Isolation, Characterization, and Compositional Analysis of Polysaccharides from Pinot Noir Wines: An Exploratory Study
Abstract
:1. Introduction
2. Results and Discussion
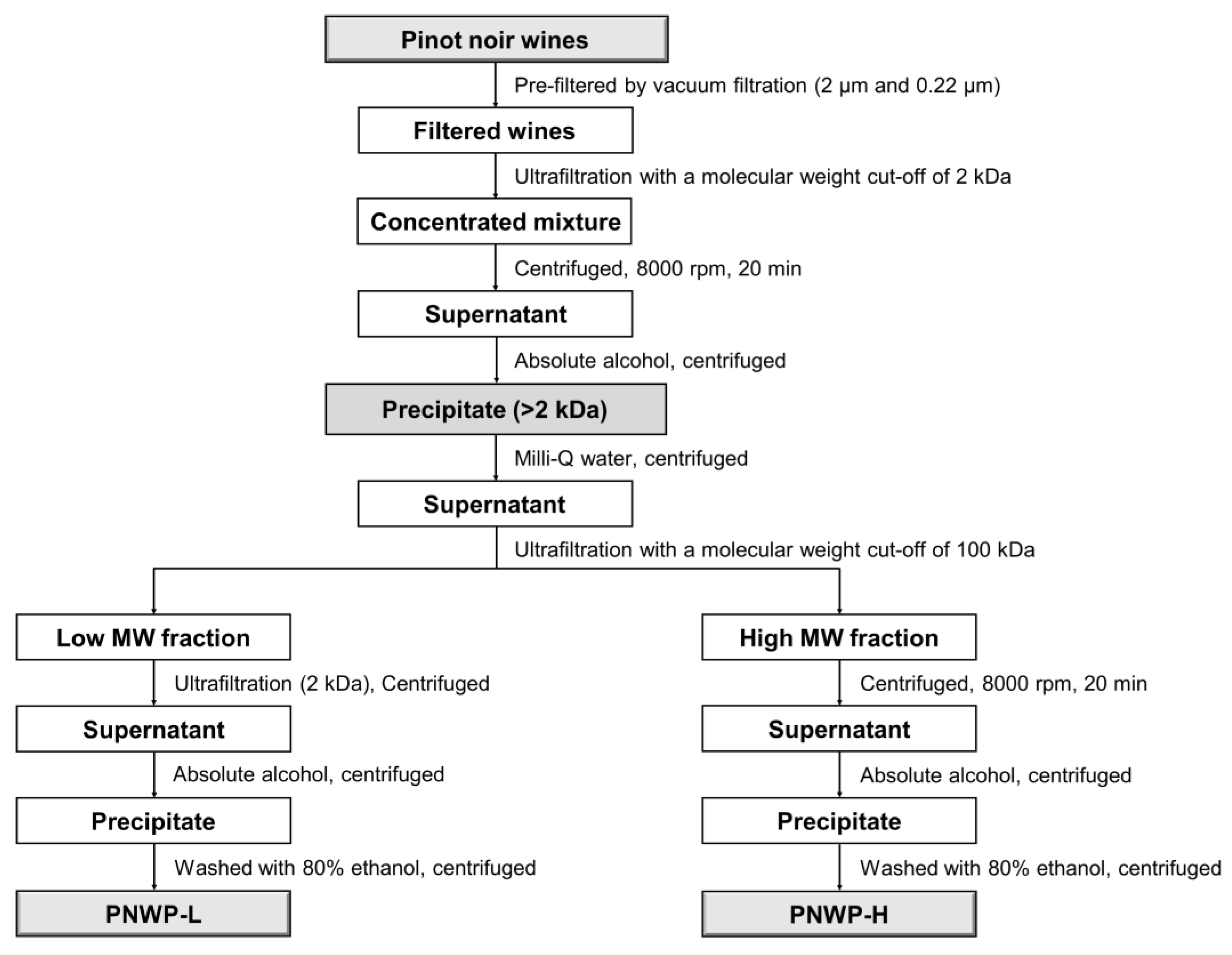
2.1. Pinot Noir Wine Polysaccharides Isolation and Fractionation
2.2. Yield and Impurity Analysis of PNWPs
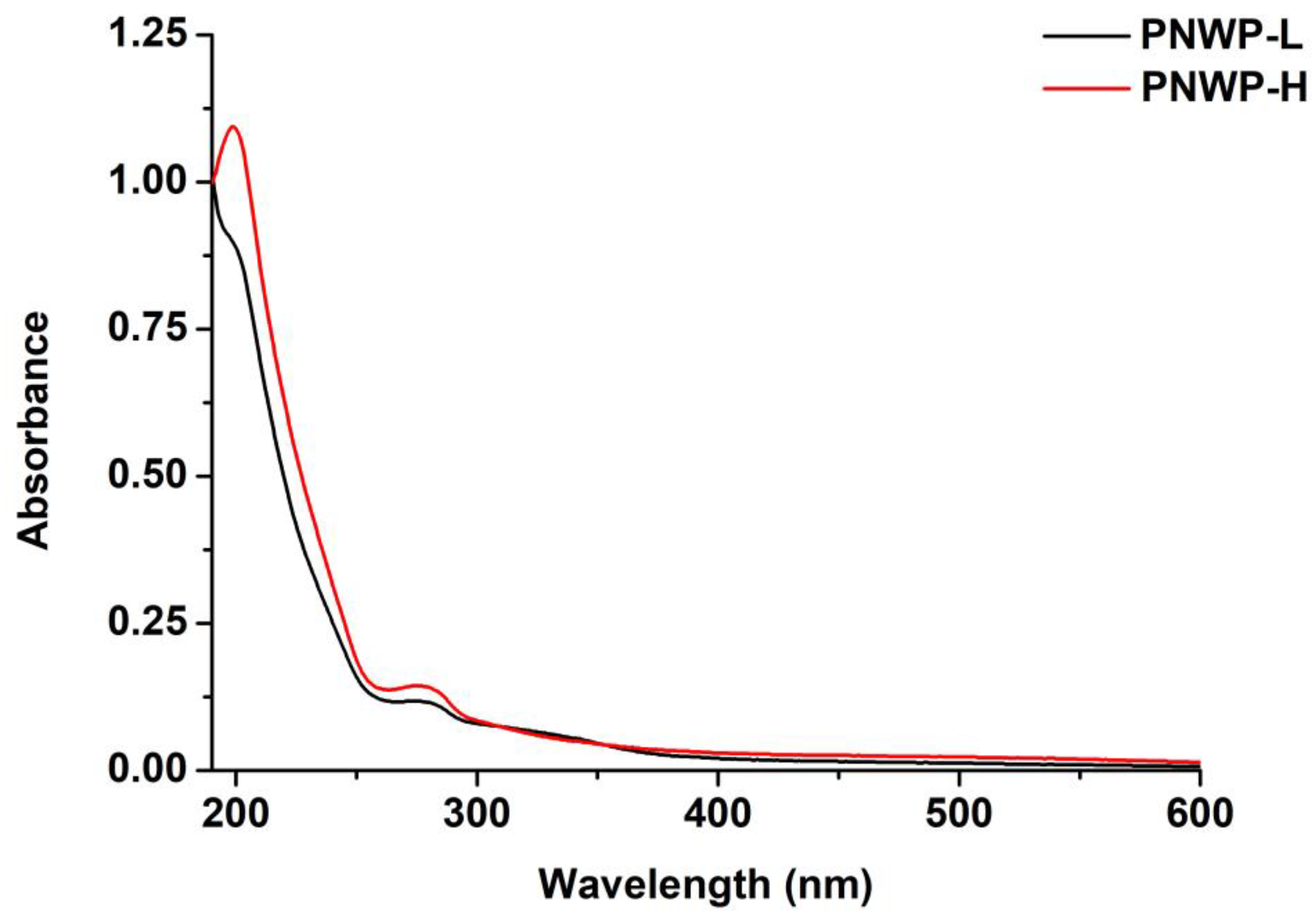
2.3. Ultraviolet-Visible Spectroscopy Analysis of PNWPs
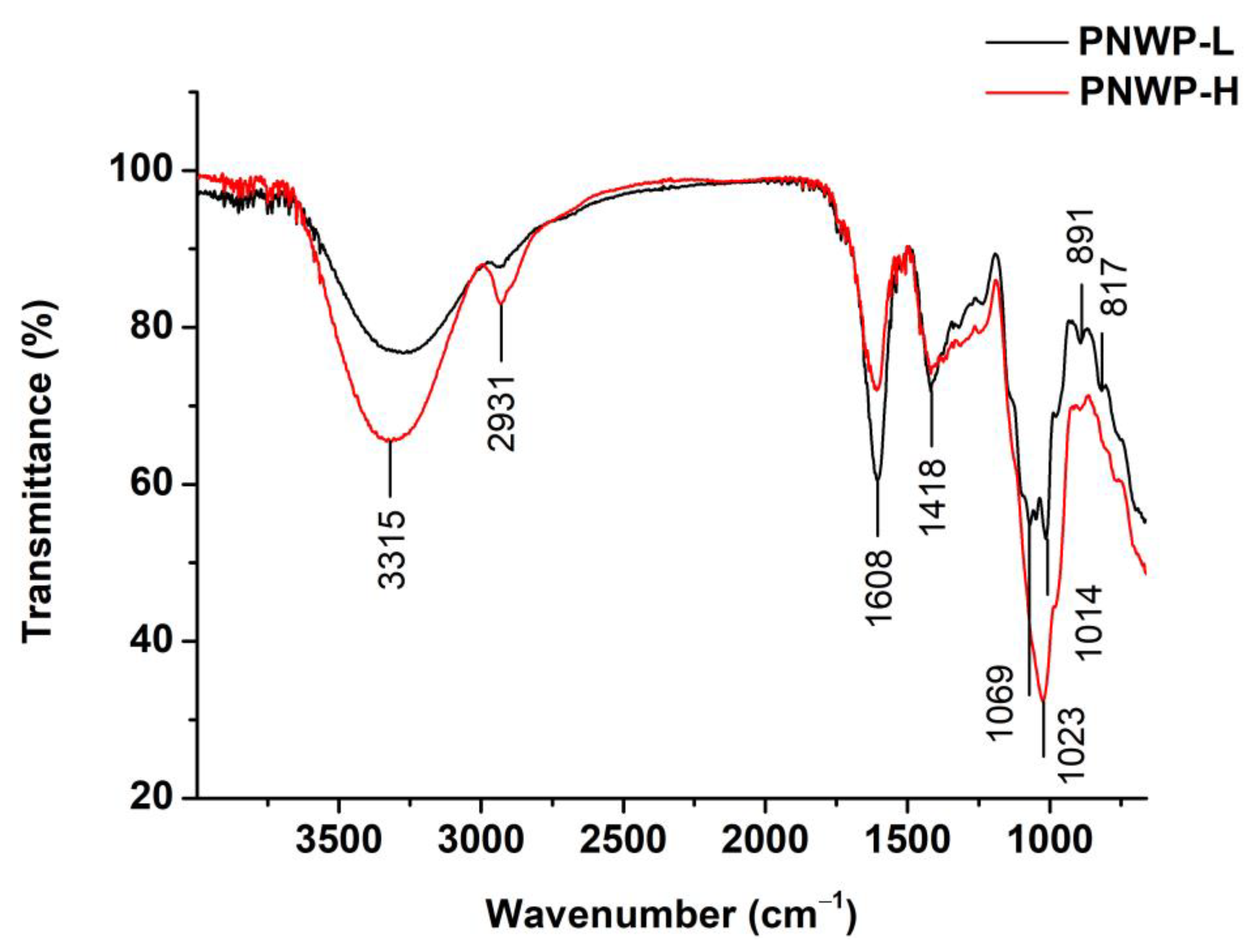
2.4. Fourier-Transform Infrared Spectroscopy Analysis of PNWPs
2.5. Molecular Weight Distribution of PNWPs
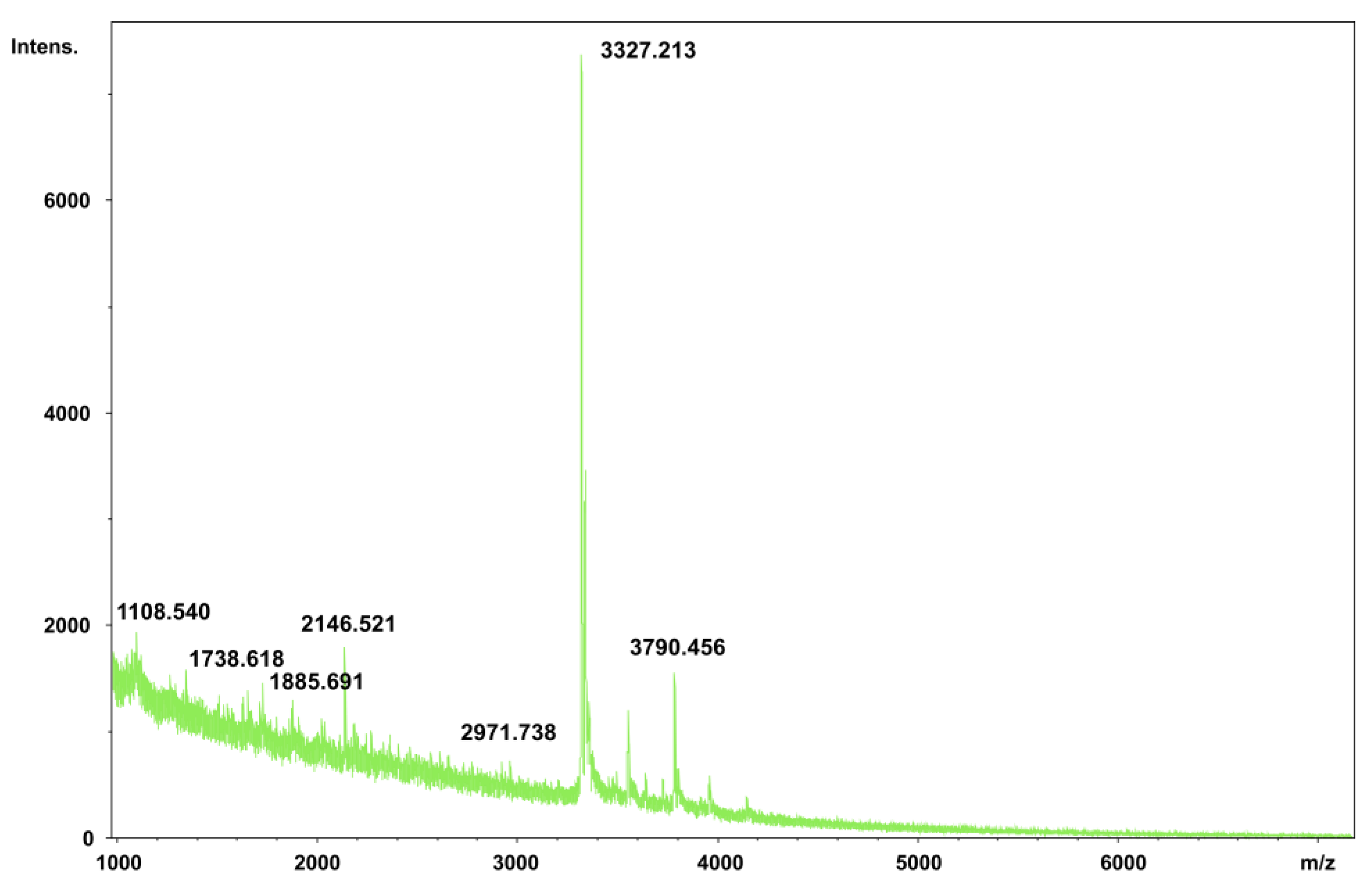
2.6. Matrix-Assisted Laser Desorption/Ionization-Time of Flight Mass Spectrometry (MALDI-TOF) Analysis of PNWP-L
2.7. Monosaccharide Composition Analysis of the PNWPs
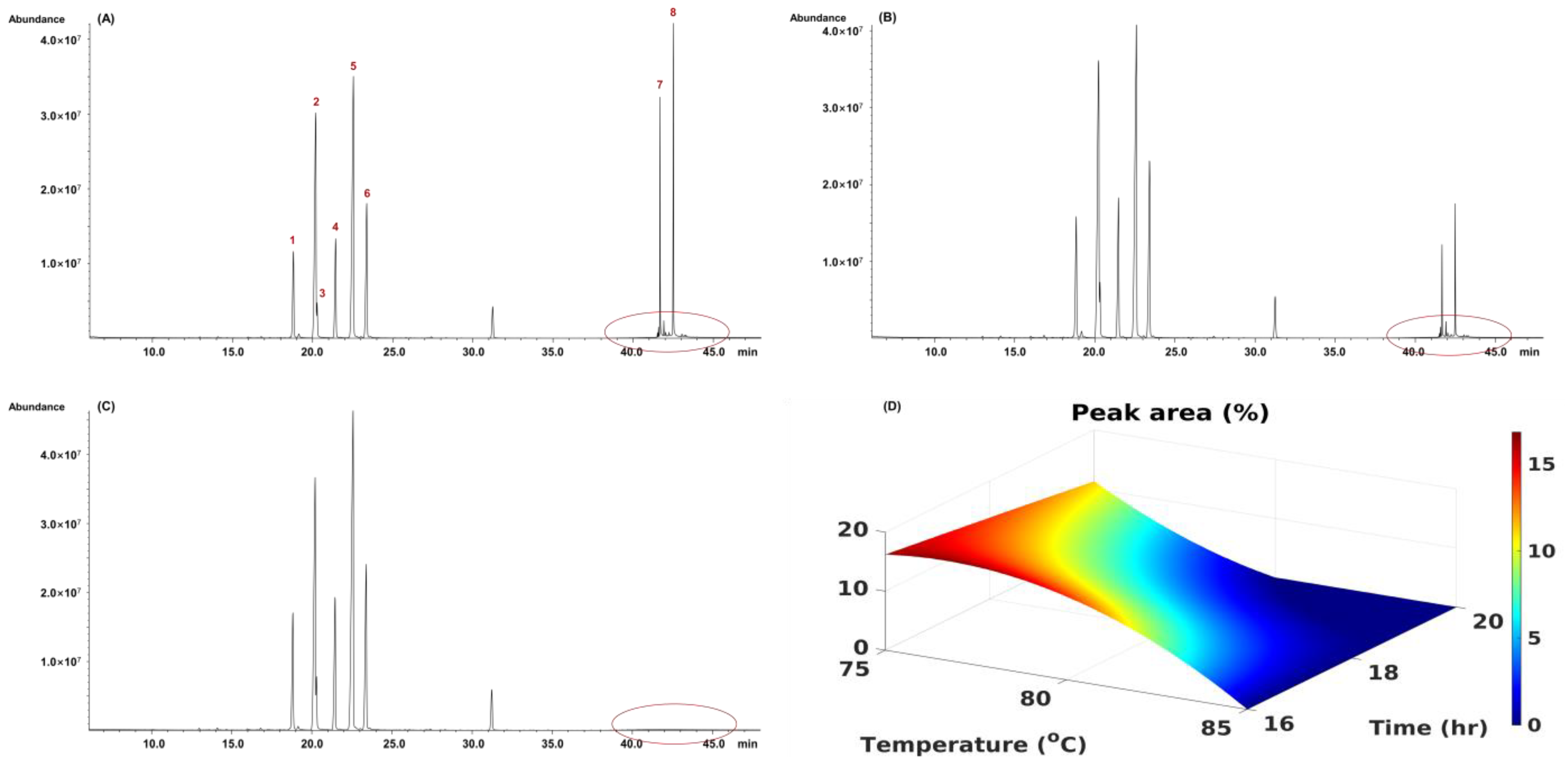
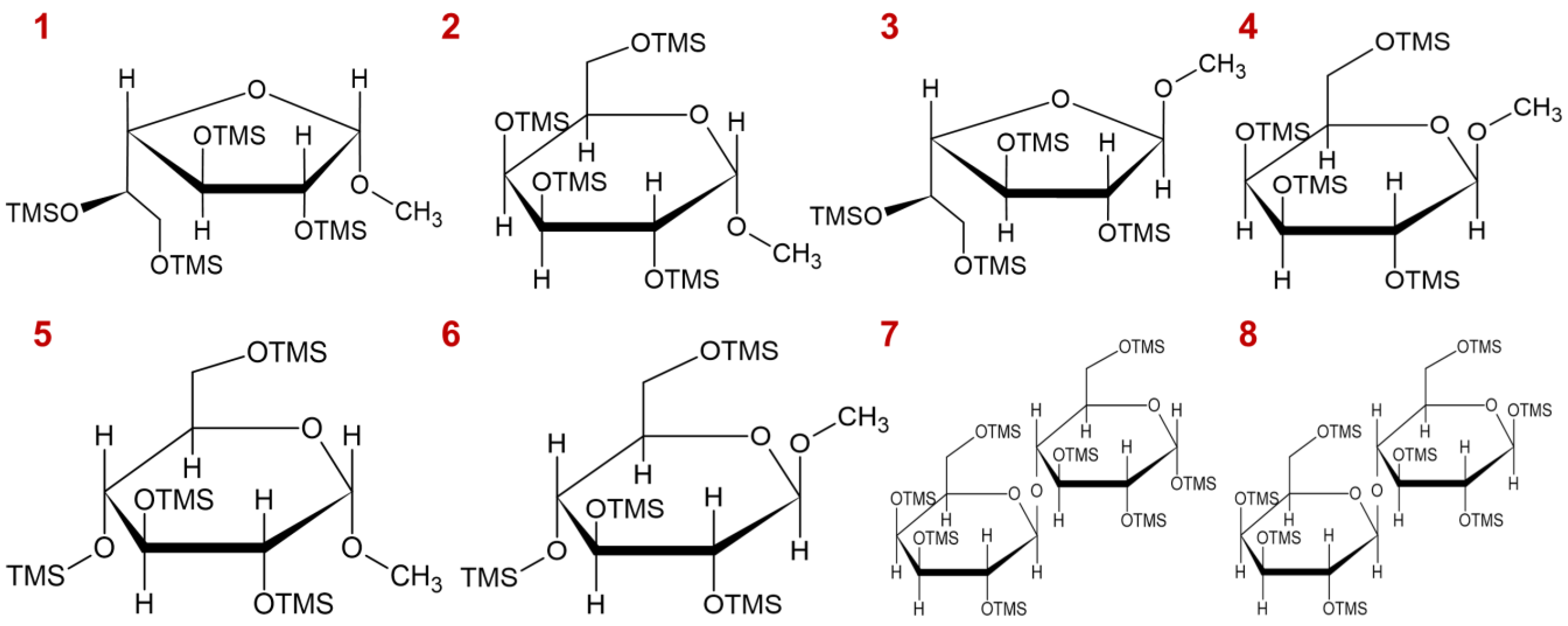
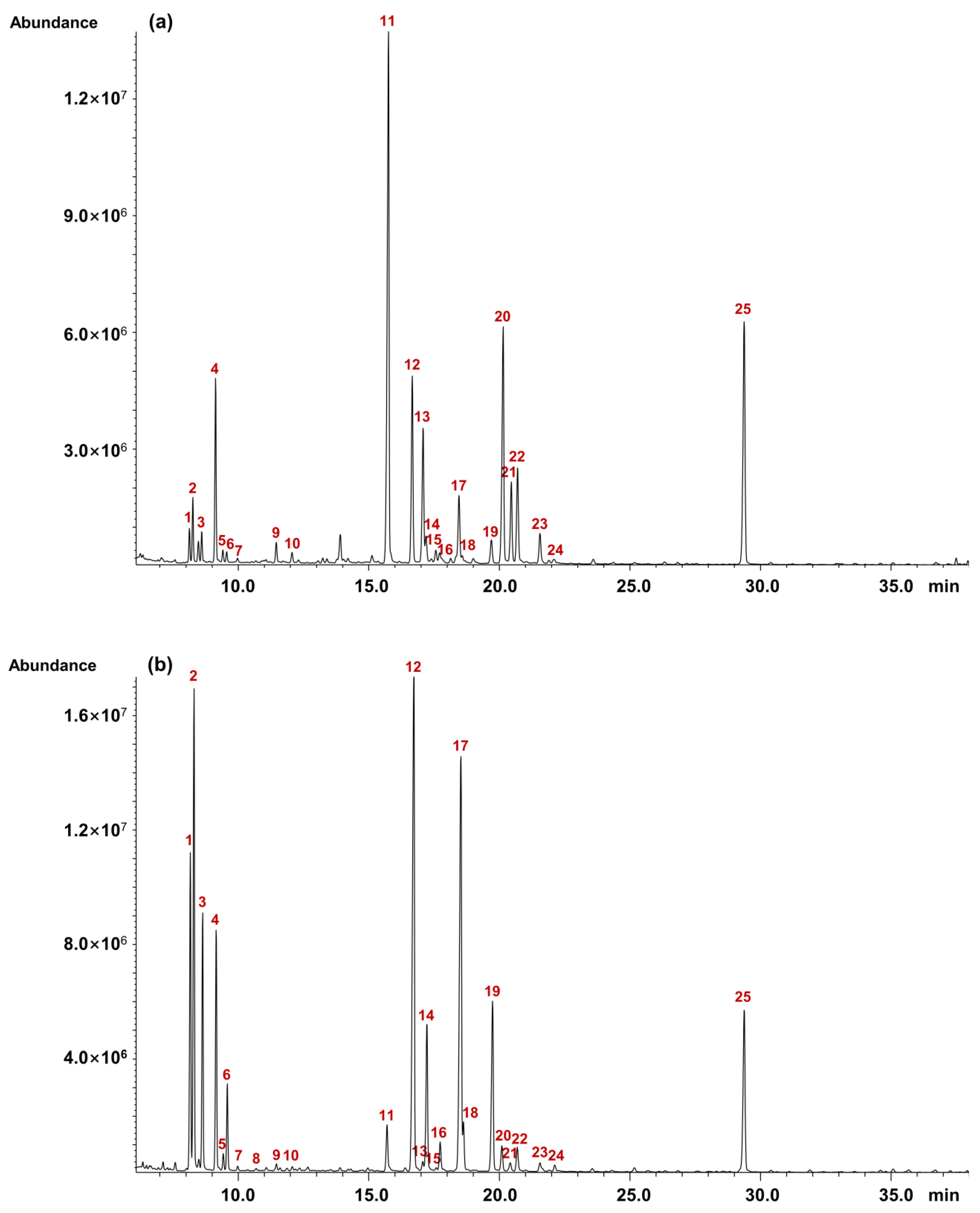
2.7.1. The Methanolysis Parameter Optimization
2.7.2. Monosaccharide Composition of Pinot Noir Wine Polysaccharides
3. Materials and Methods
3.1. Materials and Reagents
3.2. PNWPs Extraction
3.3. Protein and Total Phenol Analysis
3.4. Ultraviolet-Visible Spectroscopy Analysis
3.5. Fourier Transform Infrared Spectroscopy Analysis
3.6. Molecular Weight Analysis
3.7. Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry (MALDI-TOF) Analysis
3.8. Monosaccharide Composition Analysis
3.9. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Perez-Jimenez, M.; Sherman, E.; Pozo-Bayon, M.A.; Pinu, F.R. Application of untargeted volatile profiling and data driven approaches in wine flavoromics research. Food Res. Int. 2021, 145, 110392. [Google Scholar] [CrossRef]
- Jones-Moore, H.R.; Jelley, R.E.; Marangon, M.; Fedrizzi, B. The polysaccharides of winemaking: From grape to wine. Trends Food Sci. Technol. 2021, 111, 731–740. [Google Scholar] [CrossRef]
- Jones-Moore, H.R.; Jelley, R.E.; Marangon, M.; Fedrizzi, B. The interactions of wine polysaccharides with aroma compounds, tannins, and proteins, and their importance to winemaking. Food Hydrocoll. 2022, 123, 107150. [Google Scholar] [CrossRef]
- Mitropoulou, A.; Hatzidimitriou, E.; Paraskevopoulou, A. Aroma release of a model wine solution as influenced by the presence of non-volatile components. Effect of commercial tannin extracts, polysaccharides and artificial saliva. Food Res. Int. 2011, 44, 1561–1570. [Google Scholar] [CrossRef]
- Garrido-Banuelos, G.; Buica, A.; Schuckel, J.; Zietsman, A.J.J.; Willats, W.G.T.; Moore, J.P.; Du Toit, W.J. Investigating the relationship between grape cell wall polysaccharide composition and the extractability of phenolic compounds into Shiraz wines. Part I: Vintage and ripeness effects. Food Chem. 2019, 278, 36–46. [Google Scholar] [CrossRef]
- Martinez-Lapuente, L.; Apolinar-Valiente, R.; Guadalupe, Z.; Ayestaran, B.; Perez-Magarino, S.; Williams, P.; Doco, T. Polysaccharides, oligosaccharides and nitrogenous compounds change during the ageing of Tempranillo and Verdejo sparkling wines. J. Sci. Food Agric. 2018, 98, 291–303. [Google Scholar] [CrossRef]
- Guadalupe, Z.; Palacios, A.; Ayestarán, B. Maceration enzymes and mannoproteins: A possible strategy to increase colloidal stability and color extraction in red wines. J. Agric. Food Chem. 2007, 55, 4854–4862. [Google Scholar] [CrossRef]
- Filipe-Ribeiro, L.; Milheiro, J.; Guise, R.; Vilamarim, R.; Fraga, J.B.; Martins-Gomes, C.; Nunes, F.M.; Cosme, F. Efficiency of carboxymethylcellulose in red wine tartaric stability: Effect on wine phenolic composition, chromatic characteristics and colouring matter stability. Food Chem. 2021, 360, 129996. [Google Scholar] [CrossRef]
- Kassara, S.; Li, S.; Smith, P.; Blando, F.; Bindon, K. Pectolytic enzyme reduces the concentration of colloidal particles in wine due to changes in polysaccharide structure and aggregation properties. Int. J. Biol. Macromol. 2019, 140, 546–555. [Google Scholar] [CrossRef]
- Laguna, L.; Bartolomé, B.; Moreno-Arribas, M.V. Mouthfeel perception of wine: Oral physiology, components and instrumental characterization. Trends Food Sci. Technol. 2017, 59, 49–59. [Google Scholar] [CrossRef]
- Chong, H.H.; Cleary, M.T.; Dokoozlian, N.; Ford, C.M.; Fincher, G.B. Soluble cell wall carbohydrates and their relationship with sensory attributes in Cabernet Sauvignon wine. Food Chem. 2019, 298, 124745. [Google Scholar] [CrossRef]
- Dufour, C.; Bayonove, C.L. Influence of wine structurally different polysaccharides on the volatility of aroma substances in a model system. J. Agric. Food Chem. 1999, 47, 671–677. [Google Scholar] [CrossRef]
- Gil, M.; Kontoudakis, N.; Gonzalez, E.; Esteruelas, M.; Fort, F.; Canals, J.M.; Zamora, F. Influence of grape maturity and maceration length on color, polyphenolic composition, and polysaccharide content of Cabernet Sauvignon and Tempranillo wines. J. Agric. Food Chem. 2012, 60, 7988–8001. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Lapuente, L.; Apolinar-Valiente, R.; Guadalupe, Z.; Ayestaran, B.; Perez-Magarino, S.; Williams, P.; Doco, T. Influence of Grape Maturity on Complex Carbohydrate Composition of Red Sparkling Wines. J. Agric. Food Chem. 2016, 64, 5020–5030. [Google Scholar] [CrossRef]
- Apolinar-Valiente, R.; Romero-Cascales, I.; Williams, P.; Gómez-Plaza, E.; López-Roca, J.M.; Ros-García, J.M.; Doco, T. Effect of winemaking techniques on polysaccharide composition of Cabernet Sauvignon, Syrah and Monastrell red wines. Aust. J. Grape Wine Res. 2014, 20, 62–71. [Google Scholar] [CrossRef]
- Martinez Lapuente, L.; Guadalupe, Z.; Ayestaran, B.; Perez-Porras, P.; Bautista-Ortin, A.B.; Gomez-Plaza, E. Ultrasound treatment of crushed grapes: Effect on the must and red wine polysaccharide composition. Food Chem. 2021, 356, 129669. [Google Scholar] [CrossRef]
- Ayestarán, B.; Guadalupe, Z.; León, D. Quantification of major grape polysaccharides (Tempranillo v.) released by maceration enzymes during the fermentation process. Anal. Chim. Acta 2004, 513, 29–39. [Google Scholar] [CrossRef]
- Doco, T.; Williams, P.; Cheynier, V. Effect of flash release and pectinolytic enzyme treatments on wine polysaccharide composition. J. Agric. Food Chem. 2007, 55, 6643–6649. [Google Scholar] [CrossRef]
- Guadalupe, Z.; Ayestarán, B. Polysaccharide profile and content during the vinification and aging of Tempranillo red wines. J. Agric. Food Chem. 2007, 55, 10720–10728. [Google Scholar] [CrossRef]
- Apolinar-Valiente, R.; Williams, P.; Romero-Cascales, I.; Gomez-Plaza, E.; Lopez-Roca, J.M.; Ros-Garcia, J.M.; Doco, T. Polysaccharide composition of Monastrell red wines from four different Spanish terroirs: Effect of wine-making techniques. J. Agric. Food Chem. 2013, 61, 2538–2547. [Google Scholar] [CrossRef]
- Jeong, H.K.; Lee, D.; Kim, H.P.; Baek, S.H. Structure analysis and antioxidant activities of an amylopectin-type polysaccharide isolated from dried fruits of Terminalia chebula. Carbohydr. Polym. 2019, 211, 100–108. [Google Scholar] [CrossRef] [PubMed]
- Boulet, J.C.; Williams, P.; Doco, T. A Fourier transform infrared spectroscopy study of wine polysaccharides. Carbohydr. Polym. 2007, 69, 79–85. [Google Scholar] [CrossRef]
- Baca-Bocanegra, B.; Martinez-Lapuente, L.; Nogales-Bueno, J.; Hernandez-Hierro, J.M.; Ferrer-Gallego, R. Feasibility study on the use of ATR-FTIR spectroscopy as a tool for the estimation of wine polysaccharides. Carbohydr. Polym. 2022, 287, 119365. [Google Scholar] [CrossRef]
- Gheribi, R.; Habibi, Y.; Khwaldia, K. Prickly pear peels as a valuable resource of added-value polysaccharide: Study of structural, functional and film forming properties. Int. J. Biol. Macromol. 2019, 126, 238–245. [Google Scholar] [CrossRef] [PubMed]
- Sharma, M.; Aguado, R.; Murtinho, D.; Valente, A.J.M.; Ferreira, P.J.T. Novel approach on the synthesis of starch betainate by transesterification. Int. J. Biol. Macromol. 2021, 182, 1681–1689. [Google Scholar] [CrossRef]
- Dong, X.; Zhu, C.-P.; Huang, G.-Q.; Xiao, J.-X. Fractionation and structural characterization of polysaccharides derived from red grape pomace. Process Biochem. 2021, 109, 37–45. [Google Scholar] [CrossRef]
- Kostalova, Z.; Hromadkova, Z. Structural characterisation of polysaccharides from roasted hazelnut skins. Food Chem. 2019, 286, 179–184. [Google Scholar] [CrossRef]
- Hong, T.; Yin, J.Y.; Nie, S.P.; Xie, M.Y. Applications of infrared spectroscopy in polysaccharide structural analysis: Progress, challenge and perspective. Food Chem. X 2021, 12, 100168. [Google Scholar] [CrossRef]
- Liu, J.; Liu, C.; Zheng, X.; Chen, M.; Tang, K. Soluble soybean polysaccharide/nano zinc oxide antimicrobial nanocomposite films reinforced with microfibrillated cellulose. Int. J. Biol. Macromol. 2020, 159, 793–803. [Google Scholar] [CrossRef]
- Zhang, J.; Wen, C.; Gu, J.; Ji, C.; Duan, Y.; Zhang, H. Effects of subcritical water extraction microenvironment on the structure and biological activities of polysaccharides from Lentinus edodes. Int. J. Biol. Macromol. 2019, 123, 1002–1011. [Google Scholar] [CrossRef]
- Yang, B.; Wu, Q.; Luo, Y.; Yang, Q.; Wei, X.; Kan, J. High-pressure ultrasonic-assisted extraction of polysaccharides from Hovenia dulcis: Extraction, structure, antioxidant activity and hypoglycemic. Int. J. Biol. Macromol. 2019, 137, 676–687. [Google Scholar] [CrossRef]
- Chen, X.; Jin, J.; Tang, J.; Wang, Z.; Wang, J.; Jin, L.; Lu, J. Extraction, purification, characterization and hypoglycemic activity of a polysaccharide isolated from the root of Ophiopogon japonicus. Carbohydr. Polym. 2011, 83, 749–754. [Google Scholar] [CrossRef]
- Del Barrio-Galán, R.; Pérez-Magariño, S.; Ortega-Heras, M.; Guadalupe, Z.; Ayestarán, B. Polysaccharide characterization of commercial dry yeast preparations and their effect on white and red wine composition. LWT Food Sci. Technol. 2012, 48, 215–223. [Google Scholar] [CrossRef]
- Cordeiro Caillot, A.R.; de Lacerda Bezerra, I.; Palhares, L.; Santana-Filho, A.P.; Chavante, S.F.; Sassaki, G.L. Structural characterization of blackberry wine polysaccharides and immunomodulatory effects on LPS-activated RAW 264.7 macrophages. Food Chem. 2018, 257, 143–149. [Google Scholar] [CrossRef] [PubMed]
- Ducasse, M.-A.; Canal-Llauberes, R.-M.; de Lumley, M.; Williams, P.; Souquet, J.-M.; Fulcrand, H.; Doco, T.; Cheynier, V. Effect of macerating enzyme treatment on the polyphenol and polysaccharide composition of red wines. Food Chem. 2010, 118, 369–376. [Google Scholar] [CrossRef]
- Lei, X.; Zhu, Y.; Wang, X.; Zhao, P.; Liu, P.; Zhang, Q.; Chen, T.; Yuan, H.; Guo, Y. Wine polysaccharides modulating astringency through the interference on interaction of flavan-3-ols and BSA in model wine. Int. J. Biol. Macromol. 2019, 139, 896–903. [Google Scholar] [CrossRef]
- Watrelot, A.A.; Schulz, D.L.; Kennedy, J.A. Wine polysaccharides influence tannin-protein interactions. Food Hydrocoll. 2017, 63, 571–579. [Google Scholar] [CrossRef]
- Carvalho, E.; Mateus, N.; Plet, B.; Pianet, I.; Dufourc, E.; De Freitas, V. Influence of wine pectic polysaccharides on the interactions between condensed tannins and salivary proteins. J. Agric. Food Chem. 2006, 54, 8936–8944. [Google Scholar] [CrossRef]
- Wan, M.; Wang, M.; Zhao, Y.; Deng, H.; Tan, C.; Lin, S.; Kong, Y.; Tong, Y.; Meng, X. Extraction of mannoprotein from Saccharomyces cerevisiae and analysis of its chemical composition and molecular structure. Int. J. Biol. Macromol. 2021, 193, 2252–2259. [Google Scholar] [CrossRef]
- Guadalupe, Z.; Martínez-Pinilla, O.; Garrido, Á.; Carrillo, J.D.; Ayestarán, B. Quantitative determination of wine polysaccharides by gas chromatography–mass spectrometry (GC–MS) and size exclusion chromatography (SEC). Food Chem. 2012, 131, 367–374. [Google Scholar] [CrossRef]
- Canalejo, D.; Guadalupe, Z.; Martinez-Lapuente, L.; Ayestaran, B.; Perez-Magarino, S.; Doco, T. Characterization of polysaccharide extracts recovered from different grape and winemaking products. Food Res. Int. 2022, 157, 111480. [Google Scholar] [CrossRef] [PubMed]
- Vidal, S.; Williams, P.; Doco, T.; Moutounet, M.; Pellerin, P. The polysaccharides of red wine: Total fractionation and characterization. Carbohydr. Polym. 2003, 54, 439–447. [Google Scholar] [CrossRef]
- Eder, S.; Zueblin, P.; Diener, M.; Peydayesh, M.; Boulos, S.; Mezzenga, R.; Nystrom, L. Effect of Polysaccharide Conformation on Ultrafiltration Separation Performance. Carbohydr. Polym. 2021, 260, 117830. [Google Scholar] [CrossRef] [PubMed]
- Lan, H.; Nunes, C.; Lopes, G.R.; Wang, K.; Zhao, L.; Coimbra, M.A.; Hu, Z. In vitro immunomodulatory activity of water-soluble glucans from fresh and dried Longan (Dimocarpus longan Lour.). Carbohydr. Polym. 2021, 266, 118106. [Google Scholar] [CrossRef]
- Mohammed, J.K.; Mahdi, A.A.; Ahmed, M.I.; Ma, M.; Wang, H. Preparation, deproteinization, characterization, and antioxidant activity of polysaccharide from Medemia argun fruit. Int. J. Biol. Macromol. 2020, 155, 919–926. [Google Scholar] [CrossRef]
- Lin, X.; Ji, X.; Wang, M.; Yin, S.; Peng, Q. An alkali-extracted polysaccharide from Zizyphus jujuba cv. Muzao: Structural characterizations and antioxidant activities. Int. J. Biol. Macromol. 2019, 136, 607–615. [Google Scholar] [CrossRef]
- Wang, L.; Li, X.; Wang, B. Synthesis, characterization and antioxidant activity of selenium modified polysaccharides from Hohenbuehelia serotina. Int. J. Biol. Macromol. 2018, 120, 1362–1368. [Google Scholar] [CrossRef]
- Wei, W.; Li, J.; Qi, X.; Zhong, Y.; Zuo, G.; Pan, X.; Su, T.; Zhang, J.; Dong, W. Synthesis and characterization of a multi-sensitive polysaccharide hydrogel for drug delivery. Carbohydr. Polym. 2017, 177, 275–283. [Google Scholar] [CrossRef]
- Canalejo, D.; Guadalupe, Z.; Martinez-Lapuente, L.; Ayestaran, B.; Perez-Magarino, S. Optimization of a method to extract polysaccharides from white grape pomace by-products. Food Chem. 2021, 365, 130445. [Google Scholar] [CrossRef]
- Sran, K.S.; Sundharam, S.S.; Krishnamurthi, S.; Roy Choudhury, A. Production, characterization and bio-emulsifying activity of a novel thermostable exopolysaccharide produced by a marine strain of Rhodobacter johrii CDR-SL 7Cii. Int. J. Biol. Macromol. 2019, 127, 240–249. [Google Scholar] [CrossRef]
- Liu, D.; Tang, W.; Yin, J.-Y.; Nie, S.-P.; Xie, M.-Y. Monosaccharide composition analysis of polysaccharides from natural sources: Hydrolysis condition and detection method development. Food Hydrocoll. 2021, 116, 106641. [Google Scholar] [CrossRef]
- Wang, Z.; Zeng, Y.; Luo, D. Structure elucidation of a non-branched and entangled heteropolysaccharide from Tremella sanguinea Peng and its antioxidant activity. Carbohydr. Polym. 2016, 152, 33–40. [Google Scholar] [CrossRef] [PubMed]

| Samples | Yield (g/g) # | Protein Content (%) | Total Phenolic Content (%) |
|---|---|---|---|
| PNWP-L | 0.24 | 0.3% ± 0.1% | 1.7% ± 0.0% |
| PNWP-H | 0.41 | 5.3% ± 0.4% | 2.2% ± 0.0% |
| Samples | Peak No. | Peak MW (kDa) | MW Distribution (kDa) | Area Account (%) |
|---|---|---|---|---|
| PNWP-L | 1 | 15.9 ± 0.0 | 12–25 | 4.9 |
| 2 | 6.2 ± 0.0 | 5–12 | 95.1 | |
| PNWP-H | 1 | 66.5 ± 0.3 | 25–670 | 72.3 |
| 2 | 16.2 ± 0.0 | 12–25 | 22.1 | |
| 3 | 6.2 ± 0.0 | 5–12 | 5.6 |
| Peak No. | Composition | Monosaccharide Composition Percentage (%) | p Value # | |
|---|---|---|---|---|
| PNWP-L | PNWP-H | |||
| 1 | α-Araf | 1.3 ± 0.0 l | 8.4 ± 0.1 d | * |
| 2 | α-Arap | 2.4 ± 0.1 i | 13.1 ± 0.2 c | * |
| 3 | ß-Arap | 1.2 ± 0.0 l | 6.5 ± 0.1 f | * |
| 4 | α-Rhap | 7.6 ± 0.2 d | 6.6 ± 0.1 f | * |
| 5 | ß-Rhap | 0.6 ± 0.0 no | 0.5 ± 0.0 l | - |
| 6 | ß-Araf | 0.5 ± 0.0 o | 2.4 ± 0.1 h | * |
| 7 | α-Fucp | 0.2 ± 0.0 p | 0.1 ± 0.0 n | - |
| 8 | ß-Fucp | -q | 0.1 ± 0.0 n | * |
| 9 | α-Xylp | 1.0 ± 0.1 m | 0.2 ± 0.0 mn | * |
| 10 | ß-Xylp | 0.5 ± 0.0 o | 0.1 ± 0.0 n | * |
| 11 | α-GalfA | 31.8 ± 0.3 a | 2.1 ± 0.3 i | * |
| 12 | α-Manp | 9.9 ± 0.2 c | 21.6 ± 0.4 a | * |
| 13 | ß-GalfA | 7.3 ± 0.1 e | 0.4 ± 0.1 lm | * |
| 14 | α-Galf/α-Glcf | 1.5 ± 0.0 k | 5.8 ± 0.2 g | * |
| 15 | ß-Glcf | 0.7 ± 0.0 n | 0.1 ± 0.0 n | * |
| 16 | ß-Manp | 0.9 ± 0.0 m | 1.2 ± 0.0 k | * |
| 17 | α-Galp | 4.3 ± 0.1 h | 18.7 ± 0.2 b | * |
| 18 | ß-Galf | 0.5 ± 0.0 o | 1.6 ± 0.2 j | * |
| 19 | ß-Galp | 1.5 ± 0.0 k | 6.9 ± 0.1 e | * |
| 20 | α-GalpA | 13.8 ± 0.2 b | 1.1 ± 0.1 k | * |
| 21 | ß-GalpA | 4.5 ± 0.0 g | 0.4 ± 0.0 lm | * |
| 22 | α-Glcp | 5.7 ± 0.0 f | 1.0 ± 0.0 k | * |
| 23 | ß-Glcp/α-GlcpA | 1.9 ± 0.0 j | 0.5 ± 0.0 l | * |
| 24 | ß-GlcpA | 0.3 ± 0.0 p | 0.2 ± 0.0 mn | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, D.; Alcazar-Magana, A.; Qian, Y.P.; Tao, Y.; Qian, M.C. Isolation, Characterization, and Compositional Analysis of Polysaccharides from Pinot Noir Wines: An Exploratory Study. Molecules 2022, 27, 8330. https://doi.org/10.3390/molecules27238330
Zhu D, Alcazar-Magana A, Qian YP, Tao Y, Qian MC. Isolation, Characterization, and Compositional Analysis of Polysaccharides from Pinot Noir Wines: An Exploratory Study. Molecules. 2022; 27(23):8330. https://doi.org/10.3390/molecules27238330
Chicago/Turabian StyleZhu, Danye, Armando Alcazar-Magana, Yan Ping Qian, Yongsheng Tao, and Michael C. Qian. 2022. "Isolation, Characterization, and Compositional Analysis of Polysaccharides from Pinot Noir Wines: An Exploratory Study" Molecules 27, no. 23: 8330. https://doi.org/10.3390/molecules27238330
APA StyleZhu, D., Alcazar-Magana, A., Qian, Y. P., Tao, Y., & Qian, M. C. (2022). Isolation, Characterization, and Compositional Analysis of Polysaccharides from Pinot Noir Wines: An Exploratory Study. Molecules, 27(23), 8330. https://doi.org/10.3390/molecules27238330









