Application of Deep-Learning Algorithm Driven Intelligent Raman Spectroscopy Methodology to Quality Control in the Manufacturing Process of Guanxinning Tablets
Abstract
1. Introduction
2. Results and Discussion
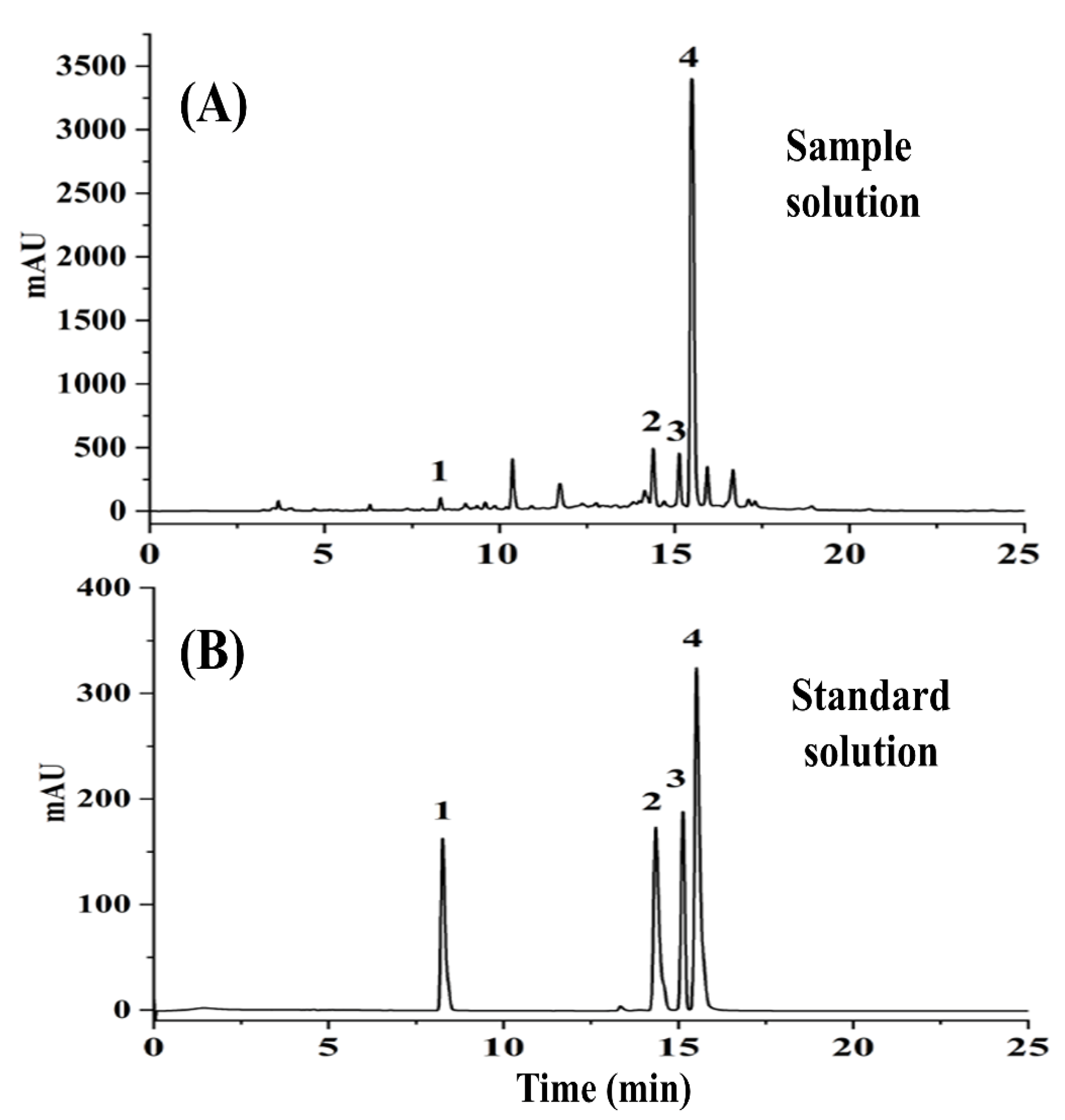
2.1. Determination of Bioactive Ingredients by HPLC-DAD
2.2. Determination of Soluble Solid by an Oven-Drying Method
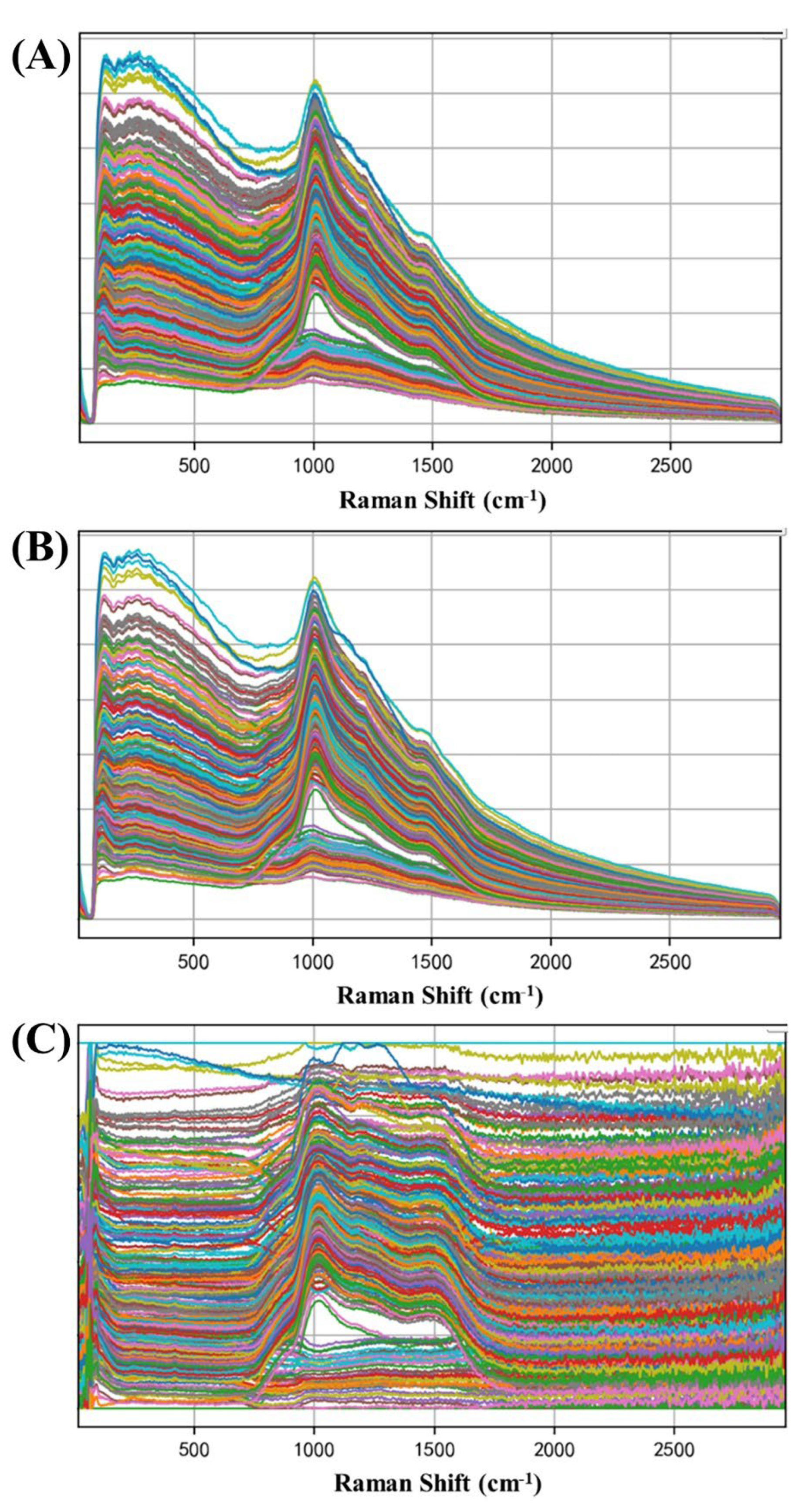
2.3. Pretreatment of Raman Spectra
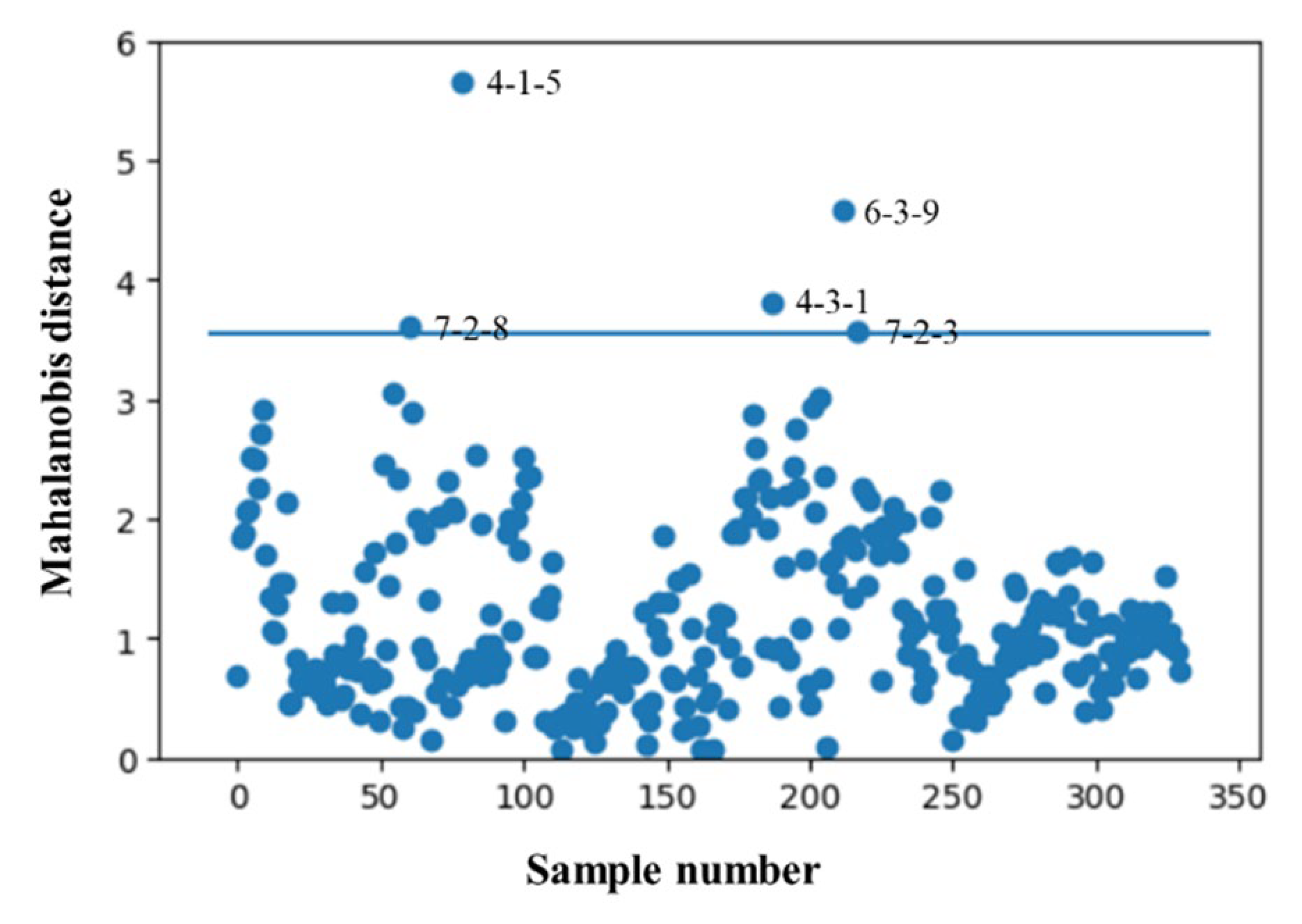
2.4. Removal of Abnormal Spectra
2.5. Determination of Variable Selection Methods
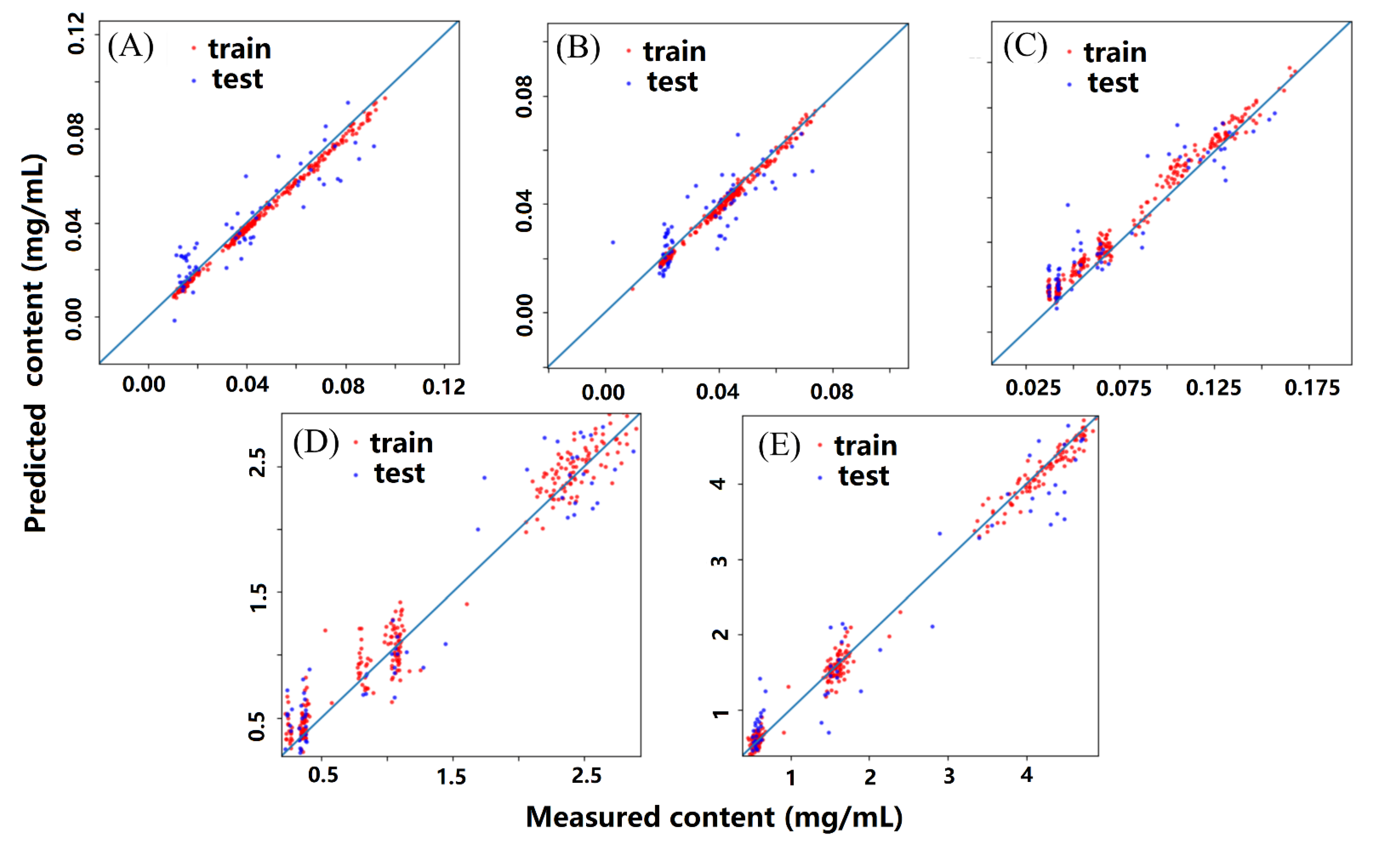
2.6. Comparison of Different Calibration Models
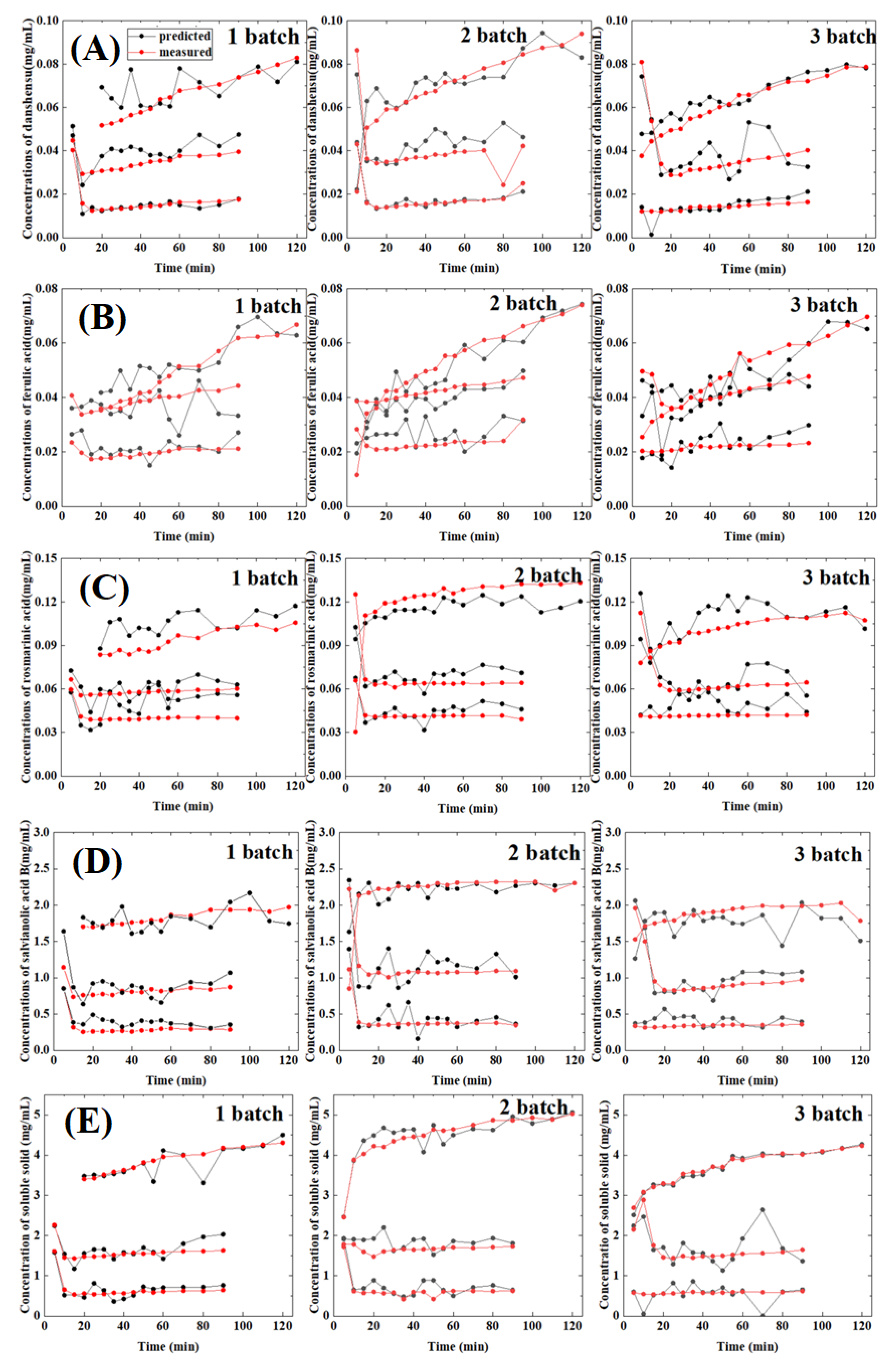
2.7. Application to Three Batches of Unknown Samples
3. Materials and Methods
3.1. Sample Collection
3.2. HPLC-DAD Analysis
3.3. Oven-Drying Method
3.4. Raman Spectra Acquisition
3.5. Removal of Abnormal Samples
3.6. Feature Band Filtering
3.7. Determination of Variable Selection Methods
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
Abbreviations
References
- Hou, Y.; Cai, X.; Miao, P.; Li, S.; Shu, C.; Li, P.; Li, W.; Li, Z. A feasibility research on the application of machine vision technology in appearance quality inspection of Xuesaitong dropping pills. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2021, 258, 119787. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhang, Y.; Wang, L.; Cui, T.; Wang, Y.; Chen, J.; Li, W. On-line screening of natural antioxidants and the antioxidant activity prediction for the extracts from flowers of Chrysanthemum morifolium ramat. J. Ethnopharmacol. 2022, 294, 115336. [Google Scholar] [CrossRef] [PubMed]
- Food and Drug Administration. Guidance for Industry: PAT-A Framework for Innovative Pharmaceutical Development, Manufacturing and Quality Assurance; Food and Drug Administration: Washington, DC, USA, 2004. [Google Scholar]
- Guo, F.L. Pharmacological effects of Salvia miltiorrhiza and its clinical application. Chin. J. Hosp. Pharm. 1999, 19, 363–365. [Google Scholar]
- Wang, C.S.; Yang, J.R.; Gui, C.Q.; Ding, B.P.; Song, J.G. Effects of Salvia miltiorrhiza injection on acute myocardial ischemia and hemorheology models of rat. Chin. J. Clin. Pharmacol. Ther. 2002, 1, 30–32. [Google Scholar]
- Chen, M.L.; Shou, Q.Y.; Pan, Y.M.; Zhang, J.B.; Sang, R.; Guan, M.W.; Wu, B.Y. Inhibitive and protective effectd of guanxinnning tablets on platelet aggregation and vascular endothelium in qi stagnation and blood tsasis rats. Chin. J. Clin. Pharmacol. Ther. 2005, 5, 586–589. [Google Scholar]
- Jurašeková, Z.; Fabriciová, G.; Silveira, L.F.; Lee, Y.N.; Gutak, J.M.; Ataabadi, M.M.; Kundrát, M. Raman Spectra and Ancient Life: Vibrational ID Profiles of Fossilized (Bone) Tissues. Int. J. Mol. Sci. 2022, 23, 10689. [Google Scholar] [CrossRef] [PubMed]
- Alali, H.; Ai, Y.; Pan, Y.L.; Videen, G.; Wang, C. A Collection of Molecular Fingerprints of Single Aerosol Particles in Air for Potential Identification and Detection Using Optical Trapping-Raman Spectroscopy. Molecules 2022, 27, 5966. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Xu, X.; Feng, S.; Zhang, H.; Xu, L.; Jiang, H.; Sun, B.; Meng, Y.; Chen, W. Rapid identification of salmonella serovars by using Raman spectroscopy and machine learning algorithm. Talanta 2022, 253, 123807. [Google Scholar] [CrossRef] [PubMed]
- Palermo, A.; Sodo, A.; Naciu, A.M.; Gioacchino, M.D.; Paolucci, A.; Masi, A.D.; Maggi, D.; Crucitti, P.; Longo, F.; Perrella, E.; et al. Clinical use of Raman spectroscopy improves diagnostic accuracy for indeterminate thyroid nodules. J. Clin. Endocrinol. Metab. 2022, 107, dgac537. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Ji, N.; Yin, L.; Wang, J. A non-invasive method for the determination of liquid injectables by Raman spectroscopy. AAPS PharmSciTech 2015, 16, 914–921. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Ma, X.; Shen, Y.T.; Nan, N.; Cao, L.M.; Yi, L.H. Non-Invasive determination of pethidine hydrochloride injection by Raman spectroscopy. Chin. Pharm. J. 2016, 51, 925–929. [Google Scholar]
- Zhang, R.; Wu, X.; Chen, Y.; Xiang, Y.; Liu, D.; Bian, X. Grey Wolf Optimizer for Variable Selection in Quantification of Quaternary Edible Blend Oil by Ultraviolet-Visible Spectroscopy. Molecules 2022, 27, 5141. [Google Scholar] [CrossRef] [PubMed]
- Aguilar-Hernández, I.; Afseth, N.K.; López-Luke, T.; Contreras-Torres, F.F.; Wold, J.P.; Ornelas-Soto, N. Surface enhanced Raman spectroscopy of phenolic antioxidants: A systematic evaluation of ferulic acid, p-coumaric acid, caffeic acid and sinapic acid. Vib. Spectrosc. 2017, 89, 113–122. [Google Scholar] [CrossRef]
- Sidoryk, K.; Filip, K.; Cmoch, P.; Łaszcz, M.; Cybulski, M. Efficient synthesis and physicochemical characterization of natural danshensu, its S isomer and intermediates thereof. J. Mol. Struct. 2018, 1153, 135–148. [Google Scholar] [CrossRef]
- Li, H.-D.; Xu, Q.-S.; Liang, Y.-Z. libPLS: An integrated library for partial least squares regression and linear discriminant analysis. Chemometr. Intell. Lab. 2018, 176, 34–43. [Google Scholar] [CrossRef]
- Liu, J.; Osadchy, M.; Ashton, L.; Foster, M.; Solomon, C.J.; Gibson, S.J. Deep convolutional neural networks for Raman spectrum recognition: A unified solution. Analyst 2017, 142, 4067–4074. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Zhang, S.; Fu, H.; Qu, H. Combining convolutional neural networks and on-line Raman spectroscopy for monitoring the Cornu Caprae Hircus hydrolysis process. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2020, 226, 117589. [Google Scholar] [CrossRef]
- Acquarelli, J.; van Laarhoven, T.; Gerretzen, J.; Tran, T.N.; Buydens, L.M.C.; Marchiori, E. Convolutional neural networks for vibrational spectroscopic data analysis. Anal. Chim. Acta 2017, 954, 22–31. [Google Scholar] [CrossRef]
- Chan, C.O.; Chu, C.C.; Mok, D.K.; Chau, F.T. Analysis of berberine and total alkaloid content in cortex phellodendri by near infrared spectroscopy (NIRS) compared with high-performance liquid chromatography coupled with ultra-visible spectrometric detection. Anal. Chim. Acta 2007, 592, 121–131. [Google Scholar] [CrossRef] [PubMed]




| Algorithms | Objectives | Calibration | Cross-Validation | Prediction | |||
|---|---|---|---|---|---|---|---|
| RMSEC | RMSECV | RMSEP | |||||
| SPA-SVR | Danshensu | −1.3462 | 0.0205 | −1.6672 | 0.0176 | −1.4600 | 0.0265 |
| Ferulic acid | −4.6801 | 0.0145 | −5.2301 | 0.0423 | −0.0021 | 0.0158 | |
| Rosmarinic acid | 0.5132 | 0.0424 | 0.4102 | 0.0644 | 0.3652 | 0.0448 | |
| Salvianolic acid B | 0.8718 | 0.2929 | 0.7189 | 0.2034 | 0.8100 | 0.3580 | |
| Soluble solid | 0.8185 | 0.6091 | 0.7033 | 0.7651 | 0.7189 | 0.7647 | |
| CARS-PLSR | Danshensu | 0.9979 | 0.0011 | 0.9223 | 0.0031 | 0.6382 | 0.0163 |
| Ferulic acid | 0.9843 | 0.0045 | 0.9456 | 0.0025 | 0.8483 | 0.0057 | |
| Rosmarinic acid | 0.9904 | 0.1539 | 0.9754 | 0.1634 | 0.9457 | 0.0092 | |
| Salvianolic acid B | 0.9958 | 0.0584 | 0.9192 | 0.1345 | 0.8696 | 0.3749 | |
| Soluble solid | 0.9904 | 0.1539 | 0.9312 | 0.2745 | 0.9282 | 0.4512 | |
| CNN | Danshensu | 0.8694 | 0.0086 | 0.8423 | 0.0035 | 0.8212 | 0.0096 |
| Ferulic acid | 0.8893 | 0.1197 | 0.7356 | 0.0768 | 0.7163 | 0.0213 | |
| Rosmarinic acid | 0.9285 | 0.0098 | 0.8749 | 0.0167 | 0.8450 | 0.0143 | |
| Salvianolic acid B | 0.8906 | 0.2988 | 0.8876 | 0.3758 | 0.8544 | 0.3371 | |
| Soluble solid | 0.8857 | 0.5389 | 0.8831 | 0.5775 | 0.8554 | 0.5812 | |
| CARS-CNN | Danshensu | 0.9893 | 0.0024 | 0.9423 | 0.0051 | 0.8458 | 0.0094 |
| Ferulic acid | 0.9875 | 0.0016 | 0.9335 | 0.0035 | 0.8667 | 0.0084 | |
| Rosmarinic acid | 0.9551 | 0.0078 | 0.9464 | 0.0079 | 0.8491 | 0.0145 | |
| Salvianolic acid B | 0.9943 | 0.1953 | 0.9213 | 0.1886 | 0.9246 | 0.2528 | |
| Soluble solid | 0.9526 | 0.1197 | 0.9384 | 0.3188 | 0.9415 | 0.3861 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tao, Y.; Bao, J.; Liu, Q.; Liu, L.; Zhu, J. Application of Deep-Learning Algorithm Driven Intelligent Raman Spectroscopy Methodology to Quality Control in the Manufacturing Process of Guanxinning Tablets. Molecules 2022, 27, 6969. https://doi.org/10.3390/molecules27206969
Tao Y, Bao J, Liu Q, Liu L, Zhu J. Application of Deep-Learning Algorithm Driven Intelligent Raman Spectroscopy Methodology to Quality Control in the Manufacturing Process of Guanxinning Tablets. Molecules. 2022; 27(20):6969. https://doi.org/10.3390/molecules27206969
Chicago/Turabian StyleTao, Yi, Jiaqi Bao, Qing Liu, Li Liu, and Jieqiang Zhu. 2022. "Application of Deep-Learning Algorithm Driven Intelligent Raman Spectroscopy Methodology to Quality Control in the Manufacturing Process of Guanxinning Tablets" Molecules 27, no. 20: 6969. https://doi.org/10.3390/molecules27206969
APA StyleTao, Y., Bao, J., Liu, Q., Liu, L., & Zhu, J. (2022). Application of Deep-Learning Algorithm Driven Intelligent Raman Spectroscopy Methodology to Quality Control in the Manufacturing Process of Guanxinning Tablets. Molecules, 27(20), 6969. https://doi.org/10.3390/molecules27206969






