Targeted Nanoparticles for the Binding of Injured Vascular Endothelium after Percutaneous Coronary Intervention
Abstract
1. Introduction
2. Results
2.1. Production of Anti-CD34 HuscFv
2.2. Binding of the Anti-CD34 HuscFv to rCD34 and CD34-Positive Cells
2.3. Physical Characterization, EE and CE of Conjugated NPs
2.4. Biocompatibility of Conjugated NPs
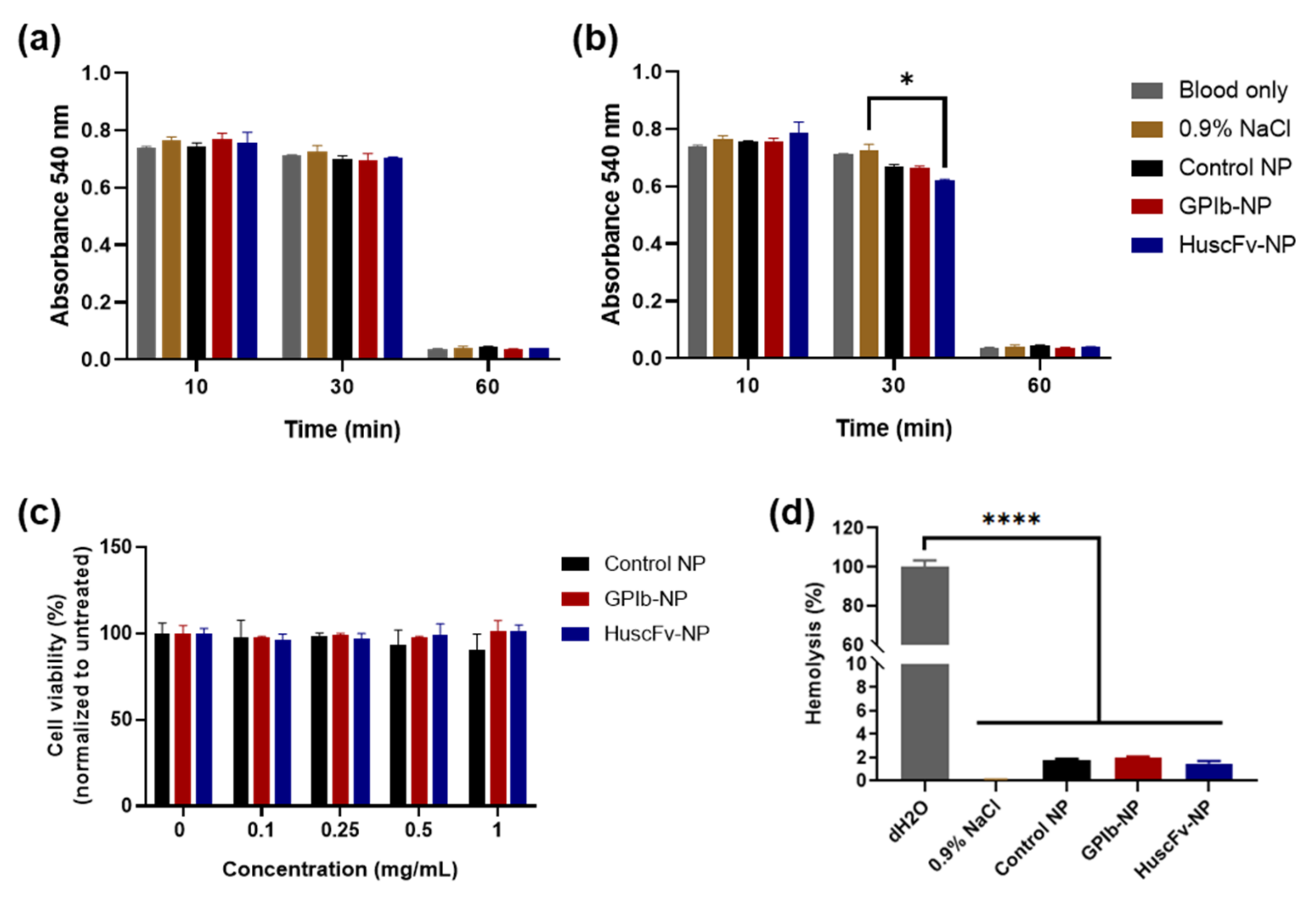
2.4.1. Blood Clotting Test
2.4.2. Hemolysis Test
2.4.3. Cytocompatibility
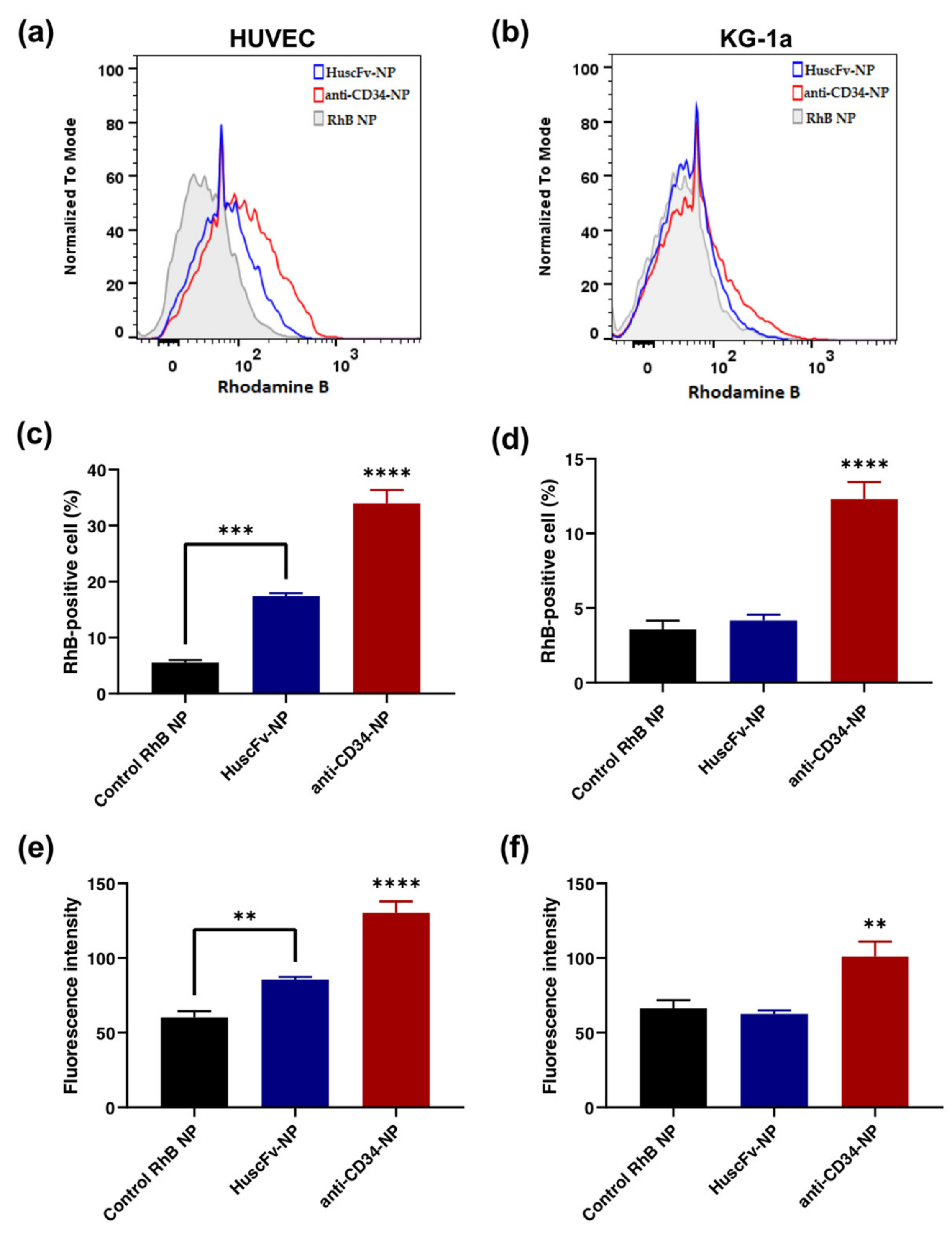
2.5. Binding of HuscFv-NPs to CD34-Positive Cells
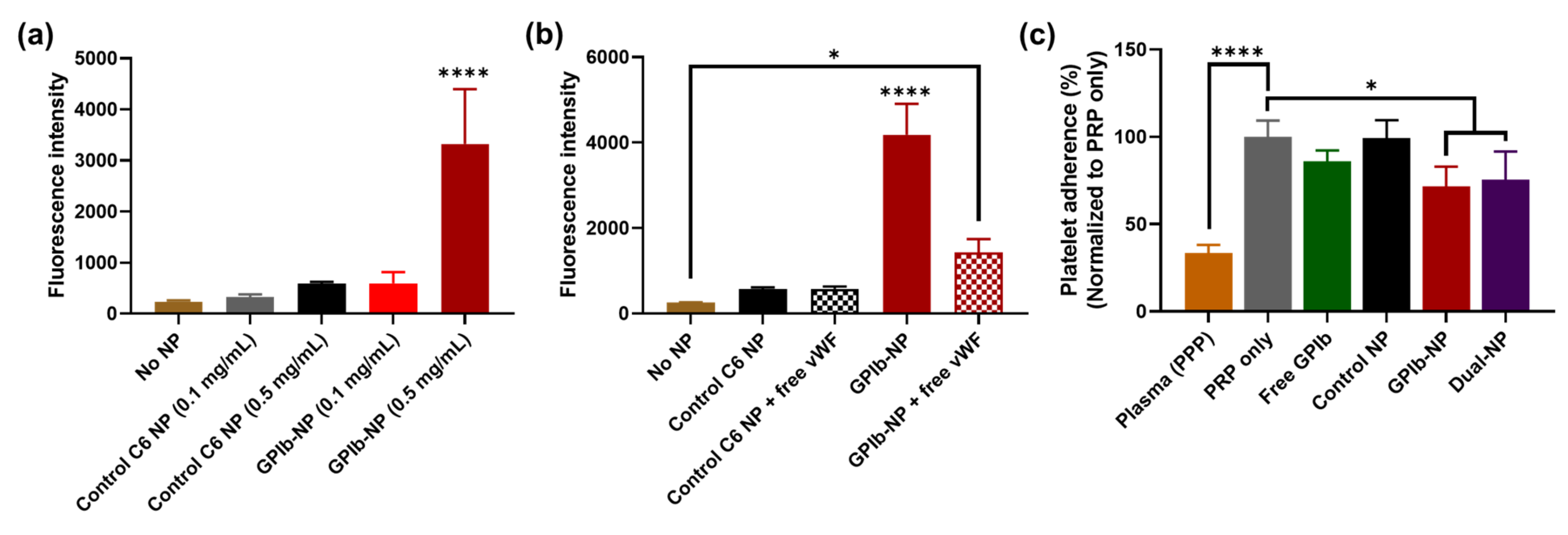
2.6. Binding of GPIb-NPs with vWF
2.7. GPIb-NPs Prevention Platelet Adherence to vWF Measure
3. Discussion
4. Materials and Methods
4.1. Cell Lines and Reagents
4.2. Production of HuscFv That Bound to Recombinant CD34 (rCD34)
4.3. Binding Test of Crude HuscFv to rCD34
4.4. Purification of Soluble HuscFv with His-Tag Affinity Resins
4.5. Binding Test of Purified Soluble HuscFv to CD34-Positive Cells by Flow Cytometry
4.6. Preparation of Drug-Loaded Polymeric NPs
4.7. Fabrication of Conjugated Polymeric Nanoparticle
4.8. Nanoparticle Characterization
4.9. Biocompatibility Study
4.10. Binding of HuscFv-Conjugated NPs (HuscFv-NPs) to CD34-Positive Cells
4.11. Binding of GPIb-Conjugated NPs (GPIb-NPs) to vWF
4.12. Platelet Binding Prevention Test
4.13. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Malakar, A.K.; Choudhury, D.; Halder, B.; Paul, P.; Uddin, A.; Chakraborty, S. A review on coronary artery disease, its risk factors, and therapeutics. J. Cell Physiol. 2019, 234, 16812–16823. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Alvarez, M.M.; Zanetti, D.; Carreras-Torres, R.; Moral, P.; Athanasiadis, G. A survey of sub-Saharan gene flow into the Mediterranean at risk loci for coronary artery disease. Eur. J. Hum. Genet. 2017, 25, 472–476. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Cardiovascular Diseases (CVDs). Available online: https://www.who.int/en/news-room/fact-sheets/detail/cardiovascular-diseases-(cvds) (accessed on 8 May 2022).
- Khan, S.Q.; Ludman, P.F. Percutaneous coronary intervention. Medicine 2022, 50, 437–444. [Google Scholar] [CrossRef]
- Cassar, A.; Holmes, D.R., Jr.; Rihal, C.S.; Gersh, B.J. Chronic Coronary Artery Disease: Diagnosis and Management. Mayo. Clin. Proc. 2009, 84, 1130–1146. [Google Scholar] [CrossRef] [PubMed]
- Alexandrescu, D.-M.; Mitu, O.; Costache, I.I.; Macovei, L.; Mitu, I.; Alexandrescu, A.; Arsenescu Georgescu, C. Risk factors associated with intra-stent restenosis after percutaneous coronary intervention. Exp. Ther. Med. 2021, 22, 1141. [Google Scholar] [CrossRef] [PubMed]
- Toutouzas, K.; Colombo, A.; Stefanadis, C. Inflammation and restenosis after percutaneous coronary interventions. Eur. Heart J. 2004, 25, 1679–1687. [Google Scholar] [CrossRef] [PubMed]
- Su, L.C.; Xu, H.; Tran, R.T.; Tsai, Y.T.; Tang, L.; Banerjee, S.; Yang, J.; Nguyen, K.T. In situ re-endothelialization via multifunctional nanoscaffolds. ACS Nano 2014, 8, 10826–10836. [Google Scholar] [CrossRef] [PubMed]
- Kraft, P.; De Meyer, S.F.; Kleinschnitz, C. Next-generation antithrombotics in ischemic stroke: Preclinical perspective on “bleeding-free antithrombosis”. J. Cereb. Blood Flow Metab. 2012, 32, 1831–1840. [Google Scholar] [CrossRef] [PubMed]
- Jukema, J.W.; Verschuren, J.J.W.; Ahmed, T.A.N.; Quax, P.H.A. Restenosis after PCI. Part 1: Pathophysiology and risk factors. Nat. Rev. Cardiol. 2012, 9, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Newby, A.C.; Zaltsman, A.B. Molecular mechanisms in intimal hyperplasia. J. Pathol. 2000, 190, 300–309. [Google Scholar] [CrossRef]
- Zolpi, E.; Filipetto, C.; Bertipaglia, B.; Taiani, J.; Gasparotto, L.; Chiavegato, A.; Gamba, P.; Sartore, S. Role of platelet activation in catheter-induced vascular wall injury. J. Endovasc. Ther. 2004, 11, 196–210. [Google Scholar] [CrossRef] [PubMed]
- Davies, M.G.; Hagen, P.O. Pathobiology of intimal hyperplasia. Br. J. Surg. 1994, 81, 1254–1269. [Google Scholar] [CrossRef] [PubMed]
- Bendas, G.; Schlesinger, M. The GPIb-IX complex on platelets: Insight into its novel physiological functions affecting immune surveillance, hepatic thrombopoietin generation, platelet clearance and its relevance for cancer development and metastasis. Exp. Hematol. Oncol. 2022, 11, 19. [Google Scholar] [CrossRef] [PubMed]
- Clemetson, K.J.; Clemetson, J.M. Platelet GPIb-V-IX complex structure, function, physiology, and pathology. Semin. Thromb. Hemost. 1995, 21, 130–136. [Google Scholar] [CrossRef]
- Wang, S.; Guo, X.; Ren, L.; Wang, B.; Hou, L.; Zhou, H.; Gao, Q.; Gao, Y.; Wang, L. Targeting and deep-penetrating delivery strategy for stented coronary artery by magnetic guidance and ultrasound stimulation. Ultrason. Sonochem. 2020, 67, 105188. [Google Scholar] [CrossRef]
- Giacoppo, D.; Alfonso, F.; Xu, B.; Claessen, B.E.P.M.; Adriaenssens, T.; Jensen, C.; Pérez-Vizcayno, M.J.; Kang, D.-Y.; Degenhardt, R.; Pleva, L.; et al. Paclitaxel-coated balloon angioplasty vs. drug-eluting stenting for the treatment of coronary in-stent restenosis: A comprehensive, collaborative, individual patient data meta-analysis of 10 randomized clinical trials (DAEDALUS study). Eur. Heart J. 2019, 41, 3715–3728. [Google Scholar] [CrossRef]
- Morice, M.C.; Serruys, P.W.; Sousa, J.E.; Fajadet, J.; Hayashi, E.B.; Perin, M.; Colombo, A.; Schuler, G.; Barragan, P.; Guagliumi, G.; et al. A Randomized comparison of a sirolimus-eluting stent with a standard stent for coronary revascularization. N. Engl. J. Med. 2002, 346, 1773–1780. [Google Scholar] [CrossRef]
- Stone, G.W.; Ellis, S.G.; Cox, D.A.; Hermiller, J.; O’Shaughnessy, C.; Mann, J.T.; Turco, M.; Caputo, R.; Bergin, P.; Greenberg, J.; et al. A polymer-based, paclitaxel-eluting stent in patients with coronary artery disease. N. Engl. J. Med. 2004, 350, 221–231. [Google Scholar] [CrossRef]
- Cockerill, I.; See, C.W.; Young, M.L.; Wang, Y.; Zhu, D. Designing Better Cardiovascular Stent Materials: A Learning Curve. Adv. Funct. Mater. 2021, 31, 2005361. [Google Scholar] [CrossRef]
- McFadden, E.P.; Stabile, E.; Regar, E.; Cheneau, E.; Ong, A.T.L.; Kinnaird, T.; Suddath, W.O.; Weissman, N.J.; Torguson, R.; Kent, K.M.; et al. Late thrombosis in drug-eluting coronary stents after discontinuation of antiplatelet therapy. Lancet 2004, 364, 1519–1521. [Google Scholar] [CrossRef]
- Ong, A.T.L.; McFadden, E.P.; Regar, E.; de Jaegere, P.P.T.; van Domburg, R.T.; Serruys, P.W. Late angiographic stent thrombosis (LAST) events with drug-eluting stents. J. Am. Coll. Cardiol. 2005, 45, 2088–2092. [Google Scholar] [CrossRef]
- Joner, M.; Finn, A.V.; Farb, A.; Mont, E.K.; Kolodgie, F.D.; Ladich, E.; Kutys, R.; Skorija, K.; Gold, H.K.; Virmani, R. Pathology of drug-eluting stents in humans: Delayed healing and late thrombotic risk. J. Am. Coll. Cardiol. 2006, 48, 193–202. [Google Scholar] [CrossRef]
- Kou, F.; Zhu, C.; Wan, H.; Xue, F.; Wang, J.; Xiang, L.; Li, J. Endothelial progenitor cells as the target for cardiovascular disease prediction, personalized prevention, and treatments: Progressing beyond the state-of-the-art. EPMA J. 2020, 11, 629–643. [Google Scholar] [CrossRef]
- Chen, J.; Li, Q.; Xu, J.; Zhang, L.; Maitz, M.F.; Li, J. Thromboresistant and rapid-endothelialization effects of dopamine and staphylococcal protein A mediated anti-CD34 coating on 316L stainless steel for cardiovascular devices. J. Mater. Chem. B 2015, 3, 2615–2623. [Google Scholar] [CrossRef]
- Doevendans, E.; Schellekens, H. Immunogenicity of Innovative and Biosimilar Monoclonal Antibodies. Antibodies 2019, 8, 21. [Google Scholar] [CrossRef]
- Kandari, D.; Bhatnagar, R. Antibody engineering and its therapeutic applications. Int. Rev. Immunol. 2021. [Google Scholar] [CrossRef]
- Shin, C.; Kim, S.S.; Jo, Y.H. Extending traditional antibody therapies: Novel discoveries in immunotherapy and clinical applications. Mol. Ther. Oncolytics 2021, 22, 166–179. [Google Scholar] [CrossRef]
- Ahmad, Z.A.; Yeap, S.K.; Ali, A.M.; Ho, W.Y.; Alitheen, N.B.; Hamid, M. scFv antibody: Principles and clinical application. Clin. Dev. Immunol. 2012, 2012, 980250. [Google Scholar] [CrossRef]
- Szmitko, P.E.; Kutryk, M.J.; Stewart, D.J.; Strauss, M.H.; Verma, S. Endothelial progenitor cell-coated stents under scrutiny. Can. J. Cardiol. 2006, 22, 1117. [Google Scholar] [CrossRef]
- Aoki, J.; Serruys, P.W.; van Beusekom, H.; Ong, A.T.; McFadden, E.P.; Sianos, G.; van der Giessen, W.J.; Regar, E.; de Feyter, P.J.; Davis, H.R.; et al. Endothelial progenitor cell capture by stents coated with antibody against CD34: The HEALING-FIM (healthy endothelial accelerated lining inhibits neointimal growth-first in man) registry. J. Am. Coll. Cardiol. 2005, 45, 1574–1579. [Google Scholar] [CrossRef]
- Guedj, A.S.; Kell, A.J.; Barnes, M.; Stals, S.; Goncalves, D.; Girard, D.; Lavigne, C. Preparation, characterization, and safety evaluation of poly(lactide-co-glycolide) nanoparticles for protein delivery into macrophages. Int. J. Nanomed. 2015, 10, 5965–5979. [Google Scholar]
- Hasan, A.; Morshed, M.; Memic, A.; Hassan, S.; Webster, T.J.; Marei, H.E. Nanoparticles in tissue engineering: Applications, challenges and prospects. Int. J. Nanomed. 2018, 13, 5637–5655. [Google Scholar] [CrossRef] [PubMed]
- Yoo, J.; Park, C.; Yi, G.; Lee, D.; Koo, H. Active Targeting Strategies Using Biological Ligands for Nanoparticle Drug Delivery Systems. Cancers 2019, 11, 640. [Google Scholar] [CrossRef] [PubMed]
- Chames, P.; van Regenmortel, M.; Weiss, E.; Baty, D. Therapeutic antibodies: Successes, limitations and hopes for the future. Br. J. Pharmacol. 2009, 157, 220–233. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Su, X.; Federoff, H.J. Single-chain fragment variable passive immunotherapies for neurodegenerative diseases. Int. J. Mol. Sci. 2013, 14, 19109–19127. [Google Scholar] [CrossRef]
- Scott, A.M.; Wolchok, J.D.; Old, L.J. Antibody therapy of cancer. Nat. Rev. Cancer 2012, 12, 278–287. [Google Scholar] [CrossRef]
- McCafferty, J.; Griffiths, A.D.; Winter, G.; Chiswellt, D.J. Phage antibodies: Filamentous phage displaying antibody variable domains. Nature 1990, 348, 552–554. [Google Scholar] [CrossRef]
- Rahbarnia, L.; Farajnia, S.; Babaei, H.; Majidi, J.; Akbari, B.; Ahdi Khosroshahi, S. Development of a novel human single chain antibody against EGFRVIII antigen by phage display technology. Adv. Pharm. Bull 2016, 6, 563–571. [Google Scholar] [CrossRef][Green Version]
- Kaewchim, K.; Glab-ampai, K.; Mahasongkram, K.; Chulanetra, M.; Seesuay, W.; Chaicumpa, W.; Sookrung, N. Engineered Fully Human Single-Chain Monoclonal Antibodies to PIM2 Kinase. Molecules 2021, 26, 6436. [Google Scholar] [CrossRef]
- Santajit, S.; Seesuay, W.; Mahasongkram, K.; Sookrung, N.; Ampawong, S.; Reamtong, O.; Diraphat, P.; Chaicumpa, W.; Indrawattana, N. Human single-chain antibodies that neutralize Pseudomonas aeruginosa-exotoxin A-mediated cellular apoptosis. Sci. Rep. 2019, 9, 14928. [Google Scholar] [CrossRef]
- Panyajai, P.; Amnajphook, N.; Keawsangthongcharoen, S.; Chiampanichayakul, S.; Tima, S.; Anuchapreeda, S. Study of leukemic stem cell population (CD34+/CD38−) and WT1 protein expression in human leukemic cell lines. J. Assoc. Med. Sci. 2018, 51, 38–44. [Google Scholar]
- Siemerink, M.J.; Klaassen, I.; Vogels, I.M.; Griffioen, A.W.; van Noorden, C.J.; Schlingemann, R.O. CD34 marks angiogenic tip cells in human vascular endothelial cell cultures. Angiogenesis 2012, 15, 151–163. [Google Scholar] [CrossRef]
- Muller, A.M.; Hermanns, M.I.; Skrzynski, C.; Nesslinger, M.; Muller, K.M.; Kirkpatrick, C.J. Expression of the endothelial markers PECAM-1, vWf, and CD34 In Vivo and In Vitro. Exp. Mol. Pathol. 2002, 72, 221–229. [Google Scholar] [CrossRef]
- Yu, B.; Ni, M.; Li, W.-H.; Lei, P.; Xing, W.; Xiao, D.-W.; Huang, Y.; Tang, Z.-J.; Zhu, H.-F.; Shen, G.-X. Human scFv antibody fragments specific for hepatocellular carcinoma selected from a phage display library. World J. Gastroenterol. 2005, 11, 3985. [Google Scholar] [CrossRef]
- Zhuang, Y.; Zhang, C.; Cheng, M.; Huang, J.; Liu, Q.; Yuan, G.; Lin, K.; Yu, H. Challenges and strategies for in situ endothelialization and long-term lumen patency of vascular grafts. Bioact. Mater. 2021, 6, 1791–1809. [Google Scholar] [CrossRef]
- Kona, S.; Dong, J.F.; Liu, Y.; Tan, J.; Nguyen, K.T. Biodegradable nanoparticles mimicking platelet binding as a targeted and controlled drug delivery system. Int. J. Pharm. 2012, 423, 516–524. [Google Scholar] [CrossRef]
- Pardeshi, S.R.; Nikam, A.; Chandak, P.; Mandale, V.; Naik, J.B.; Giram, P.S. Recent advances in PLGA based nanocarriers for drug delivery system: A state of the art review. Int. J. Polym. Mater. Polym. Biomater. 2021, 72, 49–78. [Google Scholar] [CrossRef]
- Zhu, H.; Kong, L.; Zhu, X.; Ran, T.; Ji, X. pH-Responsive Nanoparticles for Delivery of Paclitaxel to the Injury Site for Inhibiting Vascular Restenosis. Pharmaceutics 2022, 14, 535. [Google Scholar] [CrossRef]
- Zago, A.C.; Raudales, J.C.; Attizzani, G.; Matte, B.S.; Yamamoto, G.I.; Balvedi, J.A.; Nascimento, L.; Kosachenco, B.G.; Centeno, P.R.; Zago, A.J. Local delivery of sirolimus nanoparticles for the treatment of in-stent restenosis. Catheter. Cardiovasc. Interv. 2013, 81, E124–E129. [Google Scholar] [CrossRef]
- Song, C.; Labhasetwar, V.; Cui, X.; Underwood, T.; Levy, R.J. Arterial uptake of biodegradable nanoparticles for intravascular local drug delivery: Results with an acute dog model. J. Control. Release 1998, 54, 201–211. [Google Scholar] [CrossRef]
- Lee, N.K.; Wang, C.-P.J.; Lim, J.; Park, W.; Kwon, H.-K.; Kim, S.-N.; Kim, T.-H.; Park, C.G. Impact of the conjugation of antibodies to the surfaces of polymer nanoparticles on the immune cell targeting abilities. Nano Converg. 2021, 8, 24. [Google Scholar] [CrossRef] [PubMed]
- Swider, E.; Maharjan, S.; Houkes, K.; van Riessen, N.K.; Figdor, C.; Srinivas, M.; Tagit, O. Förster Resonance Energy Transfer-Based Stability Assessment of PLGA Nanoparticles In Vitro and In Vivo. ACS Appl. Bio Mater. 2019, 2, 1131–1140. [Google Scholar] [CrossRef] [PubMed]
- Greineder, C.F.; Hood, E.D.; Yao, A.; Khoshnejad, M.; Brenner, J.S.; Johnston, I.H.; Poncz, M.; Gottstein, C.; Muzykantov, V.R. Molecular engineering of high affinity single-chain antibody fragment for endothelial targeting of proteins and nanocarriers in rodents and humans. J. Control. Release 2016, 226, 229–237. [Google Scholar] [CrossRef] [PubMed]
- Balmert, S.C.; Zmolek, A.C.; Glowacki, A.J.; Knab, T.D.; Rothstein, S.N.; Wokpetah, J.M.; Fedorchak, M.V.; Little, S.R. Positive Charge of “Sticky” Peptides and Proteins Impedes Release From Negatively Charged PLGA Matrices. J. Mater. Chem. B 2015, 3, 4723–4734. [Google Scholar] [CrossRef]
- Win, K.Y.; Feng, S.-S. Effects of particle size and surface coating on cellular uptake of polymeric nanoparticles for oral delivery of anticancer drugs. Biomaterials 2005, 26, 2713–2722. [Google Scholar] [CrossRef]
- Pillai, G.J.; Greeshma, M.M.; Menon, D. Impact of poly(lactic-co-glycolic acid) nanoparticle surface charge on protein, cellular and haematological interactions. Colloids Surf. B Biointerfaces 2015, 136, 1058–1066. [Google Scholar] [CrossRef]
- Mei, H.; Pang, Z.; Hu, Y.; Shi, W.; Wang, H.; Deng, J.; Guo, T.; Jiang, X. Effect of EGF1 peptides in directing nanoparticles to thrombi. Chin. Sci. Bull. 2010, 55, 3424–3429. [Google Scholar] [CrossRef]
- Brito, L.; Amiji, M. Nanoparticulate carriers for the treatment of coronary restenosis. Int. J. Nanomed. 2007, 2, 143. [Google Scholar]
- Caldwell, J.; Taladriz-Blanco, P.; Lehner, R.; Lubskyy, A.; Ortuso, R.D.; Rothen-Rutishauser, B.; Petri-Fink, A. The micro-, submicron-, and nanoplastic hunt: A review of detection methods for plastic particles. Chemosphere 2022, 293, 133514. [Google Scholar] [CrossRef]
- Patel, J.; Amrutiya, J.; Bhatt, P.; Javia, A.; Jain, M.; Misra, A. Targeted delivery of monoclonal antibody conjugated docetaxel loaded PLGA nanoparticles into EGFR overexpressed lung tumour cells. J. Microencapsul. 2018, 35, 204–217. [Google Scholar] [CrossRef]
- Mittal, P.; Vardhan, H.; Ajmal, G.; Bonde, G.V.; Kapoor, R.; Mittal, A.; Mishra, B. Formulation, optimization, hemocompatibility and pharmacokinetic evaluation of PLGA nanoparticles containing paclitaxel. Drug Dev. Ind. Pharm. 2019, 45, 365–378. [Google Scholar] [CrossRef]
- Wang, H.; Zhao, Y.; Wang, H.; Gong, J.; He, H.; Shin, M.C.; Yang, V.C.; Huang, Y. Low-molecular-weight protamine-modified PLGA nanoparticles for overcoming drug-resistant breast cancer. J. Control. Release 2014, 192, 47–56. [Google Scholar] [CrossRef]
- Ito, T.; Watanabe, J.; Takai, M.; Konno, T.; Iwasaki, Y.; Ishihara, K. Dual mode bioreactions on polymer nanoparticles covered with phosphorylcholine group. Colloids Surf. B 2006, 50, 55–60. [Google Scholar] [CrossRef]
- Xin, L.; Zhang, H.-T.; Yang, W.-F.; Li, Y.-F.; Liu, C. Evaluation of METase-pemetrexed-loaded PEG–PLGA nanoparticles modified with anti-CD133–scFV for treatment of gastric carcinoma. Biosci. Rep. 2018, 38, BSR20171001. [Google Scholar] [CrossRef]
- Le, T.T.D.; Pham, T.H.; Nguyen, T.N.; Ngo, T.H.G.; Hoang, T.M.N.; Le, Q.H. Evaluation of anti-HER2 scFv-conjugated PLGA–PEG nanoparticles on 3D tumor spheroids of BT474 and HCT116 cancer cells. Adv. Nat. Sci. Nanosci. Nanotechnol. 2016, 7, 025004. [Google Scholar] [CrossRef][Green Version]
- Hu, T.; Yang, J.; Cui, K.; Rao, Q.; Yin, T.; Tan, L.; Zhang, Y.; Li, Z.; Wang, G. Controlled Slow-Release Drug-Eluting Stents for the Prevention of Coronary Restenosis: Recent Progress and Future Prospects. ACS Appl. Mater. Interfaces 2015, 7, 11695–11712. [Google Scholar] [CrossRef]
- Denorme, F.; Vanhoorelbeke, K.; De Meyer, S.F. von Willebrand Factor and Platelet Glycoprotein Ib: A Thromboinflammatory Axis in Stroke. Front. Immunol. 2019, 10, 2884. [Google Scholar] [CrossRef]
- Kutikhin, A.G.; Sinitsky, M.Y.; Yuzhalin, A.E.; Velikanova, E.A. Shear stress: An essential driver of endothelial progenitor cells. J. Mol. Cell. Cardiol. 2018, 118, 46–69. [Google Scholar] [CrossRef]
- Li, Q.L.; Huang, N.; Chen, C.; Chen, J.L.; Xiong, K.Q.; Chen, J.Y.; You, T.X.; Jin, J.; Liang, X. Oriented immobilization of anti-CD34 antibody on titanium surface for self-endothelialization induction. J. Biomed. Mater. Res. Part A 2010, 94, 1283–1293. [Google Scholar] [CrossRef]
- Kulkeaw, K.; Sakolvaree, Y.; Srimanote, P.; Tongtawe, P.; Maneewatch, S.; Sookrung, N.; Tungtrongchitr, A.; Tapchaisri, P.; Kurazono, H.; Chaicumpa, W. Human monoclonal ScFv neutralize lethal Thai cobra, Naja kaouthia, neurotoxin. J. Proteom. 2009, 72, 270–282. [Google Scholar] [CrossRef]
- Glab-Ampai, K.; Malik, A.A.; Chulanetra, M.; Thanongsaksrikul, J.; Thueng-In, K.; Srimanote, P.; Tongtawe, P.; Chaicumpa, W. Inhibition of HCV replication by humanized-single domain transbodies to NS4B. Biochem. Biophys. Res. Commun. 2016, 476, 654–664. [Google Scholar] [CrossRef] [PubMed]
- Punnakitikashem, P.; Truong, D.; Menon, J.U.; Nguyen, K.T.; Hong, Y. Electrospun biodegradable elastic polyurethane scaffolds with dipyridamole release for small diameter vascular grafts. Acta Biomater. 2014, 10, 4618–4628. [Google Scholar] [CrossRef] [PubMed]


| NPs | Size (nm) | Polydispersity Index | Zeta Potential (mV) | %EE | %CE |
|---|---|---|---|---|---|
| Unloaded | 192.3 ± 49.0 | 0.03 ± 0.01 | −31.3 ± 7.2 | - | - |
| C6-loaded | 188.7 ± 49.6 | 0.04 ± 0.03 | −39.4 ± 7.1 | 89.3 ± 0.3 | - |
| RhB-loaded | 198.9 ± 52.5 | 0.03 ± 0.03 | −39.3 ± 8.0 | 48.8 ± 0.3 | - |
| Control | 199.7 ± 47.8 | 0.01 ± 0.01 | −29.7 ± 9.6 | - | - |
| GPIb | 200.2 ± 50.6 | 0.02 ± 0.03 | −40.8 ± 7.8 | - | 40.1 ± 24.3 |
| HuscFv | 202.3 ± 52.0 | 0.05 ± 0.02 | −41.6 ± 6.9 | - | 38.2 ± 24.5 |
| Dual ligands | 216.6 ± 64.0 | 0.07 ± 0.01 | −38.9 ± 7.8 | - | 61.3 ± 20.2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mungchan, P.; Glab-ampai, K.; Chruewkamlow, N.; Trakarnsanga, K.; Srisawat, C.; Nguyen, K.T.; Chaicumpa, W.; Punnakitikashem, P. Targeted Nanoparticles for the Binding of Injured Vascular Endothelium after Percutaneous Coronary Intervention. Molecules 2022, 27, 8144. https://doi.org/10.3390/molecules27238144
Mungchan P, Glab-ampai K, Chruewkamlow N, Trakarnsanga K, Srisawat C, Nguyen KT, Chaicumpa W, Punnakitikashem P. Targeted Nanoparticles for the Binding of Injured Vascular Endothelium after Percutaneous Coronary Intervention. Molecules. 2022; 27(23):8144. https://doi.org/10.3390/molecules27238144
Chicago/Turabian StyleMungchan, Pennapa, Kittirat Glab-ampai, Nuttapol Chruewkamlow, Kongtana Trakarnsanga, Chatchawan Srisawat, Kytai T. Nguyen, Wanpen Chaicumpa, and Primana Punnakitikashem. 2022. "Targeted Nanoparticles for the Binding of Injured Vascular Endothelium after Percutaneous Coronary Intervention" Molecules 27, no. 23: 8144. https://doi.org/10.3390/molecules27238144
APA StyleMungchan, P., Glab-ampai, K., Chruewkamlow, N., Trakarnsanga, K., Srisawat, C., Nguyen, K. T., Chaicumpa, W., & Punnakitikashem, P. (2022). Targeted Nanoparticles for the Binding of Injured Vascular Endothelium after Percutaneous Coronary Intervention. Molecules, 27(23), 8144. https://doi.org/10.3390/molecules27238144








