Facile Synthesis of Some Coumarin Derivatives and Their Cytotoxicity through VEGFR2 and Topoisomerase II Inhibition
Abstract
1. Introduction
2. Results and Discussion
2.1. Chemistry
2.2. In Silico Studies
2.2.1. Molecular Docking
2.2.2. ADME Pharmacokinetics
2.3. Biological Assessment
2.3.1. Cytotoxicity against MCF-7 and HepG2 Cancer Cell Lines
2.3.2. Enzyme Target Activity
3. Experimental Methods
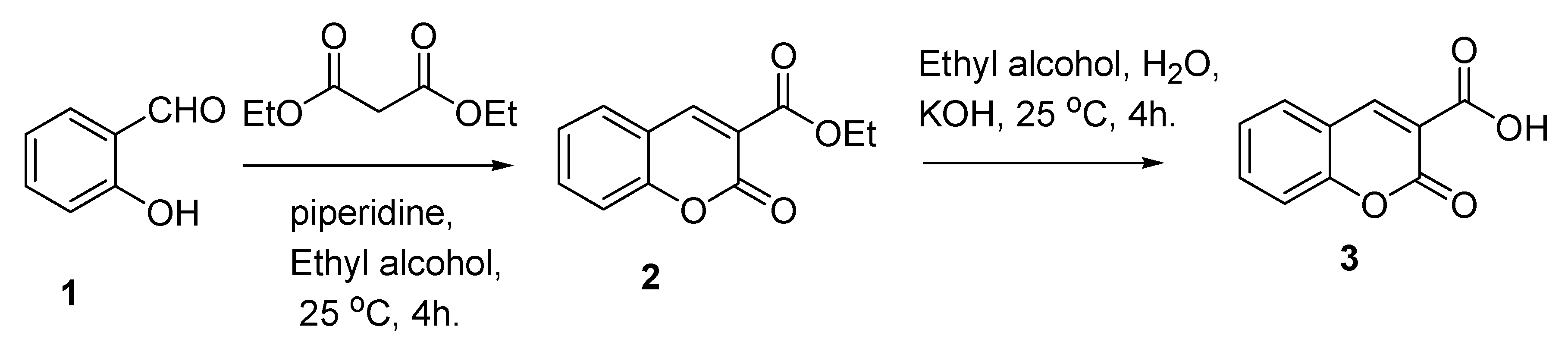
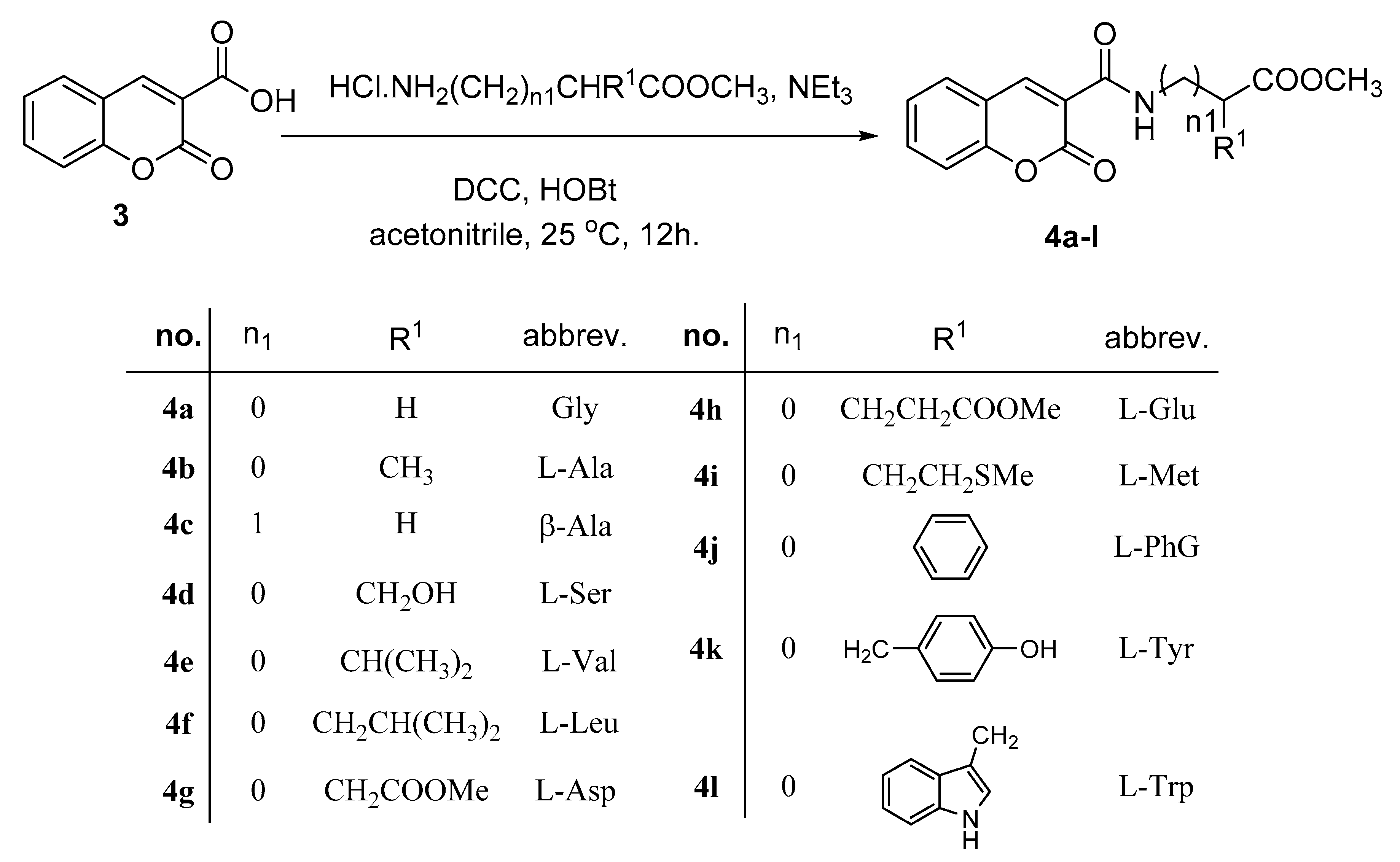
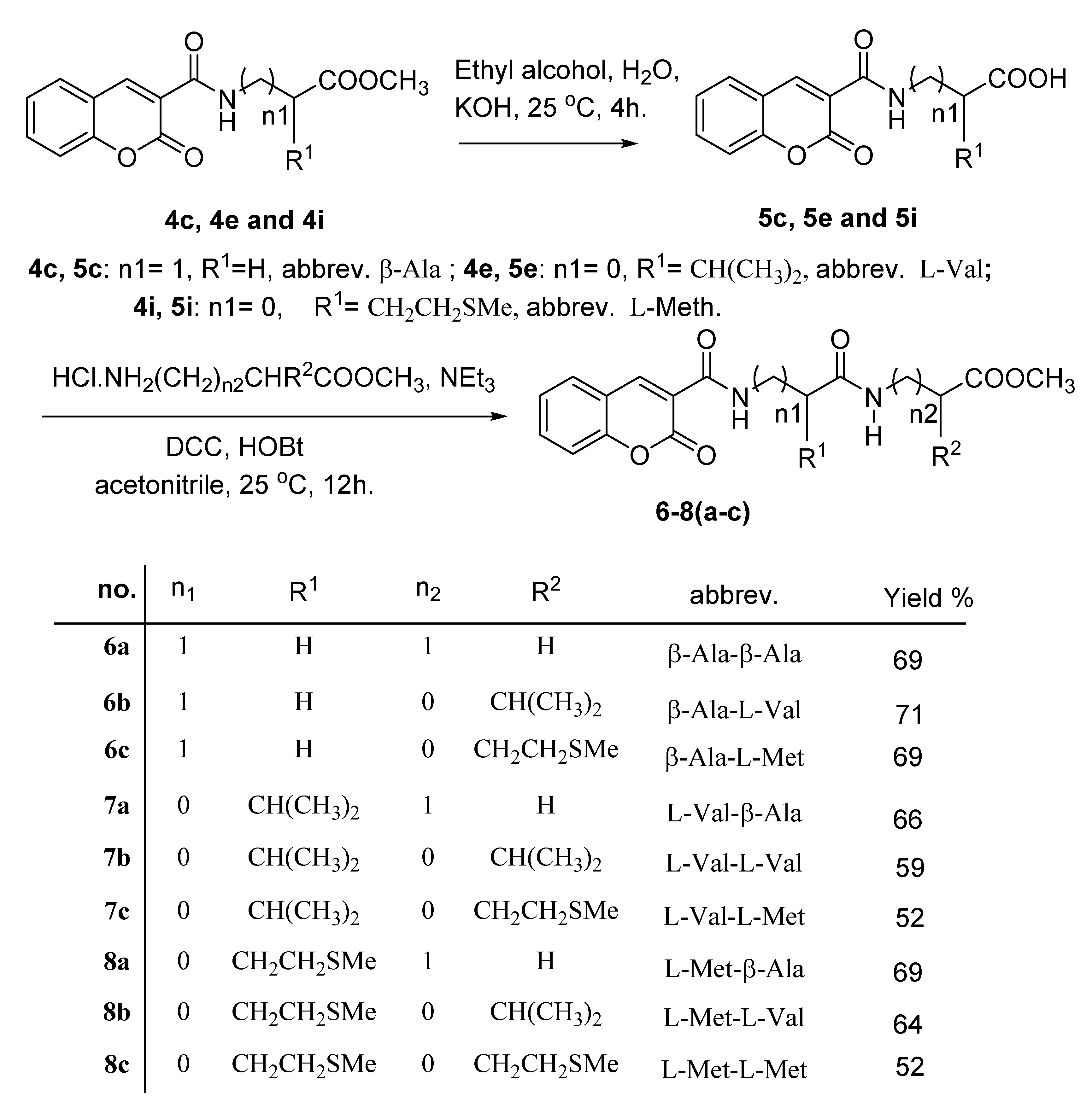
3.1. Synthesis
3.2. In Silico Studies
3.2.1. Molecular Docking
3.2.2. ADME Pharmacokinetics
3.3. Biological Assessment
3.3.1. Cytotoxicity Using MTT Assay
3.3.2. Enzyme Target Assays
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Wild, C.P.; Weiderpass, E.; Stewart, B.W. World Cancer Report: Cancer Research for Cancer Prevention; International Agency for Research on Cancer WHO: Geneva, Switzerland, 2020. [Google Scholar]
- Zhu, J.-J.; Jiang, J.-G. Pharmacological and Nutritional Effects of Natural Coumarins and Their Structure–Activity Relationships. Mol. Nutr. Food Res. 2018, 62, 1701073. [Google Scholar] [CrossRef] [PubMed]
- Rawat, A.; Reddy, A.V.B. Recent advances on anticancer activity of coumarin derivatives. Eur. J. Med. Chem. Rep. 2022, 5, 100038. [Google Scholar] [CrossRef]
- Murray, R.D.H.; Mndez, J.; Brown, S.A. The Natural Coumarins: Occurrence, Chemistry, and Biochemistry; Wiley: Chichester, UK; New York, NY, USA, 1982. [Google Scholar]
- Banerji, A.; Mallick, B.; Chatterjee, A.; Budzikiewics, H.; Breuer, M. Assafoetidin and ferocolicin, two sesquiterpenoid coumarins from ferula assafoetida regel. Tetrahedron Lett. 1988, 29, 1557–1560. [Google Scholar] [CrossRef]
- Lacy, A.; O’Kennedy, R. Studies on coumarins and coumarin-related compounds to determine their therapeutic role in the treatment of cancer. Curr. Pharm. Des. 2004, 10, 3797–3811. [Google Scholar] [CrossRef]
- Sardari, S.; Nishibe, S.; Daneshtalab, M. Coumarins, the bioactive structures with antifungal property. Stud. Nat. Prod. Chem. 2000, 23, 335–393. [Google Scholar] [CrossRef]
- Kojima, E.; Iimuro, A.; Nakajima, M.; Kinuta, H.; Asada, N.; Sako, Y.; Nakata, Z.; Uemura, K.; Arita, S.; Miki, S.; et al. Pocket-to-Lead: Structure-Based De Novo Design of Novel Non-peptidic HIV-1 Protease Inhibitors Using the Ligand Binding Pocket as a Template. J. Med. Chem. 2022, 65, 6157–6170. [Google Scholar] [CrossRef]
- Kaye, P.T.; Musa, M.A.; Nchinda, A.T.; Nocanda, X.W. Novel Heterocyclic Analogues of the HIV-1 Protease Inhibitor, Ritonavir. Synth. Commun. 2004, 34, 2575–2589. [Google Scholar] [CrossRef]
- O’Kennedy, R.; Thornes, R.D. Coumarins: Biology, Applications, and Mode of Action; John Wiley & Sons: Chichester, UK; New York, NY, USA, 1997. [Google Scholar]
- Fiedler, V.B.; Scholtholt, J. Effects of carbocromene on myocardial oxygen consumption in isolated dog hearts. J. Pharmacol. Exp. Ther. 1981, 217, 306–313. [Google Scholar] [PubMed]
- Yu, D.; Suzuki, M.; Xie, L.; Morris-Natschke, S.L.; Lee, K.-H. Recent progress in the development of coumarin derivatives as potent anti-HIV agents. Med. Res. Rev. 2003, 23, 322–345. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Kashiwada, Y.; Cosentino, L.M.; Fan, S.; Chen, C.-H.; McPhail, A.T.; Fujioka, T.; Mihashi, K.; Lee, K.-H. Anti-AIDS Agents. 15. Synthesis and Anti-HIV Activity of Dihydroseselins and Related Analogs. J. Med. Chem. 1994, 37, 3947–3955. [Google Scholar] [CrossRef]
- Pereira, N.A.; Pereira, B.M.; Nascimento, M.C.; Parente, J.P.; Mors, W.B. Pharmacological Screening of Plants Recommended by Folk Medicine as Snake Venom Antidotes; IV. Protection against Jararaca Venom by Isolated Constituents1. Planta Med. 1994, 60, 99–100. [Google Scholar] [CrossRef]
- Sarveswaran, S.; Gautam, S.C.; Ghosh, J. Wedelolactone, a medicinal plant-derived coumestan, induces caspase-dependent apoptosis in prostate cancer cells via downregulation of PKCε without inhibiting Akt. Int. J. Oncol. 2012, 41, 2191–2199. [Google Scholar] [CrossRef] [PubMed]
- García-Vilas, J.A.; Quesada, A.R.; Medina, M.Á. 4-Methylumbelliferone Inhibits Angiogenesis in Vitro and in Vivo. J. Agric. Food Chem. 2013, 61, 4063–4071. [Google Scholar] [CrossRef] [PubMed]
- Nagy, N.; Kuipers, H.F.; Frymoyer, A.R.; Ishak, H.D.; Bollyky, J.B.; Wight, T.N.; Bollyky, P.L. 4-Methylumbelliferone Treatment and Hyaluronan Inhibition as a Therapeutic Strategy in Inflammation, Autoimmunity, and Cancer. Front. Immunol. 2015, 6, 123. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharyya, S.S.; Paul, S.; Mandal, S.K.; Banerjee, A.; Boujedaini, N.; Khuda-Bukhsh, A.R. A synthetic coumarin (4-Methyl-7 hydroxy coumarin) has anti-cancer potentials against DMBA-induced skin cancer in mice. Eur. J. Pharmacol. 2009, 614, 128–136. [Google Scholar] [CrossRef]
- Duangdee, N.; Mahavorasirikul, W.; Prateeptongkum, S. Design synthesis and anti-proliferative activity of some new coumarin substituted hydrazide–hydrazone derivatives. J. Chem. Sci. 2020, 132, 66. [Google Scholar] [CrossRef]
- Chimichi, S.; Boccalini, M.; Salvador, A.; Dall’Acqua, F.; Basso, G.; Viola, G. Synthesis and Biological Evaluation of New Geiparvarin Derivatives. Chem. Med. Chem. 2009, 4, 769–779. [Google Scholar] [CrossRef] [PubMed]
- Konc, J.T.; Hejchman, E.; Kruszewska, H.; Wolska, I.; Maciejewska, D. Synthesis and pharmacological activity of O-aminoalkyl derivatives of 7-hydroxycoumarin. Eur. J. Med. Chem. 2011, 46, 2252–2263. [Google Scholar] [CrossRef]
- Ghany, L.M.A.A.; El-Dydamony, N.M.; Helwa, A.A.; Abdelraouf, S.M.; Abdelnaby, R.M. Coumarin-acetohydrazide derivatives as novel antiproliferative agents via VEGFR-2/AKT axis inhibition and apoptosis triggering. New J. Chem. 2022, 46, 17394–17409. [Google Scholar] [CrossRef]
- Batran, R.Z.; Dawood, D.H.; El-Seginy, S.A.; Ali, M.M.; Maher, T.J.; Gugnani, K.S.; Rondon-Ortiz, A.N. New Coumarin Derivatives as Anti-Breast and Anti-Cervical Cancer Agents Targeting VEGFR-2 and p38α MAPK. Archiv. Der. Pharm. 2017, 350, 1700064. [Google Scholar] [CrossRef]
- Ahmed, E.Y.; Latif, N.A.A.; El-Mansy, M.F.; Elserwy, W.S.; Abdelhafez, O.M. VEGFR-2 inhibiting effect and molecular modeling of newly synthesized coumarin derivatives as anti-breast cancer agents. Bioorganic Med. Chem. 2020, 28, 115328. [Google Scholar] [CrossRef]
- Mohamed, T.K.; Batran, R.Z.; Elseginy, S.A.; Ali, M.M.; Mahmoud, A.E. Synthesis, anticancer effect and molecular modeling of new thiazolylpyrazolyl coumarin derivatives targeting VEGFR-2 kinase and inducing cell cycle arrest and apoptosis. Bioorganic Chem. 2019, 85, 253–273. [Google Scholar] [CrossRef]
- Liu, H.; Ren, Z.-L.; Wang, W.; Gong, J.-X.; Chu, M.-J.; Ma, Q.-W.; Wang, J.-C.; Lv, X.-H. Novel coumarin-pyrazole carboxamide derivatives as potential topoisomerase II inhibitors: Design, synthesis and antibacterial activity. Eur. J. Med. Chem. 2018, 157, 81–87. [Google Scholar] [CrossRef]
- Konkoľová, E.; Hudáčová, M.; Hamuľaková, S.; Jendželovský, R.; Vargová, J.; Ševc, J.; Fedoročko, P.; Kožurková, M. Tacrine-Coumarin Derivatives as Topoisomerase Inhibitors with Antitumor Effects on A549 Human Lung Carcinoma Cancer Cell Lines. Molecules 2021, 26, 1133. [Google Scholar] [CrossRef]
- Hueso-Falcón, I.; Amesty, Á.; Anaissi-Afonso, L.; Lorenzo-Castrillejo, I.; Machín, F.; Estévez-Braun, A. Synthesis and biological evaluation of naphthoquinone-coumarin conjugates as topoisomerase II inhibitors. Bioorg. Med. Chem. Lett. 2017, 27, 484–489. [Google Scholar] [CrossRef] [PubMed]
- Holmes, K.; Roberts, O.L.; Thomas, A.M.; Cross, M.J. Vascular endothelial growth factor receptor-2: Structure, function, intracellular signalling and therapeutic inhibition. Cell Signal 2007, 19, 2003–2012. [Google Scholar] [CrossRef]
- Fontanella, C.; Ongaro, E.; Bolzonello, S.; Guardascione, M.; Fasola, G.; Aprile, G. Clinical advances in the development of novel VEGFR2 inhibitors. Ann. Transl. Med. 2014, 2, 123. [Google Scholar] [CrossRef] [PubMed]
- Reddy, N.S.; Mallireddigari, M.R.; Cosenza, S.; Gumireddy, K.; Bell, S.C.; Reddy, E.P.; Reddy, M.V.R. Synthesis of new coumarin 3-(N-aryl) sulfonamides and their anticancer activity. Bioorganic Med. Chem. Lett. 2004, 14, 4093–4097. [Google Scholar] [CrossRef] [PubMed]
- Reddy, N.S.; Gumireddy, K.; Mallireddigari, M.R.; Cosenza, S.C.; Venkatapuram, P.; Bell, S.C.; Reddy, E.P.; Reddy, M.V.R. Novel coumarin-3-(N-aryl)carboxamides arrest breast cancer cell growth by inhibiting ErbB-2 and ERK1. Bioorg. Med. Chem. 2005, 13, 3141–3147. [Google Scholar] [CrossRef]
- Pereira, T.M.; Franco, D.P.; Vitorio, F.; Kummerle, A.E. Coumarin Compounds in Medicinal Chemistry: Some Important Examples from the Last Years. Curr. Top. Med. Chem. 2018, 18, 124–148. [Google Scholar] [CrossRef]
- Naik, M.D.; Bodke, Y.D.; M, V.K.; BC, R. An efficient one-pot synthesis of coumarin-amino acid derivatives as potential anti-inflammatory and antioxidant agents. Synth. Commun. 2020, 50, 1210–1216. [Google Scholar] [CrossRef]
- Kulkarni, M.V.; Kulkarni, G.M.; Lin, C.-H.; Sun, C.-M. Recent Advances in Coumarins and 1-Azacoumarins as Versatile Biodynamic Agents. Curr. Med. Chem. 2006, 13, 2795–2818. [Google Scholar] [CrossRef]
- Al-Masoudi, N.A.; Al-Masoudi, I.A.; Ali, I.A.I.; Al-Soud, Y.A.; Saeed, B.; la Colla, P. Amino acid derivatives. Part I. Synthesis, antiviral and antitumor evaluation of new alpha-amino acid esters bearing coumarin side chain. Acta Pharm. 2006, 56, 175–188. [Google Scholar]
- Asagarasu, A.; Uchiyama, T.; Achiwa, K. Syntheses of HIV-protease inhibitors having a peptide moiety which binds to GP120. Chem. Pharm. Bull. 1998, 46, 697–703. [Google Scholar] [CrossRef] [PubMed]
- Davies, J.S.; Mohammed, A.K. Assessment of racemisation in N-alkylated amino-acid derivatives during peptide coupling in a model dipeptide system. J. Chem. Soc. Perkin Trans. 1981, 1, 2982–2990. [Google Scholar] [CrossRef]
- König, W.; Geiger, R. A new method for synthesis of peptides: Activation of the carboxyl group with dicyclohexylcarbodiimide using 1-hydroxybenzotriazoles as additives. Chem. Ber. 1970, 103, 788–798. [Google Scholar] [CrossRef]
- Strakova, I.; Petrova, M.; Belyakov, S.; Strakovs, A. Reactions of 4-chloro-3-formyl-coumarin with primary amines. Chem. Heterocycl. Compd. 2006, 42, 574–582. [Google Scholar] [CrossRef]
- Abdel-Hafez, G.A.; Mohamed, A.-M.I.; Youssef, A.F.; Simons, C.; Aboraia, A.S. Synthesis, computational study and biological evaluation of 9-acridinyl and 1-coumarinyl-1,2,3-triazole-4-yl derivatives as topoisomerase II inhibitors. J. Enzym. Inhib. Med. Chem. 2022, 37, 502–513. [Google Scholar] [CrossRef]
- Guruge, A.G.; Udawatte, C.; Weerasinghe, S.; Guruge, A.G.; Udawatte, C.; Weerasinghe, S. An In Silico Approach of Coumarin-Derived Inhibitors for Human DNA Topoisomerase I. Aust. J. Chem. 2016, 69, 1005–1015. [Google Scholar] [CrossRef]
- Khalifa, M.M.; Al-Karmalawy, A.A.; Elkaeed, E.B.; Nafie, M.S.; Tantawy, M.A.; Eissa, I.H.; Mahdy, H.A. Topo II inhibition and DNA intercalation by new phthalazine-based derivatives as potent anticancer agents: Design, synthesis, anti-proliferative, docking, and in vivo studies. J. Enzym. Inhib. Med. Chem. 2022, 37, 299–314. [Google Scholar] [CrossRef]
- Nafie, M.S.; Boraei, A.T.A. Exploration of novel VEGFR2 tyrosine kinase inhibitors via design and synthesis of new alkylated indolyl-triazole Schiff bases for targeting breast cancer. Bioorganic Chem. 2022, 122, 105708. [Google Scholar] [CrossRef] [PubMed]
- Nafie, M.S.; Tantawy, M.A.; Elmgeed, G.A. Screening of different drug design tools to predict the mode of action of steroidal derivatives as anti-cancer agents. Steroids 2019, 152, 108485. [Google Scholar] [CrossRef]
- Matin, P.; Hanee, U.; Alam, M.S.; Jeong, J.E.; Matin, M.M.; Rahman, M.R.; Mahmud, S.; Alshahrani, M.M.; Kim, B. Novel Galactopyranoside Esters: Synthesis, Mechanism, In Vitro Antimicrobial Evaluation and Molecular Docking Studies. Molecules 2022, 27, 4125. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Boraei, A.T.A.; Eltamany, E.H.; Ali, I.A.I.; Gebriel, S.M.; Nafie, M.S. Synthesis of new substituted pyridine derivatives as potent anti-liver cancer agents through apoptosis induction: In vitro, in vivo, and in silico integrated approaches. Bioorganic Chem. 2021, 111, 104877. [Google Scholar] [CrossRef]
- Nafie, M.S.; Amer, A.M.; Mohamed, A.K.; Tantawy, E.S. Discovery of novel pyrazolo [3,4-b]pyridine scaffold-based derivatives as potential PIM-1 kinase inhibitors in breast cancer MCF-7 cells. Bioorganic Med. Chem. 2020, 28, 115828. [Google Scholar] [CrossRef] [PubMed]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Nafie, M.S.; Mahgoub, S.; Amer, A.M. Antimicrobial and antiproliferative activities of novel synthesized 6-(quinolin-2-ylthio)pyridine derivatives with molecular docking study as multi-targeted JAK2/STAT3 inhibitors. Chem. Biol. Drug Des. 2021, 97, 553–564. [Google Scholar] [CrossRef]





| Tested Targets | VEGFR2 (PDB = 3U6J) a | Topoisomerase IIα (PDB = 1ZXM) b | |
|---|---|---|---|
| Compounds | |||
| Co-crystallized ligand (key interactions) | 3 HB with Asp 1064, Cys 919 and Lys 868 | 1 HB with Ser 149 | |
| 4a | - | 1 HB with Ser 149 | |
| 4b | 1HB with Lys 868 | - | |
| 4c | 1HB with Asp 1046 | - | |
| 4d | 3 HB with Tyr 916, Lys 868, and Asp 1046 | 1 HB with Ser 149 | |
| 4e | 1HB with Cys 919 | - | |
| 4f | Arene-cation with Lys 868 | 1 HB with Asn 150 | |
| 4g | 3HB with Tyr 916, Lys 868 and Asp 1046 | 2 HB with Ser 149 and Asn 91 | |
| 4h | 2 HB with Lys 868 and Cys 916 | - | |
| 4i | 2HB with Lys 868 and Cys 919 | 1 HB with Ser 149 | |
| 4j | 2HB with Thr 916 and Lys 868 | 1 HB with Ser 149 and Arene-cation with Arg 98 | |
| 4k | 1 HB with Asp 1046 and Arene-cation with Lys 868 | 3HB with Ser 149, Thr 215 and Asn 120 | |
| 4l | 1HB with Asp 1046 | 2 HB with Ser 149 and Asn 150 | |
| 5c | 2HB with Cys 919 and Lys 868 | - | |
| 5e | - | 2 HB with Asn 150 and Lys 168 | |
| 5i | 2HB with Thr 916 and Cys 919 | 1 HB with Ala 167 | |
| 6a | 2HB with Thr 916 and Asp 1046 | 1 HB with Ser 149 | |
| 6b | 1 HB with Cys 919 | - | |
| 6c | 1 HB with Cys 919 | 2 HB with Arg 98 and Ser 149 | |
| 7a | - | - | |
| 7b | 1HB with Asp 1046 | 2 HB with Ser 149 and Asn 120 | |
| 7c | - | 1 HB with Lys 157 and arene-cation Arg 98 | |
| 8a | Arene-cation Lys 868 | arene-cation Arg 98 and Lys 157 | |
| 8b | Arene-cation Lys 868 | 1 HB with Ser 149 | |
| 8c | 1 HB with Lys 920 | - | |
| # | Molinspiration 2018.10 | MolSoft | SwissADME | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| MWt (D) | MV (A3) | PSA (A2) | Log p | Nrotb | Nviolations | HBA | HBD | Solubility (mg/L) | Drug Likeness (Lipinski Pfizer Filter) | |
| 4k | 381.3 | 334.3 | 105.8 | 2.69 | 7 | 0 | 6 | 2 | 519.9 | “Yes, drug like” MW ≤ 500, Log p ≤ 4.15, HBA ≤ 10 and HDD ≤ 5 |
| 6c | 434.5 | 388.2 | 114.7 | 1.81 | 10 | 0 | 7 | 2 | 485.3 | |
| Compound | IC50 ± SD * (µM) | |
|---|---|---|
| VEGFR2 | Topoisomerase II | |
| 4k | 23.6 ± 1.19 | 4.1 ± 0.64 |
| 6c | 34.2 ± 1.06 | 8.6 ± 0.15 |
| Doxorubicin | --- | 9.65 ± 0.72 |
| Sorafenib | 30.0 ± 0.42 | ---- |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gomaa, M.S.; Ali, I.A.I.; El Enany, G.; El Ashry, E.S.H.; El Rayes, S.M.; Fathalla, W.; Ahmed, A.H.A.; Abubshait, S.A.; Abubshait, H.A.; Nafie, M.S. Facile Synthesis of Some Coumarin Derivatives and Their Cytotoxicity through VEGFR2 and Topoisomerase II Inhibition. Molecules 2022, 27, 8279. https://doi.org/10.3390/molecules27238279
Gomaa MS, Ali IAI, El Enany G, El Ashry ESH, El Rayes SM, Fathalla W, Ahmed AHA, Abubshait SA, Abubshait HA, Nafie MS. Facile Synthesis of Some Coumarin Derivatives and Their Cytotoxicity through VEGFR2 and Topoisomerase II Inhibition. Molecules. 2022; 27(23):8279. https://doi.org/10.3390/molecules27238279
Chicago/Turabian StyleGomaa, Mohamed S., Ibrahim A. I. Ali, Gaber El Enany, El Sayed H. El Ashry, Samir M. El Rayes, Walid Fathalla, Abdulghany H. A. Ahmed, Samar A. Abubshait, Haya A. Abubshait, and Mohamed S. Nafie. 2022. "Facile Synthesis of Some Coumarin Derivatives and Their Cytotoxicity through VEGFR2 and Topoisomerase II Inhibition" Molecules 27, no. 23: 8279. https://doi.org/10.3390/molecules27238279
APA StyleGomaa, M. S., Ali, I. A. I., El Enany, G., El Ashry, E. S. H., El Rayes, S. M., Fathalla, W., Ahmed, A. H. A., Abubshait, S. A., Abubshait, H. A., & Nafie, M. S. (2022). Facile Synthesis of Some Coumarin Derivatives and Their Cytotoxicity through VEGFR2 and Topoisomerase II Inhibition. Molecules, 27(23), 8279. https://doi.org/10.3390/molecules27238279






