Isolation and In Silico Prediction of Potential Drug-like Compounds with a New Dimeric Prenylated Quinolone Alkaloid from Zanthoxylum rhetsa (Roxb.) Root Extracts Targeted against SARS-CoV-2 (Mpro)
Abstract
1. Introduction
2. Results
2.1. Characterization of Compounds
2.2. Molecular Docking with MPro
2.2.1. Molecular Interaction of the Four Compounds with Mpro
2.2.2. Result Obtained from Molecular Dynamics Simulation
2.2.3. Binding Free Energy Analysis
3. Discussion
4. Materials and Methods
4.1. General Experimental Procedures
4.2. Plant Material
4.3. Extraction and Isolation
4.4. Methodologies of Molecular Docking on SARS-CoV-2 Main Proteases
4.4.1. Molecular Docking
4.4.2. Molecular Dynamics
4.4.3. Binding ENERGY Calculation MM-PBSA
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Elfiky, A.A. Natural products may interfere with SARS-CoV-2 attachment to the host cell. J. Biomol. Struct. Dyn. 2021, 39, 3194–3203. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Tiwari, S.; Deb, M.K.; Marty, J.L. Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2): A global pandemic and treatment strategies. Int. J. Antimicrob. Agents 2020, 56, 106054. [Google Scholar] [CrossRef]
- Thomford, N.E.; Senthebane, D.A.; Rowe, A.; Munro, D.; Seele; Maroyi, A.; Dzobo, K. Natural products for drug discovery in the 21st century: Innovations for novel drug discovery. Int. J. Mol. Sci. 2018, 19, 1578. [Google Scholar] [CrossRef] [PubMed]
- Prachayasittikul, V.; Worachartcheewan, A.; Shoombuatong, W.; Songtawee, N.; Simeon, S.; Prachayasittikul, V.; Nantasenamat, C. Computer-aided drug design of bioactive natural products. Curr. Top. Med. Chem. 2015, 15, 1780–1800. [Google Scholar] [CrossRef]
- Tung, C.W. Public databases of plant natural products for computational drug discovery. Curr. Comput. Aided Drug Des. 2014, 10, 191–196. [Google Scholar] [CrossRef] [PubMed]
- Atanasov, A.G.; Waltenberger, B.; Pferschy-Wenzig, E.M.; Linder, T.; Wawrosch, C.; Uhrin; Temml, V.; Wang, L.; Schwaiger, S.; Heiss, E.H.; et al. Discovery and resupply of pharmacologically active plant-derived natural products: A review. Biotechnol. Adv. 2015, 33, 1582–1614. [Google Scholar] [CrossRef]
- Yonesi, M.; Rezazadeh, A. Plants as a prospective source of natural anti-viral compounds and oral vaccines against COVID-19 coronavirus. Preprint 2020, 2020040321. [Google Scholar]
- Haque, M.M.; Begum, S.; Sohrab, M.H.; Ahsan, M.; Hasan, C.M.; Ahmed, N.; Haque, R. Secondary metabolites from the stem of Ravenia spectabilis Lindl. Pharmacogn. Mag. 2013, 9, 76. [Google Scholar]
- Fielding, B.C.; Filho, C.D.S.M.B.; Ismail, N.S.; Sousa, D.D. Alkaloids: Therapeutic potential against human coronaviruses. Molecules 2020, 25, 5496. [Google Scholar] [CrossRef] [PubMed]
- Keshavarz Valian, N.; Pourakbari, B.; Asna Ashari, K.; Hosseinpour Sadeghi, R.; Mahmoudi, S.E. Valuation of human coronavirus OC43 and SARS-CoV-2 in children with respiratory tract infection during the COVID-19 pandemic. J. Med. Virol. 2022, 94, 1450–1456. [Google Scholar] [CrossRef] [PubMed]
- Wink, M. Potential of DNA intercalating alkaloids and other plant secondary metabolites against SARS-CoV-2 causing COVID-19. Diversity 2020, 12, 175. [Google Scholar] [CrossRef]
- Kim, D.E.; Min, J.S.; Jang, M.S.; Lee, J.Y.; Shin, Y.S.; Park, C.M.; Song, J.H.; Kim, H.R.; Kim, S.; Jin, Y.H.; et al. Natural bis-benzylisoquinoline alkaloids-tetrandrine, fangchinoline, and cepharanthine, inhibit human coronavirus OC43 infection of MRC-5 human lung cells. Biomolecules 2019, 9, 696. [Google Scholar] [CrossRef]
- Quan, N.V.; Anh, L.H.; Lam, V.Q.; Takami, A.; Teschke, R.; Khanh, T.D.; Xuan, T.D. Anti-Diabetes, anti-gout, and anti-leukemia properties of essential oils from natural spices clausena indica, zanthoxylum rhetsa, and michelia tonkinensis. Molecules 2022, 27, 774. [Google Scholar] [CrossRef]
- Krieger, E.; Darden, T.; Nabuurs, S.B.; Finkelstein, A.; Vriend, G. Making optimal use of empirical energy functions: Force-field parameterization in crystal space. Proteins Struct. Funct. Bioinform. 2004, 57, 678–683. [Google Scholar] [CrossRef]
- Ngadjui, T.B.; Ayafor, J.F.; Sondengam, B.L.; Connolly, J.D.; Rycroft, D.S.; Khalid, S.A.; Waterman, G.; Brown, N.M.; Grundon, M.F.; Ramachandran, V.N. The structures of vepridimerines AD, four new dimeric prenylated quinolone alkaloids from Vepris lousii and Oricia renieri (Rutaceae). Tetrahedron Lett. 1982, 23, 2041–2044. [Google Scholar] [CrossRef]
- Ngadjui, B.T.; Ayafor, J.F.; Bilon, A.N.; Sondengam, B.L.; Connolly, J.D.; Rycroft, D.S. Synthesis of veprisine dimers and the formation of a novel cyclic tetramer from precocene I. Tetrahedron 1992, 48, 8711–8724. [Google Scholar] [CrossRef]
- Du, Y.Q.; Liu, H.; Li, C.J.; Yang, J.Z.; Ma, J.; Zhang, D.; Sun, H.; Zhang, D.M. Carbazole and amide alkaloids from the stems of Clausena lansium. J. Asian Nat. Prod. Res. 2015, 17, 1048–1053. [Google Scholar] [CrossRef] [PubMed]
- Tolkachev, O.N.; Savina, A.A.; Sheichenko, V.I.; Proskudina, V.V. 8-O-demethylchelerythrine from Macleaya cordata. Pharm. Chem. J. 1999, 33, 86–87. [Google Scholar] [CrossRef]
- Miao, F.; Yang, X.J.; Zhou, L.; Hu, H.J.; Zheng, F.; Ding, X.D.; Sun, D.M.; Zhou, C.D.; Sun, W. Structural modification of sanguinarine and chelerythrine and their antibacterial activity. Nat. Prod. Res. 2011, 25, 863–875. [Google Scholar] [CrossRef] [PubMed]
- Ishii, H.; Ichikawa, Y.; Kawanabe, E.; Ishikawa, M.; Ishikawa, T.; Kuretani, K.; Inomata, M.; Hoshi, A. Studies on the Chemical Constituents of Rutaceous Plants. LX. Development of a Versatile Method for Syntheses of the Antitumor Benzo [c] phenanthridine Alkaloids. (9). Efficient Syntheses and Antitumor Activities of Nitidine and Related Nonphenolic Benzo [c]-phenanthridine Alkaloids. Chem. Pharm. Bull. 1985, 33, 4139–4151. [Google Scholar]
- Nafiah, M.A.; Mukhtar, M.R.; Omar, H.; Ahmad, K.; Morita, H.; Litaudon, M.; Awang, K.; Hadi, A.H.A. N-Cyanomethylnorboldine: A new aporphine isolated from Alseodaphne perakensis (Lauraceae). Molecules 2011, 16, 3402–3409. [Google Scholar] [CrossRef] [PubMed]
- Zeb, M.; Khan, S.; Rahman, T.; Sajid, M.; Seloni, S. Isolation and Biological Activity of β-Sitosterol and Stigmasterol from the Roots of Indigofera heterantha. Pharm. Pharmacol. Int. J. 2017, 5, 139. [Google Scholar]
- Alexandri, E.; Ahmed, R.; Siddiqui, H.; Choudhary, M.I.; Tsiafoulis, C.G.; Gerothanassis, I.P. High resolution NMR spectroscopy as a structural and analytical tool for unsaturated lipids in solution. Molecules 2017, 22, 1663. [Google Scholar] [CrossRef]
- Fuglebakk, E.; Echave, J.; Reuter, N. Measuring and comparing structural fluctuation patterns in large protein datasets. Bioinformatics 2012, 28, 2431–2440. [Google Scholar] [CrossRef]
- MIu, L.; Bogatyreva, N.S.; Galzitskaia, O.V. Radius of gyration is indicator of compactness of protein structure. Mol. Biol. 2008, 42, 701–706. [Google Scholar]
- Stein, A.J.; Bain, G.; Prodanovich, P.; Santini, A.M.; Darlington, J.; Stelzer, N.M.; Sidhu, R.S.; Schaub, J.; Goulet, L.; Lonergan, D.; et al. Structural basis for inhibition of human autotaxin by four potent compounds with distinct modes of binding. Mol. Pharmacol. 2015, 88, 982–992. [Google Scholar] [CrossRef] [PubMed]
- Lauria, A.; Ippolito, M.; Almerico, A.M. Molecular docking approach on the Topoisomerase I inhibitors series included in the NCI anti-cancer agents mechanism database. J. Mol. Model. 2007, 13, 393–400. [Google Scholar] [CrossRef] [PubMed]
- Guex, N.; Peitsch, M.C. SWISS-MODEL and the Swiss-Pdb Viewer: An environment for comparative protein modeling. Electrophoresis 1997, 18, 2714–2723. [Google Scholar] [CrossRef]
- DeLano, W.L. The PyMOL Molecular Graphics System. 2002. Available online: http://www.pymol.org (accessed on 18 November 2022).
- Nishimasu, H.; Okudaira, S.; Hama, K.; Mihara, E.; Dohmae, N.; Inoue, A.; Ishitani, R.; Takagi, J.; Aoki, J.; Nureki, O. Crystal structure of autotaxin and insight into GPCR activation by lipid mediators. Nat. Struct. Mol. Biol. 2011, 18, 205–212. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. Software news and update AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function. Effic. Optim. Multithreading 2009, 31, 455–461. [Google Scholar]
- Ahmed, S.; Islam, N.; Shahinozzaman, M.; Fakayode, S.O.; Afrin, N.; Halim, M.A. Virtual screening, molecular dynamics, density functional theory and quantitative structure activity relationship studies to design peroxisome proliferator-activated receptor-γ agonists as anti-diabetic drugs. J. Biomol. Struct. Dyn. 2021, 39, 728–742. [Google Scholar] [CrossRef]
- Junaid, M.; Islam, N.; Hossain, M.K.; Ullah, M.O.; Halim, M.A. Metal based donepezil analogues designed to inhibit human acetylcholinesterase for Alzheimer’s disease. PLoS ONE 2019, 14, e0211935. [Google Scholar] [CrossRef] [PubMed]
- Shahinozzaman, M.; Ishii, T.; Ahmed, S.; Halim, M.A.; Tawata, S. A computational approach to explore and identify potential herbal inhibitors for the p21-activated kinase 1 (PAK1). J. Biomol. Struct. Dyn. 2020, 38, 3514–3526. [Google Scholar] [CrossRef]
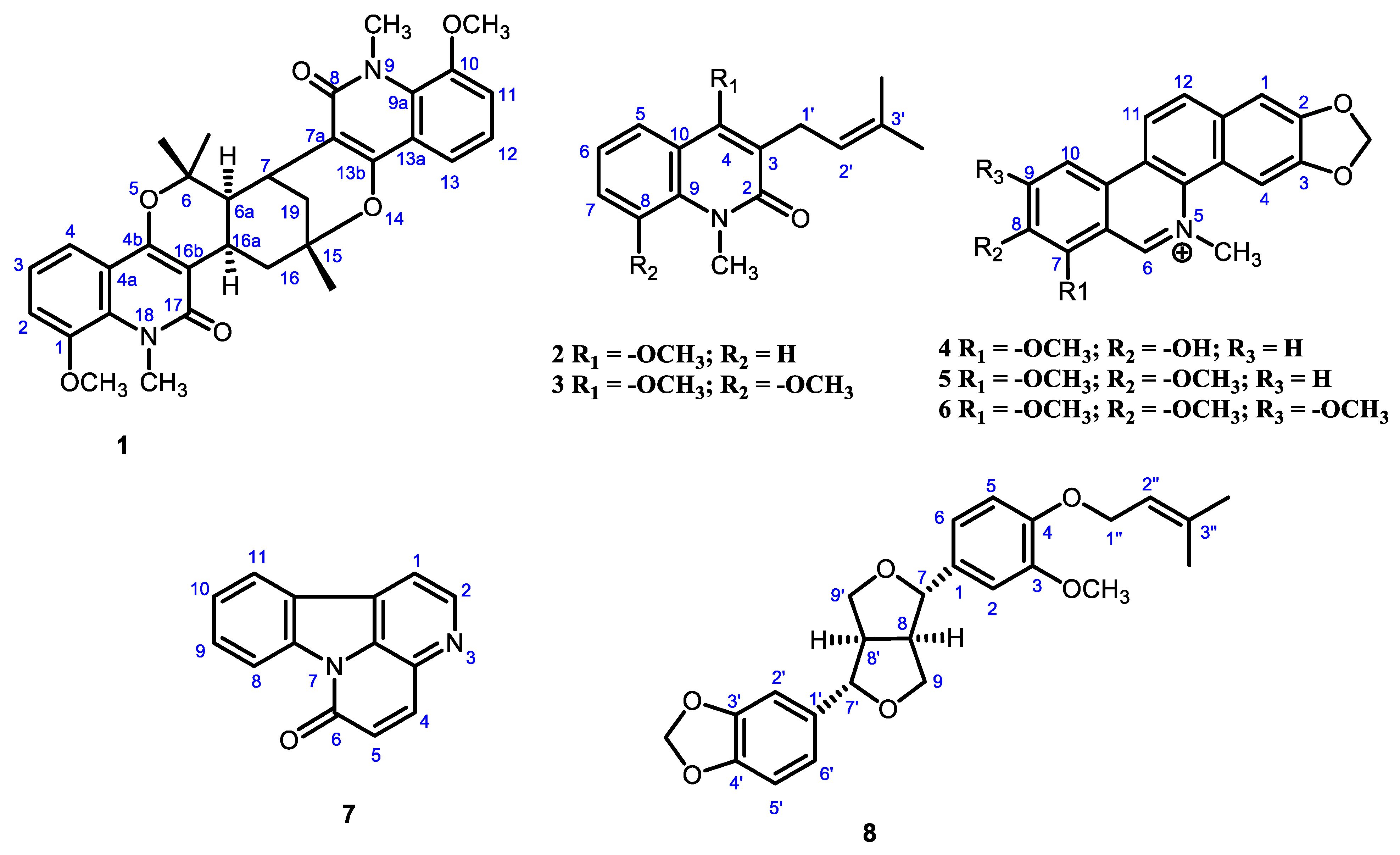






| Position | δH a | δC b | HSQC | HMBC |
|---|---|---|---|---|
| 1 | --- | 148.5 | ||
| 2 | 7.05 dd (J = 8.0, 1.3 Hz) | 114.1 | 114.1 | 130.6 (C-18a), 116.0 (C-4) |
| 3 | 7.17 dd (J = 8.0, 7.9 Hz) | 122.0 | 122.0 | 148.5 (C-1), 118.3 (C-4a) |
| 4 | 7.63 dd (J = 7.9, 1.3 Hz) | 116.0 | 116.0 | c |
| 4a | --- | 118.3 | ||
| 4b | --- | 158.1 | ||
| 6 | --- | 80.0 | ||
| 6a | 2.23 d (J = 6.3 Hz) | 43.5 | 43.5 | c |
| 7 | 3.70 br s | 27.8 | 27.8 | c |
| 7a | --- | 108.3 | ||
| 8 | --- | 163.0 | ||
| 9a | --- | 130.4 | ||
| 10 | --- | 148.3 | ||
| 11 | 7.04 dd (J = 8.0, 1.3 Hz) | 113.7 | 113.7 | 116.2 (C-13) |
| 12 | 7.14 dd (J = 8.0, 7.9 Hz) | 122.3 | 122.3 | 148.3 (C-10), 118.8 (C-13a) |
| 13 | 7.73 dd (J = 7.9, 1.3 Hz) | 116.2 | 116.2 | |
| 13a | --- | 118.8 | ||
| 13b | --- | 154.9 | ||
| 15 | --- | 77.1 | ||
| 16 | 1.57 dd (J = 14.2, 14.2 Hz) 3.20 ddd (J = 14.2, 5.5, 2.4 Hz) | 39.5 | 39.5 | |
| 16a | 3.05 ddd (J = 12.5, 6.0, 5.7 Hz) | 26.5 | 26.5 | c |
| 16b | --- | 109.7 | ||
| 17 | --- | 163.9 | ||
| 18a | --- | 130.6 | ||
| 19 | 1.74 ddd (J = 13.8, 2.4, 2.4 Hz) 2.19 dd (J = 13.8, 2.8 Hz) | 32.2 | 32.2 | c |
| Me-6 | 1.55, 3H s | 24.9 | 24.9 | 43.5 (C-6a), 29.2 (Me-6) |
| Me-6 | 1.87, 3H s | 29.2 | 29.2 | 80.0(C-6), 43.5 (C-6a), 24.9 (Me-6) |
| Me-15 | 1.55, 3H s | 28.7 | 28.7 | 77.1 (C-16), 39.5 (C-15), 32.2 (C-19) |
| OMe-1 | 3.89, 3H s | 56.9 | 56.9 | 56.9 (C-1) |
| OMe-10 | 3.88, 3H s | 56.7 | 56.7 | 56.7 (C-10) |
| NMe-9 | 3.93, 3H s | 35.2 | 35.2 | 130.4 (C-9a), 163.0 (C-8) |
| NMe-18 | 3.88, 3H s | 34.8 | 34.8 | 130.6 (C-18a), 163.9 (C-17) |
| Compound 1 | Vepridimerine A [16] | |||
|---|---|---|---|---|
| Position | δH a | δC b | δH | δC |
| Ha | 1.74 ddd (J = 13.8, 2.4, 2.4 Hz) | 32.2 | 1.70 ddd (J = 13.7, 2.8, 2.2 Hz) | 32.2 |
| Hb | 2.19 dd (J = 13.8, 2.8 Hz) | 32.2 | 2.14 dd (J = 13.7, 2.7 Hz) | 32.2 |
| Hc | 3.70 br s | 27.8 | 3.61 ddd (J = 2.8, 2.7, 1.0 Hz) | 26.2 |
| Hd | 2.23 d (J = 6.3 Hz) | 43.5 | 2.16 dd (J = 6.1, 1.0 Hz) | 43.6 |
| He | 3.05 ddd (J = 12.5, 6.0, 5.7 Hz) | 26.5 | 2.96 ddd (J = 13.4, 6.1, 5.4 Hz) | 27.6 |
| Hf | 3.20 ddd (J = 14.2, 5.5, 2.4 Hz) | 39.5 | 3.10 ddd (J = 14.2, 5.4, 2.2 Hz) | 39.5 |
| Hg | 1.57 dd (J = 14.2, 14.2 Hz) | 39.5 | 1.56 dd (J = 14.2, 13.4 Hz) | 39.5 |
| Compound | Binding Affinity | Hydrogen Bond (AA…ligand) | Hydrophobic Interaction (AA…ligand) | Electrostatic Interaction (AA…ligand) |
|---|---|---|---|---|
| 1 | GLY143 (2.601) C-H…O-C) | CYS145 (4.789) Alkyl | ||
| LEU141 (2.530) C-O…H-C) | MET49 (4.276) Alkyl | |||
| PHE140 (2.975) C-O…H-C) | MET49 (4.353) Alkyl | |||
| −8.5 | PHE140 (2.667) C-O…H-C | LEU27 (4.208) Alkyl | ||
| GLU166 (2.672) C-O…H-C | CYS145 (3.496) Alkyl | |||
| HIS163 (5.423) Pi-Alkyl | ||||
| 8 | GLU166 (2.615) C-O…H-C) | HIS41 (5.823) Pi-Pi T-shaped | ||
| LEU27 (4.72) Alkyl | ||||
| LEU27 (4.383) Alkyl | ||||
| −8.0 | CYS145 (3.967) Alkyl | |||
| HIS41 (4.504) Pi-Alkyl | ||||
| MET165 (5.479) Pi-Alkyl | ||||
| ALA191 (5.124) Pi-Alkyl | ||||
| 4 | ARG188 (2.792) C-H…O-C | GLU166 (2.687) Pi-Sigma | GLU166 (4.933) Pi-Anion | |
| HIS164 (2.549) C-O…H-C | MET165 (4.778) Pi-Alkyl | |||
| −7.8 | PHE140 (2.845) C-O…H-C | MET165 (4.741) Pi-Alkyl | ||
| GLU166 (2.613) C-O…H-C | MET49 (4.649) Pi-Alkyl | |||
| GLU166 (3.047) Pi-Donor | ||||
| 6 | ARG188 (2.784) C-H…O-C | GLU166 (2.745) Pi-Sigma | GLU166 (4.926) Pi-Anion | |
| HIS164 (2.509) C-O…H-C | MET165 (4.815) Pi-Alkyl | |||
| −7.6 | PHE140 (2.724) C-O…H-C | MET165 (4.753) Pi-Alkyl | ||
| GLU166 (2.755) C-O…H-C | MET49 (4.658) Pi-Alkyl | |||
| GLU166 (3.041) Pi-Donor | ||||
| 5 | PRO168 (2.537) Pi-Sigma | |||
| ARG188 (2.480) C-O…H-C | MET165 (4.823) Alkyl | |||
| −7.5 | THR190 (2.758) C-O…H-C | PRO168 (4.537) Pi-Alkyl | ||
| MET165 (5.086) Pi-Alkyl | ||||
| 9 | SER144 (2.737) O-H…O-C | ALA191 (3.775) Alkyl | ||
| −7.2 | LEU141 (2.757) C-O…H-O | PRO168 (4.205) Alkyl | ||
| PRO168 (3.972) Alkyl | ||||
| 10 | −6.7 | SER144 (2.710) C-H…O-C | ALA191 (4.156) Alkyl | |
| PRO168 (4.156) Alkyl | ||||
| 2 | −6.3 | HIS41 (4.917) Pi-Pi T-shaped | ||
| MET165 (4.480) Alkyl | ||||
| HIS163 (4.690) Pi-Alkyl | ||||
| HIS172 (5.434) Pi-Alkyl | ||||
| MET165 (4.759) Pi-Alkyl | ||||
| MET165 (5.048) Pi-Alkyl | ||||
| 3 | −6.1 | CYS145 (4.795) Alkyl | ||
| MET49 (5.120) Alkyl | ||||
| CYS145 (4.953) Alkyl | ||||
| HIS41 (3.736) Pi-Alkyl | ||||
| HIS163 (4.672) Pi-Alkyl | ||||
| HIS163 (4.793) Pi-Alkyl | ||||
| HIS172 (4.905) Pi-Alkyl | ||||
| MET165 (4.428) Pi-Alkyl | ||||
| MET165 (4.063) Alkyl | ||||
| HIS41 (4.707) Pi-Alkyl | ||||
| HIS41 (4.680) Pi-Alkyl |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zohora, F.T.; Azam, A.T.M.Z.; Ahmed, S.; Rahman, K.M.; Halim, M.A.; Anwar, M.R.; Sohrab, M.H.; Tabassum, F.; Hasan, C.M.; Ahsan, M. Isolation and In Silico Prediction of Potential Drug-like Compounds with a New Dimeric Prenylated Quinolone Alkaloid from Zanthoxylum rhetsa (Roxb.) Root Extracts Targeted against SARS-CoV-2 (Mpro). Molecules 2022, 27, 8191. https://doi.org/10.3390/molecules27238191
Zohora FT, Azam ATMZ, Ahmed S, Rahman KM, Halim MA, Anwar MR, Sohrab MH, Tabassum F, Hasan CM, Ahsan M. Isolation and In Silico Prediction of Potential Drug-like Compounds with a New Dimeric Prenylated Quinolone Alkaloid from Zanthoxylum rhetsa (Roxb.) Root Extracts Targeted against SARS-CoV-2 (Mpro). Molecules. 2022; 27(23):8191. https://doi.org/10.3390/molecules27238191
Chicago/Turabian StyleZohora, Fatema Tuz, A. T. M. Zafrul Azam, Sinthyia Ahmed, Khondaker Miraz Rahman, Mohammad A. Halim, Md. Rafi Anwar, Md. Hossain Sohrab, Fatema Tabassum, Choudhury Mahmood Hasan, and Monira Ahsan. 2022. "Isolation and In Silico Prediction of Potential Drug-like Compounds with a New Dimeric Prenylated Quinolone Alkaloid from Zanthoxylum rhetsa (Roxb.) Root Extracts Targeted against SARS-CoV-2 (Mpro)" Molecules 27, no. 23: 8191. https://doi.org/10.3390/molecules27238191
APA StyleZohora, F. T., Azam, A. T. M. Z., Ahmed, S., Rahman, K. M., Halim, M. A., Anwar, M. R., Sohrab, M. H., Tabassum, F., Hasan, C. M., & Ahsan, M. (2022). Isolation and In Silico Prediction of Potential Drug-like Compounds with a New Dimeric Prenylated Quinolone Alkaloid from Zanthoxylum rhetsa (Roxb.) Root Extracts Targeted against SARS-CoV-2 (Mpro). Molecules, 27(23), 8191. https://doi.org/10.3390/molecules27238191










