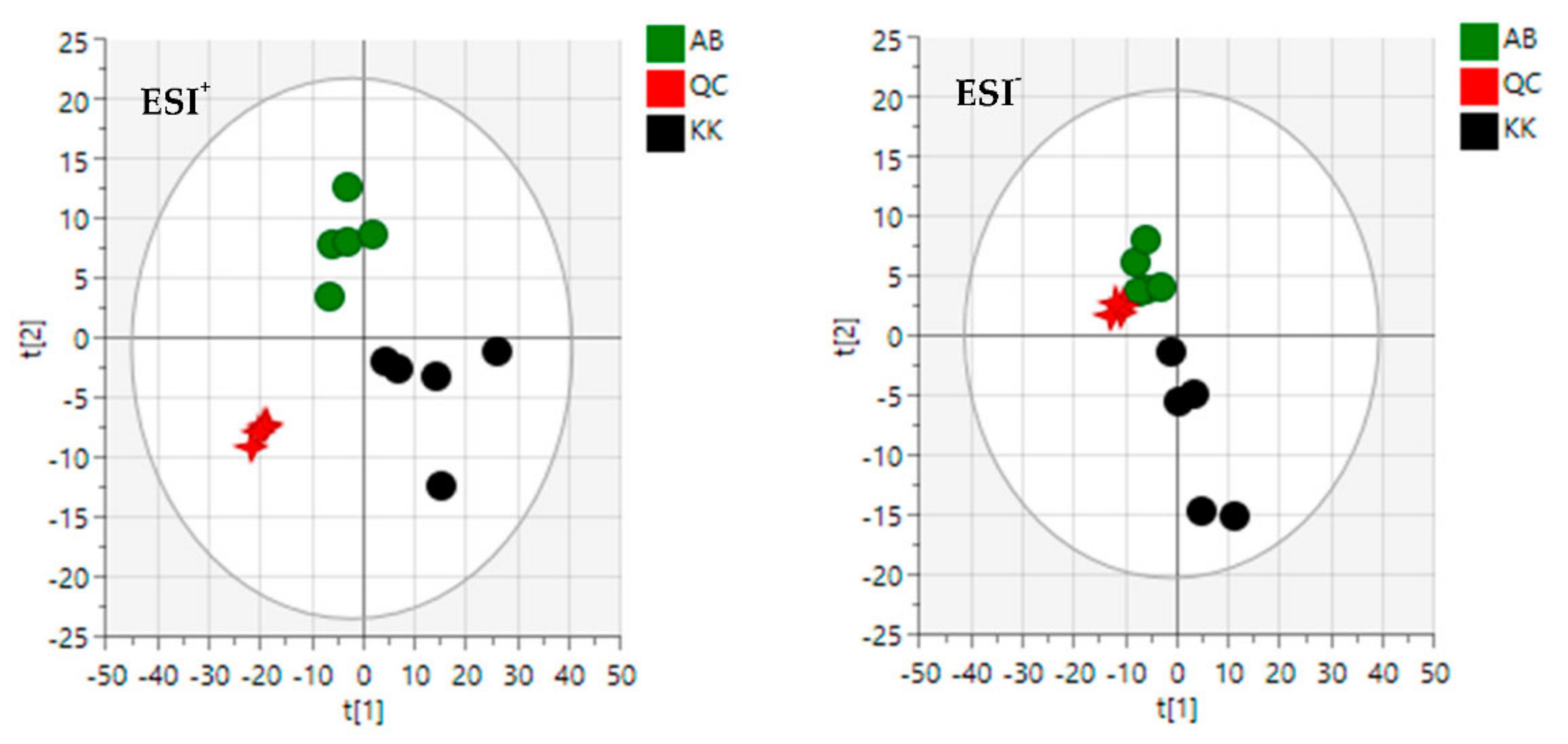
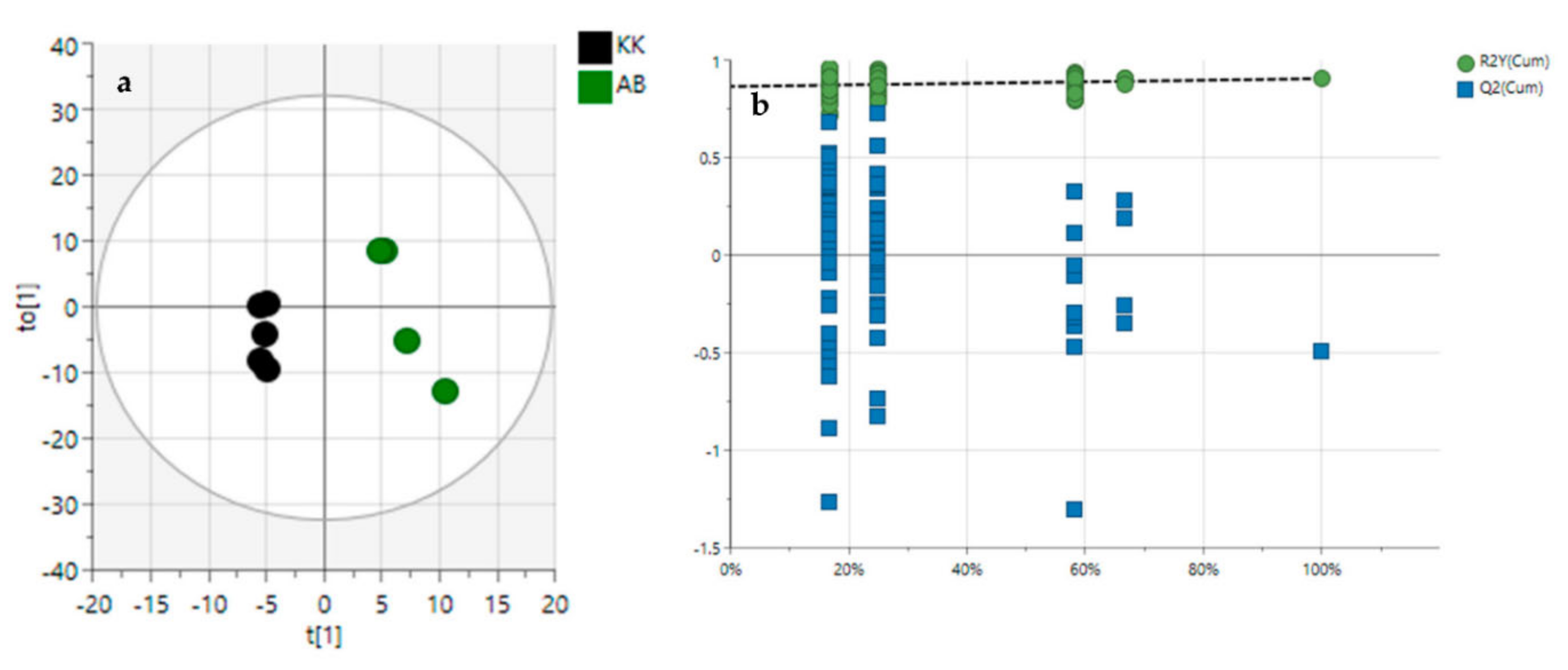
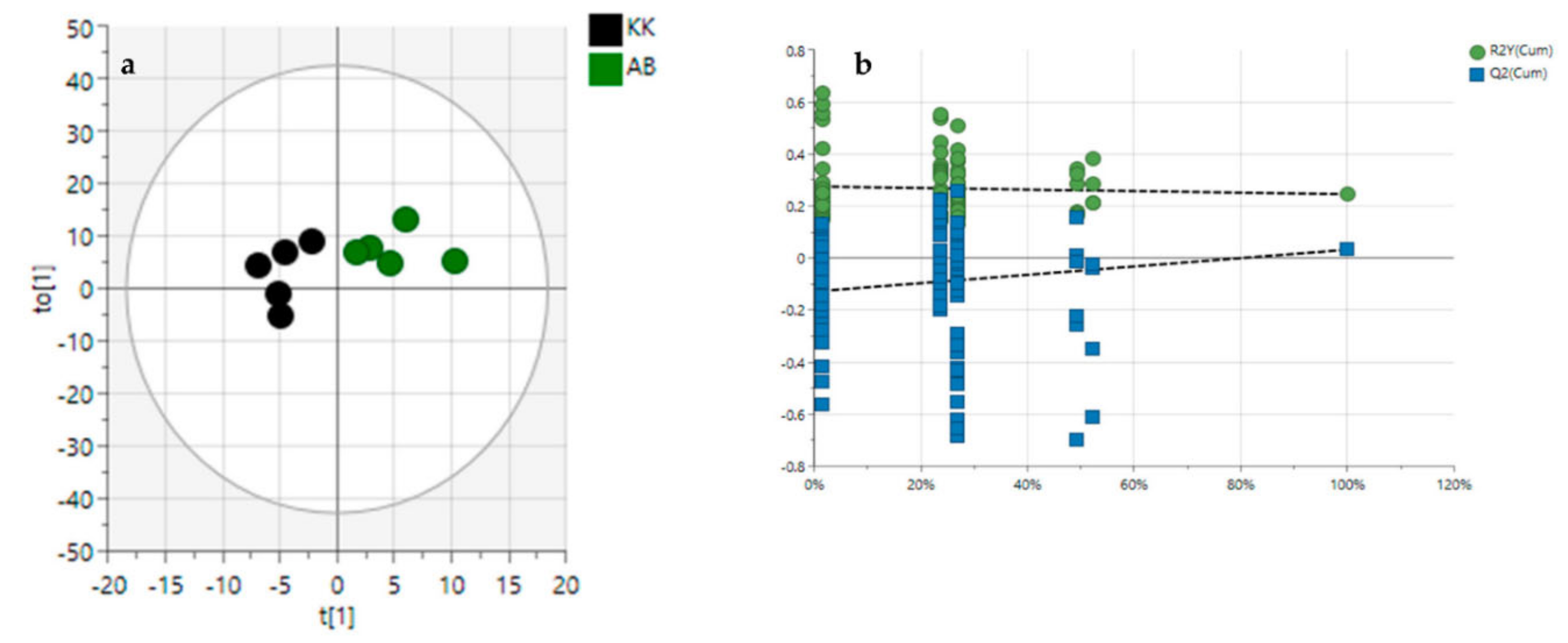
3.2. Diversity Evaluation of VAL and OGL Using Metabolomic Analysis
3.3. In Vitro Antisickling Activities of Crude Extracts
3.4. Structural Elucidation of Compounds Isolated
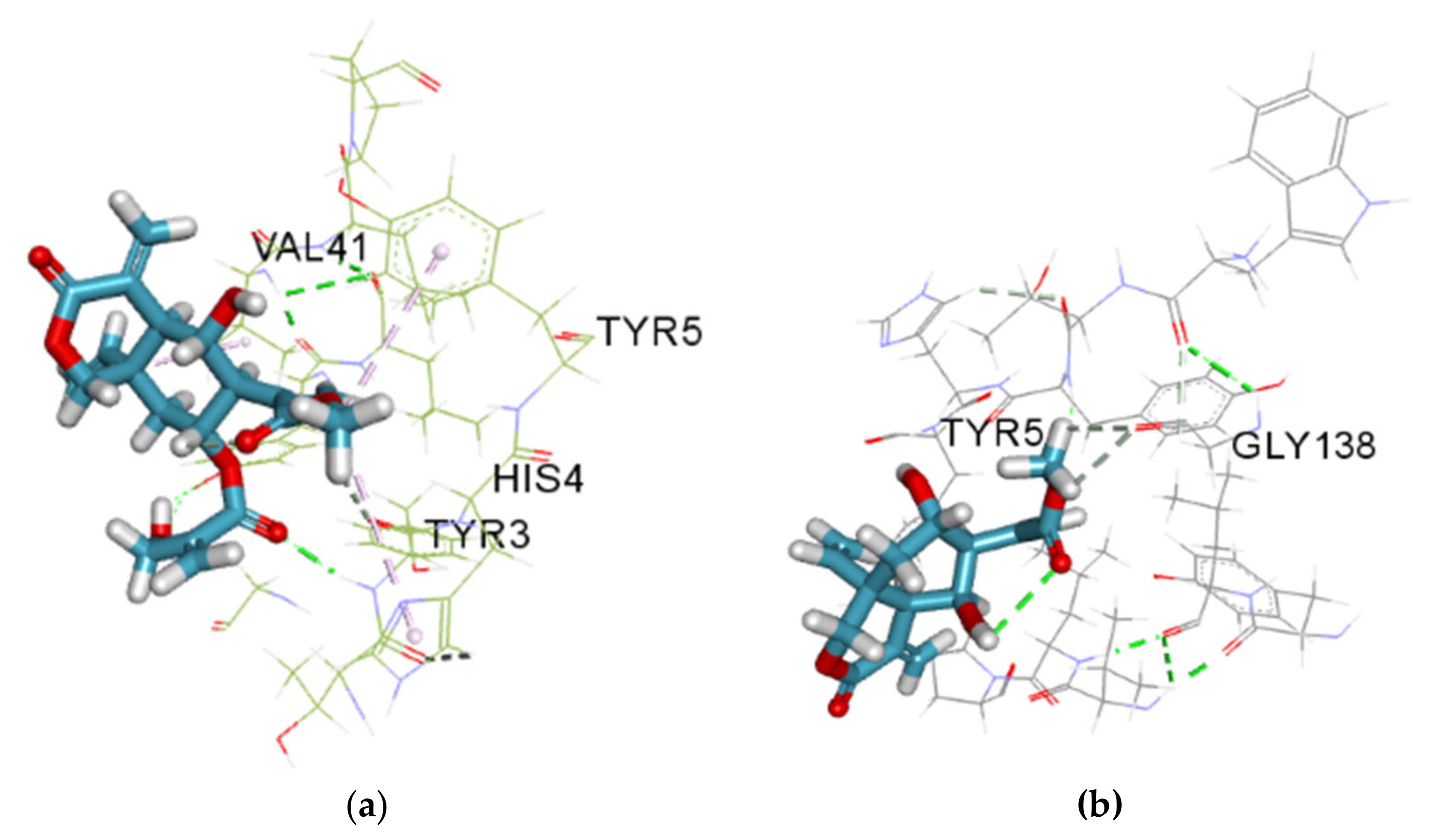
3.5. Molecular Docking Studies
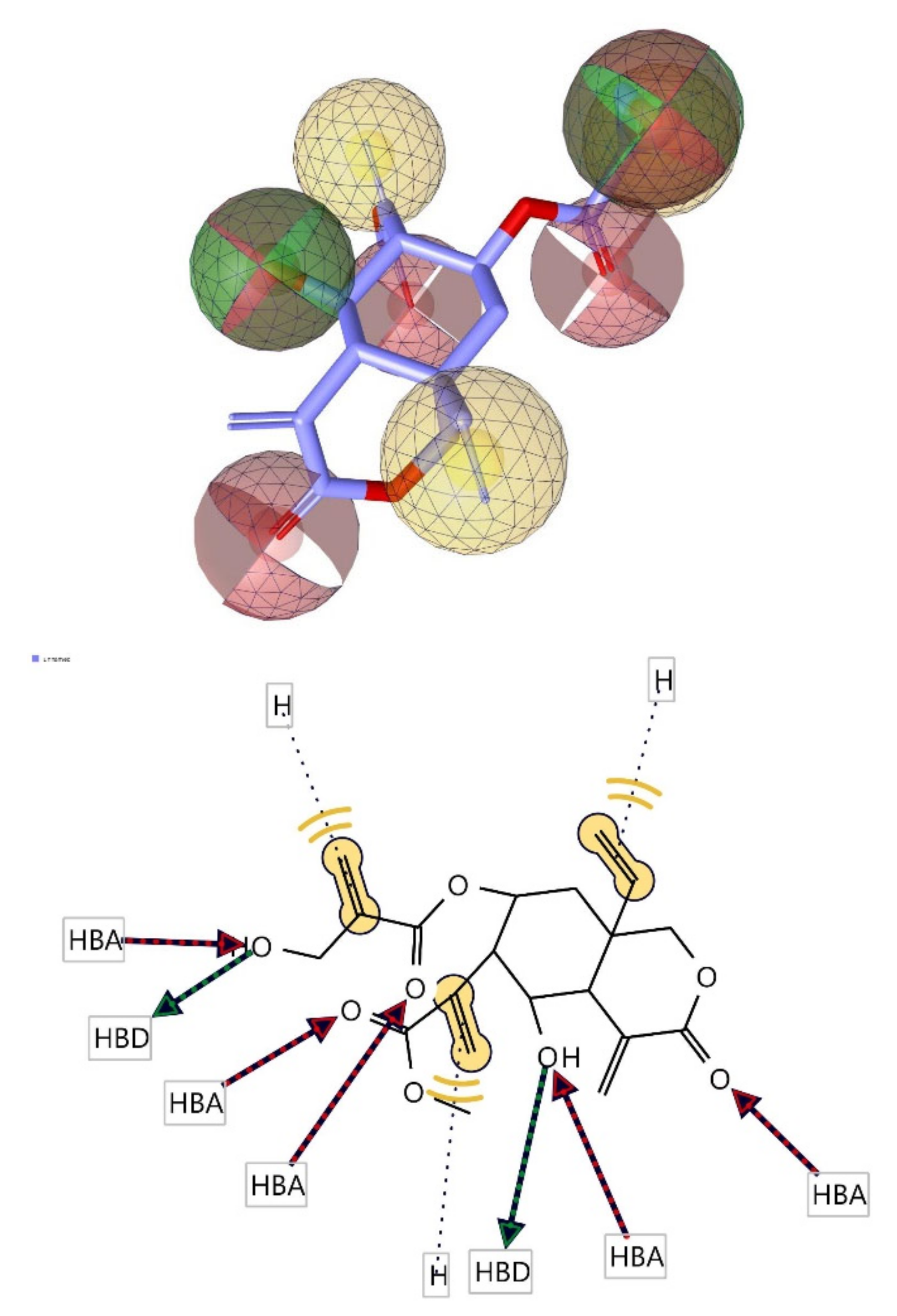
3.6. Pharmacophore Evaluation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gyasi, R.M.; Mensah, C.M.; Adjei, P.; Agyemang, S. Public Perceptions of the Role of Traditional Medicine in the Health Care Delivery System in Ghana. Glob. J. Health Sci. 2011, 3, 40–49. [Google Scholar] [CrossRef]
- Tshilanda, D.D.; Onyamboko, D.N.; Babady-Bila, P.; Ngbolua, K.-T.; Tshibangu, D.S.; Dibwe, E.D.F.; Mpiana, P.T. Anti-sickling Activity of Ursolic Acid Isolated from the Leaves of Ocimum gratissimum L. (Lamiaceae). Nat. Prod. Bioprospecting 2015, 5, 215–221. [Google Scholar] [CrossRef] [PubMed]
- Iyare, C.; Uzoigwe, J.; Okorie, P.O.; Ugwu, P.I.; Ezeh, C.O.; Iyare, E.E. Pregnancy outcome and early postnatal weights in diabetic and non-diabetic pregnant rats administered ethanolic extract of Ocimum gratissimum leaves during pregnancy. Int. J. Nurs. Midwifery 2018, 10, 26–32. [Google Scholar] [CrossRef]
- Maud, K.M.; Hannington, O.O.; Olwa, O.; Dominic, W.M. Ethno-pharmacological screening of Vernonia amygdalina and Cleome gynandra traditionally used in Childbirth in Western Uganda. In Proceedings of the 11th NAPRECA Symposium Book of Proceedings, Antananarivo, Madagascar, 9–12 August 2005. [Google Scholar]
- Adedayo, L.D.; Seun, G. Anxiolytic and explorative potentials of the methanol leaf extract of Vernonia amygdalina in male Wistar rats. Ann. Depress. Anxiety-Vol. 2018, 5, 1094. [Google Scholar] [CrossRef]
- Ofem, O.; Ani, E.; Eno, A. Effect of aqueous leaves extract of Ocimum gratissimum on hematological parameters in rats. Int. J. Appl. Basic Med. Res. 2012, 2, 38–42. [Google Scholar] [CrossRef]
- Oyeyemi, I.T.; Akinlabi, A.A.; Adewumi, A.; Aleshinloye, A.O.; Oyeyemi, O.T. Vernonia amygdalina: A folkloric herb with anthelminthic properties. Beni-Suef Univ. J. Basic Appl. Sci. 2018, 7, 43–49. [Google Scholar] [CrossRef]
- Nabukenya, I.; Rubaire-Akiiki, C.; Olila, D.; Ikwap, K.; Höglund, J. Ethnopharmacological practices by livestock farmers in Uganda: Survey experiences from Mpigi and Gulu districts. J. Ethnobiol. Ethnomedicine 2014, 10, 9. [Google Scholar] [CrossRef][Green Version]
- Adeniyi, S.A.; Orjiekwe, C.L.; Ehiagbonare, J.E.; Arimah, B.D. Preliminary phytochemical analysis and insecticidal activity of ethanolic extracts of four tropical plants (Vernonia amygdalina, Sida acuta, Ocimum gratissimum and Telfaria occidentalis) against beans weevil (Acanthscelides obtectus). Int. J. Phys. Sci. 2010, 5, 753–762. [Google Scholar]
- Okon, U.A.; Umoren, I.U. Comparison of antioxidant activity of insulin, Ocimum gratissimum L. and Vernonia amygdalina L. in type 1 diabetic rat model. J. Integr. Med. 2017, 15, 302–309. [Google Scholar] [CrossRef]
- Kadiri, O.; Olawoye, B. Vernonia amygdalina: An underutilized vegetable with nutraceutical Potentials—A Review. Turk. J. Agric. Food Sci. Technol. 2016, 4, 763–768. [Google Scholar] [CrossRef]
- Mgbeje, B.I.; Umoh, E.U.; Emmanuel-Ikpeme, C. Comparative analysis of phytochemical composition of four selected tropical medicinal plants namely: Ocimum gratissimum, Piper guineense, Gongronema latifolium and Vernonia amygdalina. J. Complement. Altern. Med. Res. 2019, 7, 1–11. [Google Scholar] [CrossRef]
- Diyaolu, O.A.; Attah, A.F.; Oluwabusola, E.T.; Moody, J.O.; Jaspars, M.; Ebel, R. Heavy Metals, Proximate Analysis and Brine Shrimp Lethality of Vernonia amygdalina and Ocimum gratissimum Growing in Crude Oil-Rich Delta State, Nigeria. Foods 2021, 10, 2913. [Google Scholar] [CrossRef] [PubMed]
- Agbogidi, O. Trace metal profile of some fruits in Kokori and Abraka Market, Delta State, Nigeria. Int. J. Sch. Res. Gate 2014, 2, 4. [Google Scholar]
- Rees, D.C.; Williams, T.N.; Gladwin, M.T. Sickle-cell disease. Lancet 2010, 376, 2018–2031. [Google Scholar] [CrossRef] [PubMed]
- Ingram, V.M. Gene mutations in human haemoglobin: The chemical difference between normal and sickle cell haemoglobin. Nature 1957, 180, 326–328. [Google Scholar] [CrossRef]
- Nurain, I.O.; Bewaji, C.O.; Johnson, J.S.; Davenport, R.D.; Zhang, Y. Potential of Three Ethnomedicinal Plants as Antisickling Agents. Mol. Pharm. 2017, 14, 172–182. [Google Scholar] [CrossRef]
- Vichinsky, E.; Hoppe, C.C.; Ataga, K.I.; Ware, R.E.; Nduba, V.; El-Beshlawy, A.; Hassab, H.; Achebe, M.M.; Alkindi, S.; Brown, R.C.; et al. A Phase 3 Randomized Trial of Voxelotor in Sickle Cell Disease. N. Engl. J. Med. 2019, 381, 509–519. [Google Scholar] [CrossRef]
- Piel, F.B.; Hay, S.I.; Gupta, S.; Weatherall, D.J.; Williams, T.N. Global burden of sickle cell anaemia in children under five, 2010–2050: Modelling based on demographics, excess mortality, and interventions. PLoS Med. 2013, 10, e1001484. [Google Scholar] [CrossRef]
- Lasch, J.; Küllertz, G.; Opalka, J.R. Separation of Erythrocytes into Age-Related Fractions by Density or Size? Counterflow Centrifugation. Clin. Chem. Lab. Med. (CCLM) 2000, 38, 629–632. [Google Scholar] [CrossRef]
- Agrawal, R.K.; Patel, R.K.; Nainiwal, L.; Trivedi, B. Hydroxyurea in sickle cell disease: Drug review. Indian J. Hematol. Blood Transfus. 2014, 30, 91–96. [Google Scholar] [CrossRef]
- Niihara, Y.; Miller, S.T.; Kanter, J.; Lanzkron, S.; Smith, W.R.; Hsu, L.L.; Vichinsky, E.P. A phase 3 trial of l-glutamine in sickle cell disease. N. Engl. J. Med. 2018, 379, 226–235. [Google Scholar] [CrossRef]
- Ataga, K.I.; Kutlar, A.; Kanter, J.; Liles, D.; Cancado, R.; Friedrisch, J.; Guthrie, T.H.; Knight-Madden, J.; Alvarez, O.A.; Gordeuk, V.R.; et al. Crizanlizumab for the Prevention of Pain Crises in Sickle Cell Disease. N. Engl. J. Med. 2017, 376, 429–439. [Google Scholar] [CrossRef]
- Herity, L.B.; Vaughan, D.M.; Rodriguez, L.R.; Lowe, D.K. Voxelotor: A Novel Treatment for Sickle Cell Disease. Ann. Pharmacother. 2021, 55, 240–2451. [Google Scholar] [CrossRef]
- Wang, C.; Zhang, N.; Wang, Z.; Qi, Z.; Zhu, H.; Zheng, B.; Li, P.; Liu, J. Nontargeted Metabolomic Analysis of Four Different Parts of Platycodon grandiflorum Grown in Northeast China. Molecules 2017, 22, 1280. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, C.; Lin, H.; Liu, Y.; Li, Y.; Zhao, Y.; Li, P.; Liu, J. Discovery of the Potential Biomarkers for Discrimination between Hedyotis diffusa and Hedyotis corymbosa by UPLC-QTOF/MS Metabolome Analysis. Molecules 2018, 23, 1525. [Google Scholar] [CrossRef]
- Adusumilli, R.; Mallick, P. Data Conversion with ProteoWizard msConvert. Methods Mol. Biol. 2017, 1550, 339–368. [Google Scholar]
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Orešič, M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform. 2010, 11, 395. [Google Scholar] [CrossRef]
- Pang, Z.; Wang, G.; Ran, N.; Lin, H.; Wang, Z.; Guan, X.; Yuan, Y.; Fang, K.; Liu, J.; Wang, F. Inhibitory Effect of Methotrexate on Rheumatoid Arthritis Inflammation and Comprehensive Metabolomics Analysis Using Ultra-Performance Liquid Chromatography-Quadrupole Time of Flight-Mass Spectrometry (UPLC-Q/TOF-MS). Int. J. Mol. Sci. 2018, 19, 2894. [Google Scholar] [CrossRef]
- Triba, M.N.; Le Moyec, L.; Amathieu, R.; Goossens, C.; Bouchemal, N.; Nahon, P.; Rutledge, D.N.; Savarin, P. PLS/OPLS models in metabolomics: The impact of permutation of dataset rows on the K-fold cross-validation quality parameters. Mol. Biosyst. 2015, 11, 13–19. [Google Scholar] [CrossRef]
- Pauline, N.; Cabral, B.N.P.; Anatole, P.C.; Jocelyne, A.M.V.; Bruno, M.; Jeanne, N.Y. The in vitro antisickling and antioxidant effects of aqueous extracts Zanthoxyllum heitzii on sickle cell disorder. BMC Complement. Altern. Med. 2013, 13, 162. [Google Scholar] [CrossRef]
- Harrington, D.J.; Adachi, K.; Royer, W.E., Jr. The high resolution crystal structure of deoxyhemoglobin S. J. Mol. Biol. 1997, 272, 398–407. [Google Scholar] [CrossRef] [PubMed]
- Somers, W.S.; Tang, J.; Shaw, G.D.; Camphausen, R.T. Insights into the molecular basis of leukocyte tethering and rolling revealed by structures of P-and E-selectin bound to SLeX and PSGL-1. Cell 2000, 103, 467–479. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Shapovalov, M.V.; Dunbrack, R.L. A Smoothed Backbone-Dependent Rotamer Library for Proteins Derived from Adaptive Kernel Density Estimates and Regressions. Structure 2011, 19, 844–858. [Google Scholar] [CrossRef] [PubMed]
- Dallakyan, S.; Olson, A.J. Small-molecule library screening by docking with pyrx. Methods Mol. Biol. 2015, 1263, 243–250. [Google Scholar]
- Koul, J.L.; Koul, S.; Singh, C.; Taneja, S.C.; Shanmugavel, M.; Kampasi, H.; Qazi, G.N. In vitro cytotoxic elemanolides from Vernonia lasiopus. Planta Med. 2003, 69, 164–166. [Google Scholar] [CrossRef]
- Wolber, G.; Thierry, L. LigandScout: 3-D pharmacophores derived from protein-bound ligands and their use as virtual screening filters. J. Chem. Inf. Model 2005, 45, 160–169. [Google Scholar] [CrossRef]
- Jisaka, M.; Ohigashi, H.; Takegawa, K.; Hirota, M.; Irie, R.; Huffman, M.A.; Koshimįzu, K. Steroid glucosides from Vernonia amygdalina, a possible chimpanzee medicinal plant. Phytochemistry 1993, 34, 409–413. [Google Scholar] [CrossRef]
- Heavisides, E.; Rouger, C.; Reichel, A.F.; Ulrich, C.; Wenzel-Storjohann, A.; Sebens, S.; Tasdemir, D. Seasonal variations in the metabolome and bioactivity profile of Fucus vesiculosus extracted by an optimised, pressurised liquid extraction protocol. Mar. Drugs 2018, 16, 503. [Google Scholar] [CrossRef]
- Alabi, Q.K.; Akomolafe, R.O.; Omole, J.G.; Aturamu, A.; Ige, M.S.; Kayode, O.O.; Kajewole-Alabi, D. Polyphenol-rich extract of Ocimum gratissimum leaves prevented toxic effects of cyclophosphamide on the kidney function of Wistar rats. BMC Complement. Med. Ther. 2021, 21, 274. [Google Scholar] [CrossRef]
- Chikezie, P. Studies on the anti-sickling effects of Azadirachta indica and Vernonia amygdalina on Hbss erythrocytes. Int. J. Nat. Appl. Sci. 2006, 2, 24–28. [Google Scholar] [CrossRef]
- Li, X.J.; Zhang, Q.; Zhang, A.L.; Gao, J.M. Metabolites from Aspergillus fumigatus, an endophytic fungus associated with Melia azedarach, and their antifungal, antifeedant, and toxic activities. J. Agric. Food Chem. 2012, 60, 3424–3431. [Google Scholar] [CrossRef]
- Copmans, D.; Rateb, M.; Tabudravu, J.N.; Pérez-Bonilla, M.; Dirkx, N.; Vallorani, R.; Diaz, C.; del Palacio, J.P.; Smith, A.J.; Ebel, R.; et al. Zebrafish-Based Discovery of Antiseizure Compounds from the Red Sea: Pseurotin A2 and Azaspirofuran A. ACS Chem. Neurosci. 2018, 9, 1652–1662. [Google Scholar] [CrossRef]
- Alberto, A.V.P.; Ferreira, N.C.D.S.; Soares, R.F.; Alves, L.A. Molecular Modeling Applied to the Discovery of New Lead Compounds for P2 Receptors Based on Natural Sources. Front. Pharmacol. 2020, 11, 01221. [Google Scholar] [CrossRef]
- Safo, M.K.; Abdulmalik, O.; Danso-Danquah, R.; Burnett, J.C.; Nokuri, S.; Joshi, G.S.; Musayev, F.N.; Asakura, T.; Abraham, D.J. Structural Basis for the Potent Antisickling Effect of a Novel Class of Five-Membered Heterocyclic Aldehydic Compounds. J. Med. Chem. 2004, 47, 4665–4676. [Google Scholar] [CrossRef]
- Omar, A.M.; Mahran, M.A.; Ghatge, M.S.; Chowdhury, N.; Bamane, F.H.A.; El-Araby, M.E.; Abdulmalik, O.; Safo, M.K. Identification of a novel class of covalent modifiers of hemoglobin as potential antisickling agents. Org. Biomol. Chem. 2015, 13, 6353–6370. [Google Scholar] [CrossRef]
- Wagner, D.D.; Frenette, P.S. The vessel wall and its interactions. Blood J. Am. Soc. Hematol. 2008, 111, 5271–5281. [Google Scholar] [CrossRef]
- Mayadas, T.N.; Johnson, R.C.; Rayburn, H.; Hynes, R.O.; Wagner, D.D. Leukocyte rolling and extravasation are severely compromised in P selectin-deficient mice. Cell 1993, 74, 541–554. [Google Scholar] [CrossRef]
- Telen, M.J.; Wun, T.; McCavit, T.L.; De Castro, L.M.; Krishnamurti, L.; Lanzkron, S.; Hsu, L.L.; Smith, W.R.; Rhee, S.; Magnani, J.L.; et al. Randomized Phase 2 Study of GMI-1070 in SCD: Reduction in Time to Resolution of Vaso-Occlusive Events and Decreased Opioid Use. Blood 2015, 125, 2656–2664. [Google Scholar] [CrossRef]
- Patil, R.; Das, S.; Stanley, A.; Yadav, L.; Sudhakar, A.; Varma, A.K. Optimized Hydrophobic Interactions and Hydrogen Bonding at the Target-Ligand Interface Leads the Pathways of Drug-Designing. PLoS ONE 2010, 5, e12029. [Google Scholar] [CrossRef]
- Li, Y.; Kong, D.; Fu, Y.; Sussman, M.R.; Wu, H. The effect of developmental and environmental factors on secondary metabolites in medicinal plants. Plant Physiol. Biochem. 2020, 148, 80–89. [Google Scholar] [CrossRef] [PubMed]





| # Species | Sample No. | Community | Source | Coordinates |
|---|---|---|---|---|
| VAL | 1 | Abraka | Oria | 5°45′ N 6°15′ E |
| 2 | Abraka | Urhuovie | 5°47′ N 6°15′ E | |
| 3 | Abraka | Ajalom | 5°47′ N 6°15′ E | |
| 4 | Kokori | Egbo | 5°38′ N 6°04′ E | |
| 5 | Kokori | Kokori | 5°38′ N 6°15′ E | |
| 6 | Kokori | Samagidi | 5°38′ N 6°22′ E | |
| OGL | 7 | Abraka | Oria | 5°45′ N 6°15′ E |
| 8 | Abraka | Urhuovie | 5°47′ N 6°15′ E | |
| 9 | Abraka | Ajalom | 5°47′ N 6°15′ E | |
| 10 | Kokori | Egbo | 5°38′ N 6°04′ E | |
| 11 | Kokori | Kokori | 5°38′ N 6°15′ E | |
| 12 | Kokori | Samagidi | 5°38′ N 6°22′ E |
| Target Name | PDB Code | Dimensions (Å) | Centre |
|---|---|---|---|
| deoxyhemoglobin S | 2HBS | X: 24.4838; Y: 26.6805; Z: 25.0000 | X: 7.5857; Y: 11.1820; Z: 28.1205 |
| P-selectin | 1G1S | X: 34.1555; Y: 21.0892; Z: 25.0000 | X: 43.7339; Y: 85.8462 Z: 48.1365 |
| Samples (Extracts) | % Sickling ± SD | % Sickling Reversal ± SD |
|---|---|---|
| OGL-AB | 28.43 ± 0.50 | 35.31 ± 0.30 |
| VAL-AB | 26.89 ± 0.51 | 33.52 ± 0.30 |
| OGL-KK | 26.17 ± 0.16 | 29.73 ± 0.12 |
| VAL-KK | 26.29 ± 0.46 | 30.22 ± 0.50 |
| Hy.urea | 26.36 ± 0.02 | 34.85 ± 0.16 |
| Control | 40.25 ± 0.32 | 0.00 |
| Compounds | Binding Affinity (KJ/mol) | |
|---|---|---|
| 1G1S | 2HBS | |
| Lasiopulide | −20.9200 | −23.4304 |
| Vernodalol | −21.7568 | −22.5936 |
| (4R)-2-Methylpentane-2,4-diol | −15.0624 | −17.9912 |
| Furfural | −13.8072 | −16.736 |
| 4-{2-chloro-4-[3-(1H-imidazol-2-yl)propanoyl]phenoxy}butanoic acid | NB | −31.7984 |
| Protoporphyrin IX | NB | −43.932 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Diyaolu, O.A.; Oluwabusola, E.T.; Attah, A.F.; Olori, E.O.; Fagbemi, A.A.; Preet, G.; Soldatou, S.; Moody, J.O.; Jaspars, M.; Ebel, R. Can Crude Oil Exploration Influence the Phytochemicals and Bioactivity of Medicinal Plants? A Case of Nigerian Vernonia amygdalina and Ocimum gratissimum. Molecules 2022, 27, 8372. https://doi.org/10.3390/molecules27238372
Diyaolu OA, Oluwabusola ET, Attah AF, Olori EO, Fagbemi AA, Preet G, Soldatou S, Moody JO, Jaspars M, Ebel R. Can Crude Oil Exploration Influence the Phytochemicals and Bioactivity of Medicinal Plants? A Case of Nigerian Vernonia amygdalina and Ocimum gratissimum. Molecules. 2022; 27(23):8372. https://doi.org/10.3390/molecules27238372
Chicago/Turabian StyleDiyaolu, Oluwatofunmilayo A., Emmanuel T. Oluwabusola, Alfred F. Attah, Eric O. Olori, Adeshola A. Fagbemi, Gagan Preet, Sylvia Soldatou, Jones O. Moody, Marcel Jaspars, and Rainer Ebel. 2022. "Can Crude Oil Exploration Influence the Phytochemicals and Bioactivity of Medicinal Plants? A Case of Nigerian Vernonia amygdalina and Ocimum gratissimum" Molecules 27, no. 23: 8372. https://doi.org/10.3390/molecules27238372
APA StyleDiyaolu, O. A., Oluwabusola, E. T., Attah, A. F., Olori, E. O., Fagbemi, A. A., Preet, G., Soldatou, S., Moody, J. O., Jaspars, M., & Ebel, R. (2022). Can Crude Oil Exploration Influence the Phytochemicals and Bioactivity of Medicinal Plants? A Case of Nigerian Vernonia amygdalina and Ocimum gratissimum. Molecules, 27(23), 8372. https://doi.org/10.3390/molecules27238372







