Chemical Profiling and Quantification of Potential Bioactive Components in Gandouling Pill by Ultra-High Performance Liquid Chromatography Coupled with Diode Array Detector/Quadruple-Qrbitrap Mass Spectrometry
Abstract
1. Introduction
2. Results
2.1. Optimization of the Extraction Method
2.2. Optimization of the Separation Method
2.3. Chemical Profiling of GDL Pill
2.3.1. Characterization of Alkaloids
2.3.2. Characterization of Organic Acids
2.3.3. Characterization of Phenolic Compounds
2.3.4. Characterization of Other Compounds
2.4. Absorption Components of GDL Pill in Rat Plasma
2.5. Quantitation of the Plasma-Absorption Components in GDL Pill
2.5.1. Method Validation
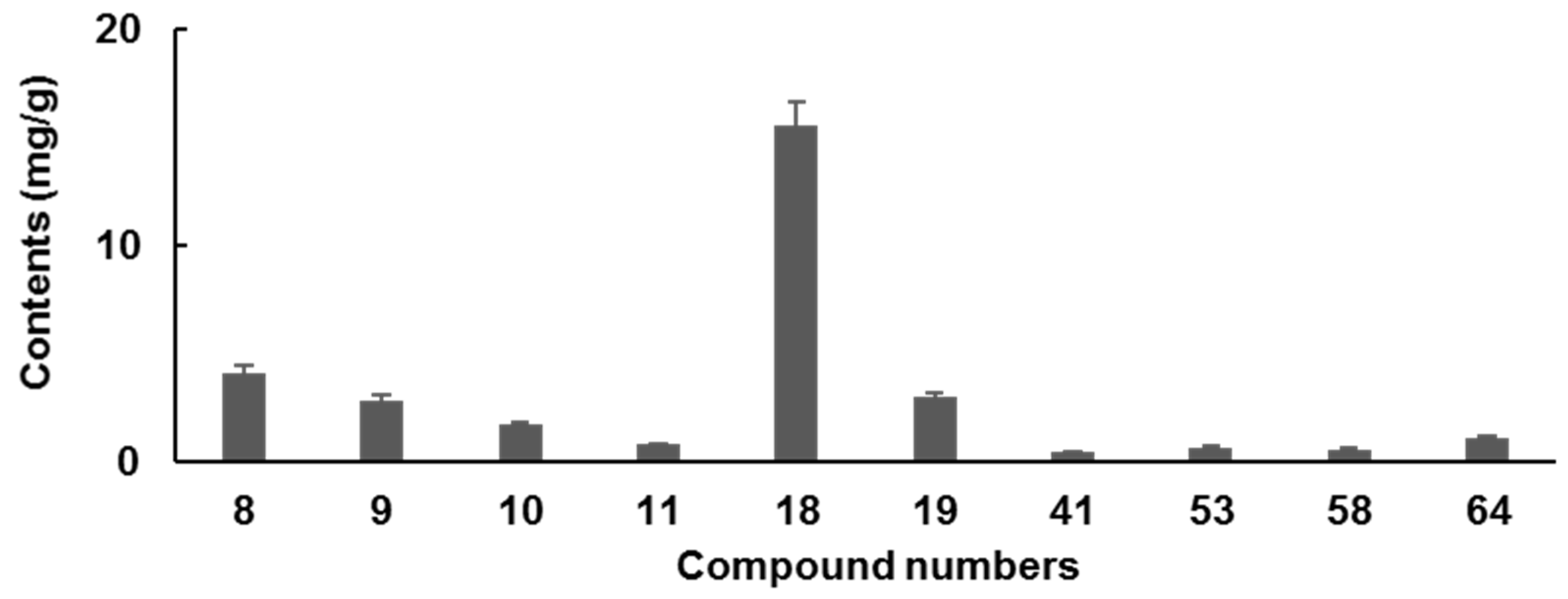
2.5.2. Sample Analysis
3. Materials and Methods
3.1. Chemicals and Reagents
3.2. Sample Solution Preparation
3.2.1. Preparation of Reference Standard Solutions
3.2.2. Preparation of Sample Solutions
3.3. Animal Experiments
3.4. Liquid Chromatography
3.5. Mass Spectrometry
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Chinese Pharmacopoeia Commission. Pharmacopoeia of the People’s Republic of China, 2015th ed.; Chinese Medical Science and Technology Press: Beijing, China, 2015; pp. 538–539. [Google Scholar]
- Qiao, X.; Li, R.; Miao, W.; Liu, J.; Chen, H.; Guo, D.; Ye, M. A targeted strategy to analyze untargeted mass spectral data: Rapid chemical profiling of Scutellaria baicalensis using ultra-high performance liquid chromatography coupled with hybrid quadrupole orbitrap mass spectrometry and key ion filtering. J. Chromatogr. A 2016, 1441, 83–95. [Google Scholar] [CrossRef] [PubMed]
- Shang, Z.; Xu, L.; Xiao, Y.; Du, W.; An, R.; Ye, M.; Qiao, X. A global profiling strategy using comprehensive two-dimensional liquid chromatography coupled with dual-mass spectrometry platforms: Chemical analysis of a multi-herb Chinese medicine formula as a case study. J. Chromatogr. A 2021, 1642, 462021. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Wu, W.Y.; Sun, G.X.; Wang, D.D.; Hou, J.J.; Yang, W.Z.; Jiang, B.H.; Liu, X.; Guo, D.A. A dynamic multiple reaction monitoring method for the multiple components quantification of complex traditional Chinese medicine preparations: Niuhuang Shangqing pill as an example. J. Chromatogr. A 2013, 1294, 58–69. [Google Scholar] [CrossRef]
- Shang, Z.; Xu, L.; Kuang, Y.; Lin, Y.; Liu, S.; Sun, L.; Bo, T.; Ye, M.; Qiao, X. Simultaneous determination of 35 constituents and elucidation of effective constituents in a multi-herb Chinese medicine formula Xiaoer-Feire-Kechuan. J. Pharm. Anal. 2021, 11, 717–725. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, B.; Zhang, J.; Tian, X.; Sun, D.; Li, Q.; Wang, R. Qualitative and quantitative analysis of Yifei Tongluo granules to identify main bioactive components using LC–DAD/MS and pharmacokinetic studies. J. Pharm. Biomed. Anal. 2019, 163, 130–136. [Google Scholar] [CrossRef] [PubMed]
- Bandmann, O.; Weiss, K.; Kaler, S. Wilson’s disease and other neurological copper disorders. Lancet Neurol. 2015, 14, 103–113. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, L.; Hao, C. Literature study on Gandouling tablet, a special preparation of Xin’an for the treatment of hepatolenticular degeneration. Chin. J. Exp. Tradit. Med. Formul. 2022, Unpublished work.
- Xu, L.; Cai, Y.; Jiang, H.; Wang, Y.; Han, H.; Hou, Z.; Ma, S.; Xu, M. Treatment of Wilson’s Disease with Intermingled Phlegm and Blood Stasis Syndrome by Gandouling Tablet. Chin. J. Exp. Tradit. Med. Form. 2017, 23, 173–177. [Google Scholar]
- Wang, Y.; Bao, Y.; Sun, M.; Yang, W.; Han, H.; Fang, X.; Zhang, J.; Xu, Z. Study of Gandouling interves wilson disease cognitive dysfunction in rats and the nerve cells apotosis of hippocampus. Chin. J. Clin. Pharmacol. 2015, 31, 2333–2336. [Google Scholar]
- Mei, Y.; Wei, L.; Tan, M.; Wang, C.; Zou, L.; Chen, J.; Cai, Z.; Yin, S.; Zhang, F.; Shan, C.; et al. Qualitative and quantitative analysis of the major constituents in Spatholobi Caulis by UFLC-Triple TOF-MS/MS and UFLC-QTRAP-MS/MS. J. Pharm. Biomed. Anal. 2020, 194, 113803. [Google Scholar] [CrossRef] [PubMed]
- Yao, M.; Li, J.; He, M.; Ouyang, H.; Ruan, L.; Huang, X.; Rao, Y.; Yang, S.; Zhou, X.; Bai, J. Investigation and identification of the multiple components of Rheum officinale Baill. Using ultra-high performance liquid chromatography coupled with quadrupole-time-of-flight tandem mass spectrometry and data mining strategy. J. Sep. Sci. 2021, 44, 681–690. [Google Scholar] [CrossRef] [PubMed]
- Qiao, X.; Wang, Q.; Song, W.; Qian, Y.; Xiao, Y.; An, R.; Guo, D.-A.; Ye, M. A chemical profiling solution for Chinese medicine formulae using comprehensive and loop-based multiple heart-cutting two-dimensional liquid chromatography coupled with quadrupole time-of-flight mass spectrometry. J. Chromatogr. A 2016, 1438, 198–204. [Google Scholar] [CrossRef] [PubMed]
- Tian, P.-P.; Zhang, X.-X.; Wang, H.-P.; Li, P.-L.; Liu, Y.-X.; Li, S.-J. Rapid Analysis of Components in Coptis chinensis Franch by Ultra-Performance Liquid Chromatography with Quadrupole Time-of-Flight Mass Spectrometry. Pharmacogn. Mag. 2017, 13, 175–179. [Google Scholar] [CrossRef] [PubMed]
- Hao, M.; Ji, D.; Li, L.; Su, L.; Zhang, J.; Wang, Q.; Gu, W.; Jiang, C.; Lu, T.; Mao, C. Metabolic Profiling Analysis of Three Processed Rhizomes of Curcuma wenyujin Y.H. Chen et C. Ling by Ultra-performance Liquid Chromatography/Time-of-Flight Mass Spectrometry. Pharmacogn. Mag. 2019, 15, 164–171. [Google Scholar]
- Chen, L.; Wang, S.; Qin, X.; Meng, M. Qualitative and Quantitative Analysis of Active Components of Copditis Rhizoma in Gandouling Pills by HPLC. Chin. J. Infor. TCM 2018, 25, 87–89. [Google Scholar]
- Sun, W.; Tong, L.; Miao, J.; Huang, J.; Li, D.; Li, Y.; Xiao, H.; Sun, H.; Bi, K. Separation and analysis of phenolic acids from Salvia miltiorrhiza and its related preparations by off-line two-dimensional hydrophilic interaction chromatography × reversed-phase liquid chromatography coupled with ion trap time-of-flight mass spectrometry. J. Chromatogr. A 2016, 1431, 79–88. [Google Scholar] [CrossRef] [PubMed]
- Jaiswal, Y.; Liang, Z.; Ho, A.; Chen, H.; Zhao, Z. Tissue-Specific Metabolite Profiling of Turmeric by Using Laser Microdissection, Ultra-High Performance Liquid Chromatography-Quadrupole Time of Fight-Mass Spectrometry and Liquid Chromatography-Tandem Mass Spectrometry. Eur. J. Mass Spectrom. 2014, 20, 383–393. [Google Scholar] [CrossRef] [PubMed]
- Xie, W.; Zhang, H.; Zeng, J.; Chen, H.; Zhao, Z.; Liang, Z. Tissues-based chemical profiling and semi-quantitative analysis of bioactive components in the root of Salvia miltiorrhiza Bunge by using laser microdissection system combined with UPLC-q-TOF-MS. Chem. Cent. J. 2016, 10, 42. [Google Scholar] [CrossRef] [PubMed]
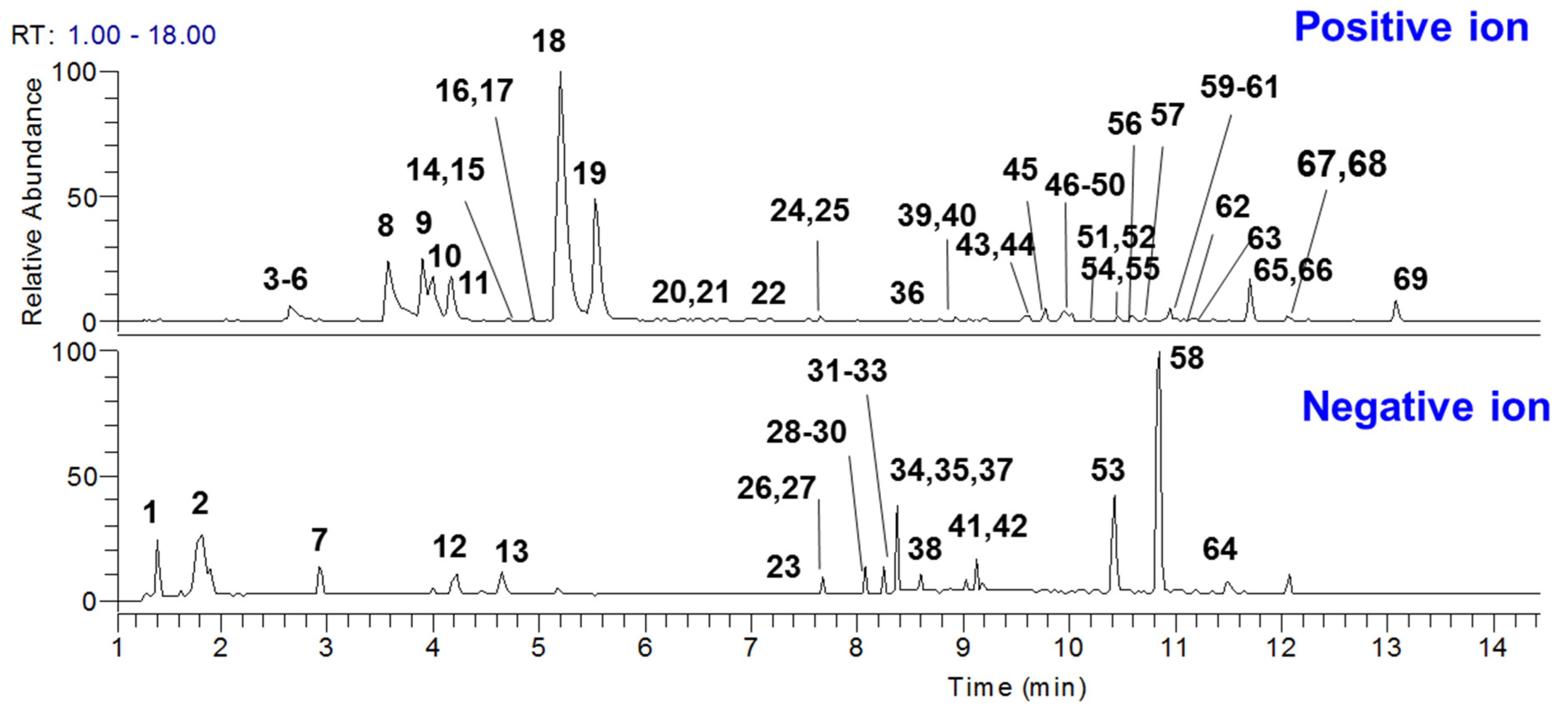
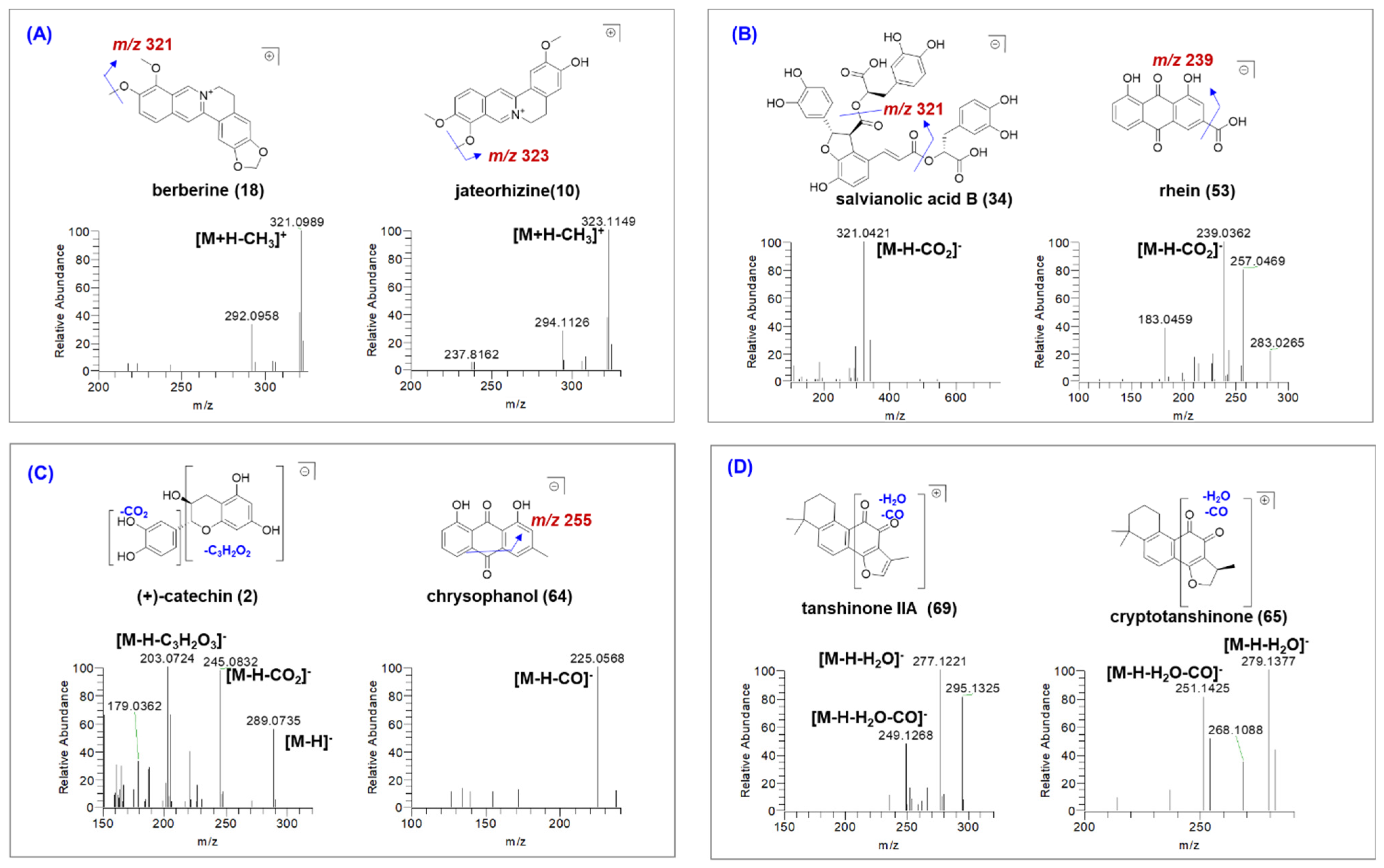
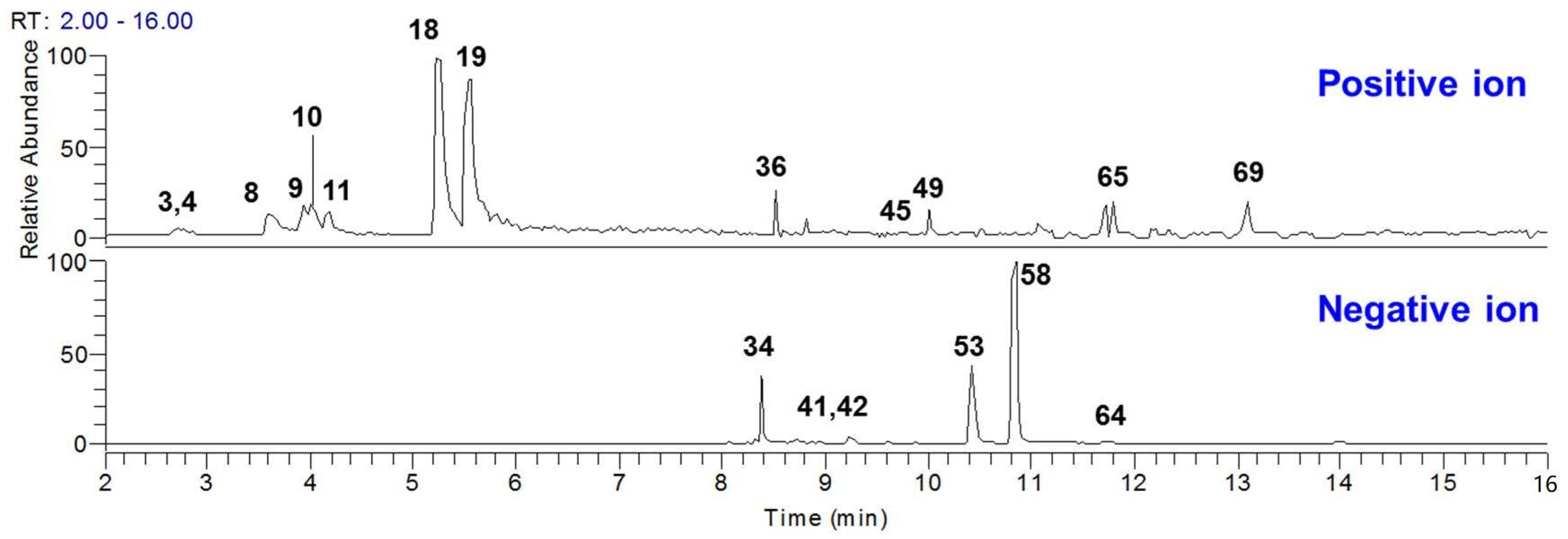
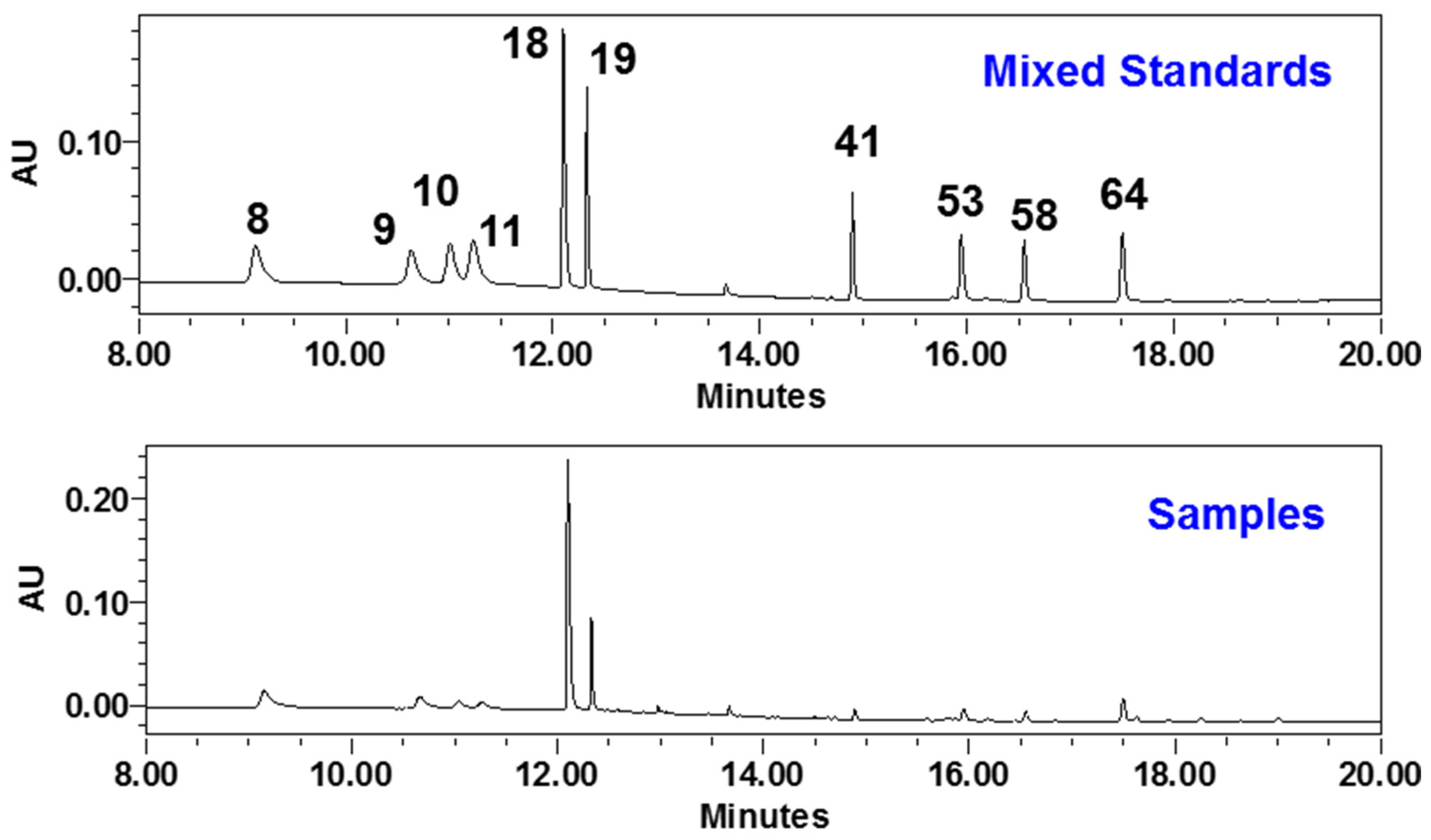
| Peak | tR | Formula | Measured [M − H]−/[M + H]+ (m/z) | Error (ppm) | Ion Mode | MS/MS Fragments | Source | Identification | Type | Plasma |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 * | 1.53 | C30H25O12 | 577.1389 | 2.4 | − | 407.0789, 289.0739, 125.0251 | JXT | procyanidin B2 [11] | phenolic | |
| 2 * | 1.88 | C15H13O6 | 289.0735 | 1.3 | − | 245.0831, 203.0724, 109.0300 | DH | (+)-catechin [12] | phenolic | |
| 3 | 2.62 | C19H18NO4 | 324.1235 | 0.7 | + | 309.0989 | HL | demethyleneberberine [13] | alkaloid | + |
| 4 | 2.65 | C19H16NO4 | 322.1077 | 1.3 | + | 307.0838, 294.1135, 279.0910 | HL | thalifendine or groenlandicine [13] | alkaloid | + |
| 5 * | 2.68 | C15H13O6 | 289.0736 | 2.3 | − | 245.0830, 203.0725, 109.0300 | JXT | epicatechin [11] | phenolic | |
| 6 | 2.76 | C19H18NO4 | 324.1231 | 0.3 | + | 309.0989 | HL | demethyleneberberine/isomer [13] | alkaloid | |
| 7 | 2.90 | C17H19O9 | 367.1053 | 3.2 | − | 193.0515, 134.0380 | HL | 5-O-feruloylquinic acid [14] | organic acid | |
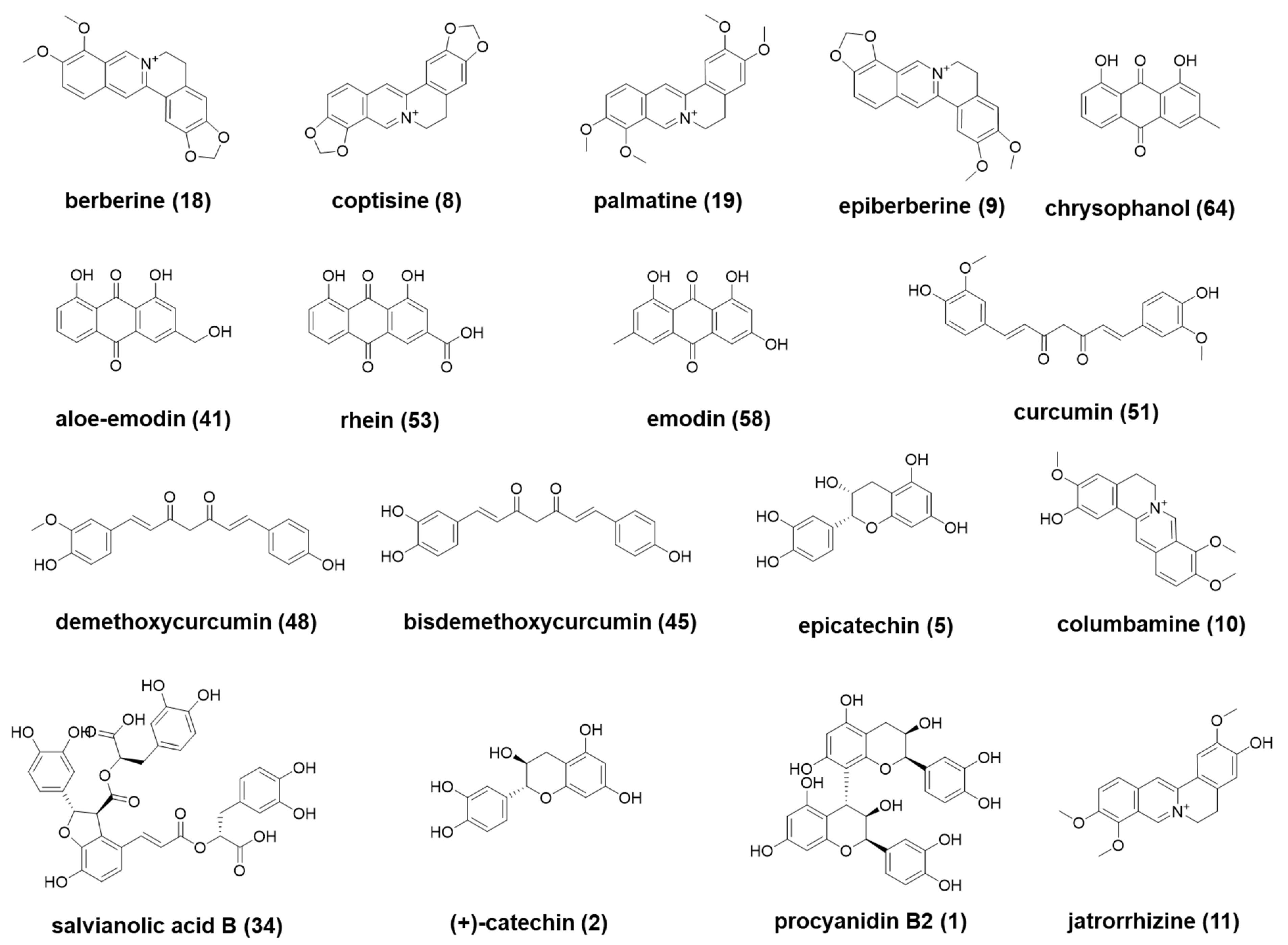
| 8 * | 3.56 | C19H14NO4 | 320.0921 | 1.4 | + | 292.0968, 236.8748 | HL | coptisine [13] | alkaloid | + |
| 9 * | 3.89 | C20H18NO4 | 336.1233 | 0.9 | + | 320.0908, 308.1270, 292.0979 | HL | epiberberine [13] | alkaloid | + |
| 10 * | 4.00 | C20H20NO4 | 338.1387 | 0.7 | + | 323.1150, 308.0917, 294.1116 | HL | columbamine [13] | alkaloid | + |
| 11 * | 4.16 | C20H20NO4 | 338.1389 | 0.9 | + | 323.1145, 308.0905, 294.1126 | HL | jatrorrhizine [13] | alkaloid | + |
| 12 | 4.17 | C17H19O9 | 367.1052 | 4.2 | − | 193.0514, 173.0463 | HL | 3-O-feruloylquinic acid [14] | organic acid | |
| 13 | 4.63 | C17H19O9 | 367.1054 | 4.3 | − | 191.0571, 173.0463 | HL | 4-O-feruloylquinic acid [14] | organic acid | |
| 14 | 4.66 | C36H38NO12 | 676.2398 | 1.4 | + | 430.4002, 334.1067 | HL | coptichine-quinic acid conjungate-CO + 2H [13] | alkaloid | |
| 15 | 4.69 | C20H16NO4 | 334.1078 | 1.3 | + | 306.1124 | HL | worenine [13] | alkaloid | |
| 16 | 4.91 | C21H20NO4 | 350.1390 | 1.8 | + | 334.1051 | HL | worenine + CH2 + 2H [13] | alkaloid | |
| 17 | 5.07 | C36H38NO12 | 676.2398 | 1.4 | + | 430.4001, 334.1066 | HL | coptichine-quinic [13] acid conjungate-CO + 2H # [13] | alkaloid | |
| 18 * | 5.18 | C20H18NO4 | 336.1234 | 1.0 | + | 321.0989, 292.0956 | HL | berberine [13] | alkaloid | + |
| 19 * | 5.51 | C21H22NO4 | 352.1546 | 0.8 | + | 337.1306, 322.1067, 308.1273 | HL | palmatine [13] | alkaloid | + |
| 20 | 6.34 | C21H20NO4 | 350.1392 | 1.5 | + | 335.1153, 306.1127 | HL | worenine + CH2 + 2H [13] | alkaloid | |
| 21 | 6.59 | C30H26NO8 | 528.1666 | −0.8 | + | 334.1072, 319.0836 | HL | demethylcoptichine/isomer [13] | alkaloid | |
| 22 | 7.17 | C15H21O2 | 233.1540 | 1.8 | + | 175.1120 | EZ | furanogermenone [15] | ketone | |
| 23 | 7.48 | C21H17O11 | 445.0800 | 1.8 | − | 283.0266, 239.0362 | DH | rhein-8-glucoside [12] | phenolic | |
| 24 | 7.64 | C30H26NO8 | 528.1663 | −1.5 | + | 334.1071, 319.0834 | HL | demethylcoptichine/isomer [13] | alkaloid | |
| 25 | 7.64 | C31H28NO9 | 558.1763 | 0.9 | + | 334.1069, 319.0836 | HL | coptichine + O [13] | alkaloid | |
| 26 | 7.68 | C22H21O11 | 461.1118 | 1.5 | − | 313.0581, 169.0150, 147.0458 | DH | rumejaposide D [12] | phenolic | |
| 27 | 7.68 | C38H17O4 | 537.1077 | −3.2 | − | 339.0527, 295.0626, 185.0252 | DS | lithospermic acid [17] | organic acid | |
| 28 | 8.08 | C22H19O12 | 475.0883 | 1.8 | − | 269.0469 | DH | endocrocin-glucoside [12] | phenolic | |
| 29 | 8.10 | C38H17O4 | 537.1071 | −3.6 | − | 295.0622, 185.0254, 109.0299 | DS | lithospermic acid/isomer [6] | organic acid | |
| 30 | 8.10 | C26H21O10 | 493.1167 | 4.3 | − | 295.0625, 185.0252, 109.0300 | DS | salvianolic acid A [17] | organic acid | |
| 31 | 8.24 | C14H23O15 | 431.1007 | −2.3 | − | 268.0391 | DH | aloe-emodin-1-glucoside/isomer [12] | phenolic | |
| 32 | 8.27 | C26H21O10 | 493.1169 | 3.4 | − | 295.0625, 185.0252, 109.0300 | DS | salvianolic acid A/isomer [17] | organic acid | |
| 33 | 8.28 | C14H23O15 | 431.1008 | −2.4 | − | 269.0470 | DH | aloe-emodin-1-glucoside/isomer [12] | phenolic | |
| 34 * | 8.38 | C36H29O16 | 717.1504 | 4.3 | − | 339.0526, 321.0421, 295.0629, 109.0301 | DS | salvianolic acid B [17] | organic acid | + |
| 35 | 8.46 | C26H19O10 | 491.1012 | −3.7 | − | 311.0581, 293.0473, 135.0459 | DS | salvianolic acid C [17] | organic acid | |
| 36 | 8.51 | C20H16NO7 | 382.0928 | 1.8 | + | 318.0754, 190.0499 | HL | dehydro-chilenine [13] | alkaloid | + |
| 37 | 8.51 | C22H19O11 | 459.0959 | 3.2 | − | 266.0597, 253.0519 | DH | 2-carboxyl chrysophanol-glc I [12] | phenolic | |
| 38 | 8.84 | C24H21O13 | 517.1014 | 2.6 | − | 269.0470 | DH | malonyl-emodin-glucoside [12] | phenolic | |
| 39 | 8.92 | C15H19O3 | 247.1330 | 0.8 | + | 139.0391, 123.0443 | EZ | zederone/isomer [15] | ketone | |
| 40 | 8.92 | C15H23O2 | 235.1697 | 2.2 | + | 189.1637, 177.1275 | EZ | curcumenone/isomer [15] | ketone | |
| 41 * | 9.24 | C15H9O5 | 269.0470 | 4.3 | − | 240.0440 | DH | aloe-emodin [12] | phenolic | |
| 42 | 9.27 | C18H13O8 | 357.0636 | 2.3 | − | 225.0569, 181.0670, 121.0301 | DS | salvianic acid C [17] | organic acid | + |
| 43 | 9.78 | C15H23O | 217.1588 | 0.6 | + | 161.0957 | EZ | furanodiene/isomer [15] | ketone | |
| 44 | 9.78 | C15H23O2 | 235.1695 | 1.2 | + | 177.1272, 161.0959 | EZ | curcumenone/isomer [15] | ketone | |
| 45 * | 9.88 | C19H17O6 | 309.1123 | 0.8 | + | 225.0910, 147.0441 | JH | bisdemethoxycurcumin [18] | phenolic | + |
| 46 | 9.95 | C15H23O | 217.1589 | 1.3 | + | 161.0964 | EZ | furanodiene/isomer [15] | ketone | |
| 47 | 9.95 | C15H23O2 | 235.1695 | 1.2 | + | 189.1639, 161.0963 | EZ | Curcumenol [15] | ketone | |
| 48 * | 10.00 | C20H19O6 | 339.1232 | 1.7 | + | 255.1016, 177.0547, 147.0441 | JH | demethoxycurcumin [18] | phenolic | |
| 49 | 10.03 | C15H19O3 | 247.1330 | 0.8 | + | 139.0390, 123.0444 | EZ | zederone [15] | ketone | + |
| 50 | 10.03 | C15H17O2 | 229.1225 | 1.2 | + | 201.1274, 123.0443 | EZ | curzeone/isomer [15] | ketone | |
| 51 * | 10.11 | C21H21O6 | 369.1338 | 1.6 | + | 285.1125, 253.0859, 177.0547 | JH | curcumin [18] | phenolic | |
| 52 | 10.22 | C15H25O2 | 237.1852 | 1.4 | + | 219.1746, 135.1169 | EZ | Neocurdione [15] | ketone | |
| 53 * | 10.41 | C15H7O6 | 283.0262 | 3.5 | − | 257.0469, 239.0362 | DH | rhein [12] | organic acid | + |
| 54 | 10.45 | C15H25O2 | 237.1852 | 1.4 | + | 219.1741, 135.1169 | EZ | curdione [15] | ketone | |
| 55 | 10.45 | C15H23O | 219.1746 | 1.3 | + | 135.1170 | EZ | germacrone/isomer [15] | ketone | |
| 56 | 10.58 | C18H15O3 | 279.1020 | 1.8 | + | 261.0909, 233.0961, 205.1009 | DS | dihydrotanshinone I [19] | tanshinone | |
| 57 | 10.70 | C15H17O2 | 229.1226 | 1.3 | + | 201.1274 | EZ | curzeone/isomer [15] | ketone | |
| 58 * | 10.83 | C15H9O5 | 269.0469 | 4.2 | − | 241.0518, 225.0569 | DH | emodin [12] | phenolic | + |
| 59 | 10.89 | C18H17O3 | 281.1174 | 0.9 | + | 263.1065, 235.1116 | DS | danshenxinkun B [19] | tanshinone | |
| 60 | 10.95 | C15H17O | 213.1275 | 0.9 | + | 198.1042, 185.1320 | EZ | Pyrocurzerenone [15] | ketone | |
| 61 | 10.95 | C15H19O2 | 231.1382 | 1.4 | + | 213.1267, 173.0959, 83.0862 | EZ | curzerenone/isomer [15] | ketone | |
| 62 | 11.17 | C15H19O2 | 231.1382 | 1.4 | + | 213.1279, 83.0860 | EZ | curzerenone/isomer [15] | ketone | |
| 63 | 11.31 | C15H19O2 | 231.1383 | 1.7 | + | 213.1273, 203.1432 | EZ | curzerenone/isomer [15] | ketone | |
| 64 * | 11.71 | C15H9O4 | 253.0519 | 3.2 | − | 225.0568 | DH | chrysophanol [12] | phenolic | + |
| 65 | 11.75 | C19H21O3 | 297.1488 | 1.0 | + | 279.1377, 251.1425 | DS | cryptotanshinone [19] | tanshinone | + |
| 66 | 11.75 | C18H13O3 | 277.0860 | 0.3 | + | 249.0904 | DS | tanshinone I [19] | tanshinone | |
| 67 | 12.37 | C15H23O | 219.1747 | 1.5 | + | 135.1167 | EZ | germacrone/isomer [15] | ketone | |
| 68 | 12.43 | C19H17O3 | 293.1174 | 0.8 | + | 275.1057, 247.1114 | DS | 1,2 -didehydrotanshinone IIA [19] | tanshinone | |
| 69 | 13.08 | C19H19O3 | 295.1332 | 1.4 | + | 277.1221, 249.1268 | DS | tanshinone IIA [19] | tanshinone | + |
| Analytes | Regression Equations | r2 | Linear Range (μg/mL) | Precious | Repeatability (n = 6) | Stability (n = 6) | Recovery (n = 6) | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Intra-Day (n = 6) | Inter-Day (n = 3) | Spiked (μg) | Found (μg) | Recovery (%) | RSD (%) | ||||||
| coptisine (8) | y = 184,80x − 1005.9 | 0.9999 | 1.56–25.0 | 0.12 | 0.52 | 1.77 | 0.60 | 2.43 | 2.50 | 97.06 | 1.24 |
| epiberberine (9) | y = 16,328x − 2759.6 | 0.9992 | 1.56–25.0 | 0.45 | 0.59 | 4.26 | 4.41 | 2.45 | 2.50 | 97.82 | 1.26 |
| columbamine (10) | y = 17,745x + 1440 | 0.9995 | 1.56–25.0 | 0.42 | 0.43 | 2.48 | 4.05 | 2.41 | 2.50 | 96.37 | 1.34 |
| jateorhizine (11) | y = 25,348x + 37,911 | 0.9973 | 1.56–25.0 | 1.03 | 1.96 | 3.57 | 2.01 | 2.59 | 2.50 | 103.43 | 1.69 |
| berberine (18) | y = 18,120x − 1359.4 | 0.9995 | 3.13–50.0 | 0.29 | 0.79 | 3.22 | 2.24 | 4.74 | 5.00 | 94.84 | 2.24 |
| palmatine (19) | y = 20,530x + 39,866 | 1.0000 | 1.56–25.0 | 1.62 | 1.33 | 3.34 | 2.33 | 2.66 | 2.50 | 106.21 | 1.70 |
| aloe-emodin (41) | y = 10,137x + 1241.4 | 0.9996 | 0.78–12.5 | 0.22 | 0.81 | 3.13 | 3.90 | 1.24 | 1.25 | 98.96 | 1.14 |
| rhein (53) | y = 14,615x + 3046.5 | 0.9993 | 0.78–12.5 | 0.32 | 1.63 | 1.07 | 1.22 | 1.21 | 1.25 | 96.58 | 0.99 |
| emodin (58) | y = 17,564x + 889.7 | 0.9999 | 0.78–12.5 | 0.34 | 0.61 | 0.99 | 1.45 | 1.23 | 1.25 | 98.04 | 1.81 |
| chrysophanol (64) | y = 11,515x + 1484.2 | 0.9997 | 0.78–12.5 | 0.74 | 0.60 | 0.42 | 0.45 | 1.23 | 1.25 | 98.62 | 4.81 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Y.; Hao, W.; Yang, Y.; Zhang, S.; Wang, H.; Wang, M.; Dong, T.; Shang, Z.; Yang, W. Chemical Profiling and Quantification of Potential Bioactive Components in Gandouling Pill by Ultra-High Performance Liquid Chromatography Coupled with Diode Array Detector/Quadruple-Qrbitrap Mass Spectrometry. Molecules 2022, 27, 8247. https://doi.org/10.3390/molecules27238247
Yang Y, Hao W, Yang Y, Zhang S, Wang H, Wang M, Dong T, Shang Z, Yang W. Chemical Profiling and Quantification of Potential Bioactive Components in Gandouling Pill by Ultra-High Performance Liquid Chromatography Coupled with Diode Array Detector/Quadruple-Qrbitrap Mass Spectrometry. Molecules. 2022; 27(23):8247. https://doi.org/10.3390/molecules27238247
Chicago/Turabian StyleYang, Yue, Wenjie Hao, Yulong Yang, Shijie Zhang, Han Wang, Meixia Wang, Ting Dong, Zhanpeng Shang, and Wenming Yang. 2022. "Chemical Profiling and Quantification of Potential Bioactive Components in Gandouling Pill by Ultra-High Performance Liquid Chromatography Coupled with Diode Array Detector/Quadruple-Qrbitrap Mass Spectrometry" Molecules 27, no. 23: 8247. https://doi.org/10.3390/molecules27238247
APA StyleYang, Y., Hao, W., Yang, Y., Zhang, S., Wang, H., Wang, M., Dong, T., Shang, Z., & Yang, W. (2022). Chemical Profiling and Quantification of Potential Bioactive Components in Gandouling Pill by Ultra-High Performance Liquid Chromatography Coupled with Diode Array Detector/Quadruple-Qrbitrap Mass Spectrometry. Molecules, 27(23), 8247. https://doi.org/10.3390/molecules27238247







