LC-HRMS/MS-Based Metabolomics Approaches Applied to the Detection of Antifungal Compounds and a Metabolic Dynamic Assessment of Orchidaceae
Abstract
1. Introduction
2. Results
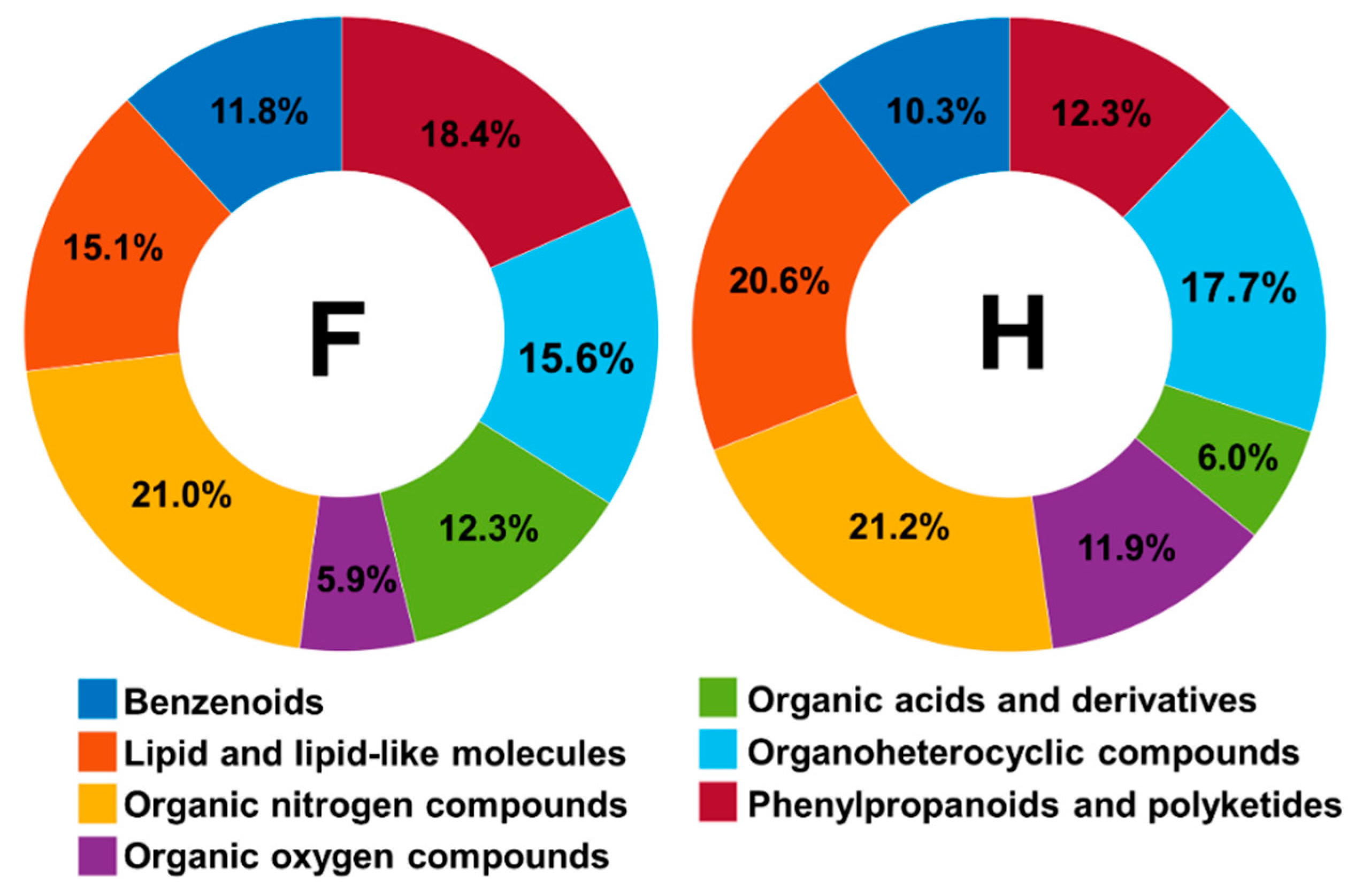
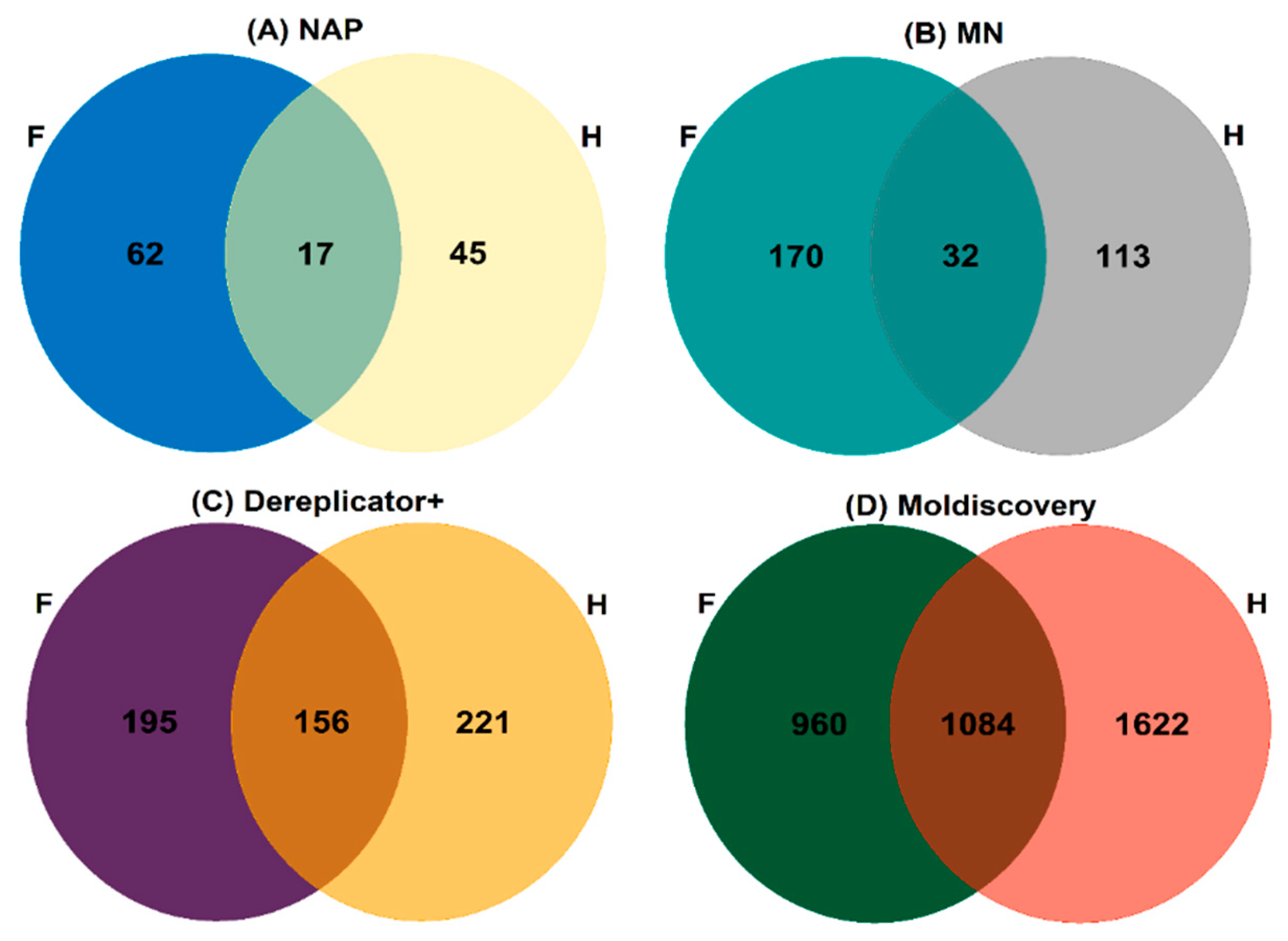
2.1. Comprehensive Structural Annotation of Orchidaceae Species Using Molecular Networking
2.2. Structural Annotation Strategy Using In Silico Fragmentation Tools to the Fingerprinting of Healthy and Fungal-Infected Plants from Orchidaceae
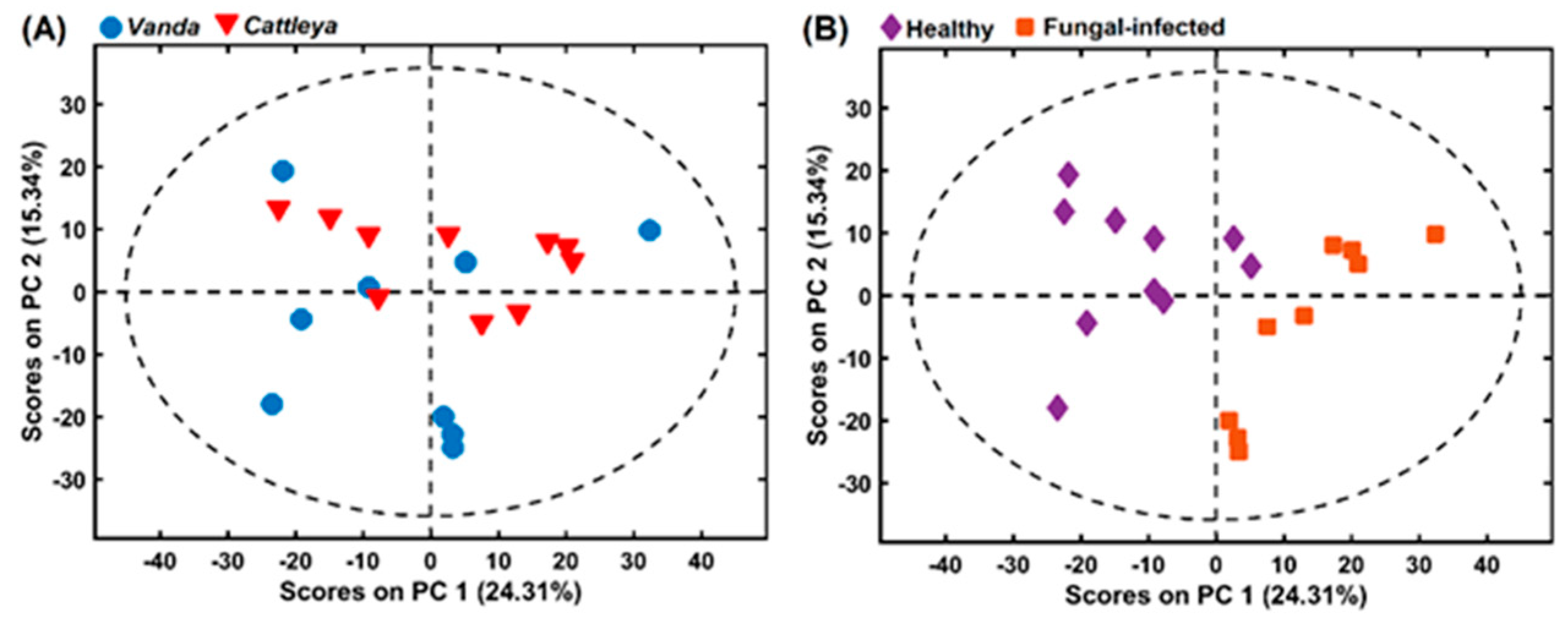
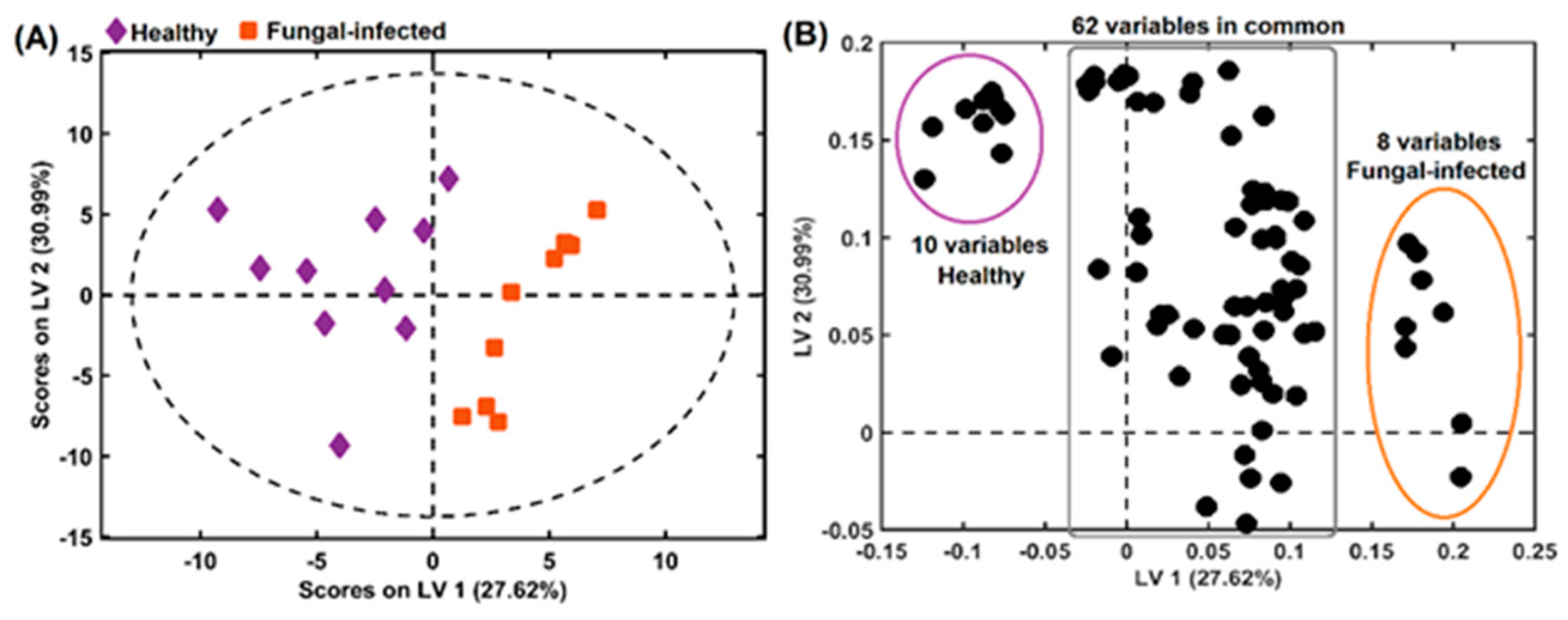
2.3. Chemometrics Methods Applied to Orchidaceae Plants Spectral Analysis
3. Materials and Methods
3.1. Chemicals and Materials
3.2. Plant Material
3.3. Sample Preparation
3.4. LC-HRMS Analysis
3.5. Compound Characterization
3.6. Chemometric Data Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gao, Y.; Ji, J.; Zhang, Y.; Yang, N.; Zhang, M. Biochemical and transcriptomic analyses of the symbiotic interaction between Cremastra appendiculata and the mycorrhizal fungus Coprinellus disseminatus. BMC Plant Biol. 2022, 22, 15. [Google Scholar] [CrossRef] [PubMed]
- Natta, S.; Mondol, S.A.; Pal, K.; Mandal, S.; Sahana, N.; Pal, R.; Pandit, G.K.; Alam, B.K.; Das, S.S.; Biswas, S.S.; et al. Chemical composition, antioxidant activity and bioactive constituents of six native endangered medicinal orchid species from north-eastern Himalayan region of India. S. Afr. J. Bot. 2022, 150, 248–259. [Google Scholar] [CrossRef]
- Gutiérrez, R.M.P. Orchids: A Review of Uses in Traditional Medicine, Its Phytochemistry and Pharmacology. J. Med. Plants Res. 2010, 4, 592–638. [Google Scholar] [CrossRef]
- Fisch, M.; Flick, B.H.; Arditti, J. Structure and antifungal activity of hircinol, loroglossol and orchinol. Phytochemistry 1973, 12, 437–441. [Google Scholar] [CrossRef]
- Auberon, F.; Olatunji, O.J.; Raminoson, D.; Muller, C.D.; Soengas, B.; Bonté, F.; Lobstein, A. Isolation of novel stilbenoids from the roots of Cyrtopodium paniculatum (Orchidaceae). Fitoterapia 2017, 116, 99–105. [Google Scholar] [CrossRef]
- Xiao, K.; Zhang, H.-J.; Xuan, L.-J.; Zhang, J.; Xu, Y.-M.; Bai, D.-L. Stilbenoids: Chemistry and Bioactivities. Stud. Nat. Prod. Chem. 2008, 34, 453–646. [Google Scholar] [CrossRef]
- Ward, E.W.B.; Unwin, C.H.; Stoells, A. Loroglossol: An Orchid Phytoalexin. Phytopathology 1975, 65, 632–633. [Google Scholar] [CrossRef]
- Ióca, L.P.; Romminger, S.; Santos, M.F.C.; Bandeira, K.F.; Rodrigues, F.T.; Kossuga, M.H.; Nicacio, K.J.; Ferreira, E.L.F.; Morais-Urano, R.P.; Passos, M.S.; et al. A strategy for the rapid identification of fungal metabolites and the discovery of the antiviral activity of pyrenocine a and harzianopyridone. Quím. Nova 2016, 39, 720–731. [Google Scholar] [CrossRef]
- Lima, N.M.; Andrade, T.J.A.S.; Silva, D.H.S. Dereplication of terpenes and phenolic compounds from Inga edulis extracts using HPLC-SPE-TT, RP-HPLC-PDA and NMR spectroscopy. Nat. Prod. Res. 2022, 36, 488–492. [Google Scholar] [CrossRef]
- Kind, T.; Fiehn, O. Seven Golden Rules for heuristic filtering of molecular formulas obtained by accurate mass spectrometry. BMC Bioinform. 2007, 8, 105. [Google Scholar] [CrossRef]
- Hubert, J.; Nuzillard, J.-M.; Renault, J.-H. Dereplication strategies in natural product research: How many tools and methodologies behind the same concept? Phytochem. Rev. 2017, 16, 55–95. [Google Scholar] [CrossRef]
- Weidt, S.; Haggarty, J.; Kean, R.; Cojocariu, C.I.; Silcock, P.J.; Rajendran, R.; Ramage, G.; Burgess, K.E.V. A novel targeted/untargeted GC-Orbitrap metabolomics methodology applied to Candida albicans and Staphylococcus aureus biofilms. Metabolomics 2016, 12, 1–10. [Google Scholar] [CrossRef]
- Wolfender, J.-L.; Terreaux, C.; Hostettmann, K. The Importance Of LC-MS And LC-NMR in the Discovery of New Lead Compounds from Plants. Pharm. Biol. 2000, 38, 41–54. [Google Scholar] [CrossRef]
- Bitzer, J.; Köpcke, B.; Stadler, M.; Hellwig, V.; Ju, Y.-M.; Seip, S.; Henkel, T. Accelerated Dereplication of Natural Products, Supported by Reference Libraries. Chimia 2007, 61, 332–338. [Google Scholar] [CrossRef]
- Carter, G.T. NP/MS since 1970: From the basement to the bench top. Nat. Prod. Rep. 2014, 31, 711–717. [Google Scholar] [CrossRef]
- Phapale, P.; Palmer, A.; Gathungu, R.M.; Kale, D.; Brügger, B.; Alexandrov, T. Public LC-Orbitrap-MS/MS Spectral Library for Metabolite Identification. bioRxiv 2020. [Google Scholar] [CrossRef]
- Kuhl, C.; Tautenhahn, R.; Böttcher, C.; Larson, T.R.; Neumann, S. CAMERA: An Integrated Strategy for Compound Spectra Extraction and Annotation of Liquid Chromatography/Mass Spectrometry Data Sets. Anal. Chem. 2012, 84, 283–289. [Google Scholar] [CrossRef]
- Wernisch, S.; Pennathur, S. Evaluation of coverage, retention patterns, and selectivity of seven liquid chromatographic methods for metabolomics. Anal. Bioanal. Chem. 2016, 408, 6079–6091. [Google Scholar] [CrossRef]
- Brodsky, L.; Moussaieff, A.; Shahaf, N.; Aharoni, A.; Rogachev, I. Evaluation of Peak Picking Quality in LC−MS Metabolomics Data. Anal. Chem. 2010, 82, 9177–9187. [Google Scholar] [CrossRef]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef]
- Mohimani, H.; Gurevich, A.; Shlemov, A.; Mikheenko, A.; Korobeynikov, A.; Cao, L.; Shcherbin, E.; Nothias, L.-F.; Dorrestein, P.C.; Pevzner, P.A. Dereplication of microbial metabolites through database search of mass spectra. Nat. Commun. 2018, 9, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, R.R.; Wang, M.; Nothias, L.-F.; Van Der Hooft, J.J.J.; Caraballo-Rodríguez, A.M.; Fox, E.; Balunas, M.J.; Klassen, J.L.; Lopes, N.P.; Dorrestein, P.C. Propagating annotations of molecular networks using in silico fragmentation. PLoS Comput. Biol. 2018, 14, e1006089. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Guler, M.; Tagirdzhanov, A.; Lee, Y.-Y.; Gurevich, A.; Mohimani, H. MolDiscovery: Learning mass spectrometry fragmentation of small molecules. Nat. Commun. 2021, 12, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Reginaldo, F.P.S.; Bueno, P.C.P.; de Matos, I.C.C.; de Araújo Roque, A.R.; Fett-Neto, A.G.; Cavalheiro, A.J.; Giordani, R.B. Molecular Networking Discloses the Chemical Diversity of Flavonoids and Selaginellins in Selaginella convoluta. Planta Med. 2021, 87, 113–123. [Google Scholar] [CrossRef] [PubMed]
- Chacon, D.S.; Santos, M.D.M.; Bonilauri, B.; Vilasboa, J.; da Costa, C.T.; da Silva, I.B.; Torres, T.d.M.; de Araújo, T.F.; Roque, A.D.A.; Pilon, A.C.; et al. Non-target molecular network and putative genes of flavonoid biosynthesis in Erythrina velutina Willd., a Brazilian semiarid native woody plant. Front. Plant Sci. 2022, 13, 947558. [Google Scholar] [CrossRef]
- Yáñez-Barrientos, E.; González-Ibarra, A.A.; Wrobel, K.; Wrobel, K.; Corrales-Escobosa, A.R.; Alonso-Castro, A.J.; Carranza-Álvarez, C.; Ponce-Hernández, A.; Isiordia-Espinoza, M.A.; Ortiz-Andrade, R.; et al. Antinociceptive effects of Laelia anceps Lindl. and Cyrtopodium macrobulbon (Lex.) G.A. Romero & Carnevali, and comparative evaluation of their metabolomic profiles. J. Ethnopharmacol. 2022, 291, 115172. [Google Scholar] [CrossRef]
- Li, X.; Wang, P.; Tong, Y.; Liu, J.; Shu, G. UHPLC-Q-Exactive Orbitrap MS/MS-Based Untargeted Metabolomics and Molecular Networking Reveal the Differential Chemical Constituents of the Bulbs and Flowers of Fritillaria thunbergii. Molecules 2022, 27, 6944. [Google Scholar] [CrossRef]
- Gomes, P.W.P.; Barretto, H.; Reis, J.D.E.; Muribeca, A.; Veloso, A.; Albuquerque, C.; Teixeira, A.; Braamcamp, W.; Pamplona, S.; Silva, C.; et al. Chemical Composition of Leaves, Stem, and Roots of Peperomia pellucida (L.) Kunth. Molecules 2022, 27, 1847. [Google Scholar] [CrossRef]
- Delporte, C.; Noret, N.; Vanhaverbeke, C.; Hardy, O.J.; Martin, J.; Tremblay-franco, M.; Touboul, D.; Gorel, A.; Faes, M.; St, C.; et al. Does the Phytochemical Diversity of Wild Plants Like the Erythrophleum Genus Correlate with Geographical Origin? Molecules 2021, 26, 1668. [Google Scholar] [CrossRef]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.-M.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed minimum reporting standards for chemical analysis. Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef]
- Van der Hooft, J.J.J.; Wandy, J.; Barrett, M.P.; Burgess, K.E.V.; Rogers, S. Topic modeling for untargeted substructure exploration in metabolomics. Proc. Natl. Acad. Sci. USA 2016, 113, 13738–13743. [Google Scholar] [CrossRef]
- Ernst, M.; Kang, K.B.; Caraballo-Rodríguez, A.M.; Nothias, L.-F.; Wandy, J.; Chen, C.; Wang, M.; Rogers, S.; Medema, M.H.; Dorrestein, P.C.; et al. MolNetEnhancer: Enhanced Molecular Networks by Integrating Metabolome Mining and Annotation Tools. Metabolites 2019, 9, 144. [Google Scholar] [CrossRef]
- Nothias, L.-F.; Petras, D.; Schmid, R.; Dührkop, K.; Rainer, J.; Sarvepalli, A.; Protsyuk, I.; Ernst, M.; Tsugawa, H.; Fleischauer, M.; et al. Feature-based molecular networking in the GNPS analysis environment. Nat. Methods 2020, 17, 905–908. [Google Scholar] [CrossRef]
- Arora, M.; Mahajan, A.; Sembi, J.K. A review on phytochemical and pharmacological potential of family orchidaceae. Int. Res. J. Pharm. 2017, 8, 9–24. [Google Scholar] [CrossRef]
- Huang, L.-M.; Huang, H.; Chuang, Y.-C.; Chen, W.-H.; Wang, C.-N.; Chen, H.-H. Evolution of Terpene Synthases in Orchidaceae. Int. J. Mol. Sci. 2021, 22, 6947. [Google Scholar] [CrossRef]
- Diwan, D.; Rashid, M.; Vaishnav, A. Current understanding of plant-microbe interaction through the lenses of multi-omics approaches and their benefits in sustainable agriculture. Microbiol. Res. 2022, 265, 127180. [Google Scholar] [CrossRef]
- Blanco-Ulate, B.; Vincenti, E.; Cantu, D.; Powell, A.L.T. Ripening of Tomato Fruit and Susceptibility to Botrytis Cinerea. In Botrytis—the Fungus, the Pathogen and its Management in Agricultural Systems; Fillinger, S., Elad, Y., Eds.; Springer: Cham, Switzerland, 2016; pp. 387–412. [Google Scholar] [CrossRef]
- Amaral, J.; Valledor, L.; Alves, A.; Martín-García, J.; Pinto, G. Studying tree response to biotic stress using a multi-disciplinary approach: The pine pitch canker case study. Front. Plant Sci. 2022, 13, 916138. [Google Scholar] [CrossRef]
- Gobbo-Neto, L.; Lopes, N.P. Plantas Medicinais: Fatores de Influência No Conteúdo de Metabólitos Secundários. Quim. Nova 2007, 30, 374–381. [Google Scholar] [CrossRef]
- Mwangangi, I.M.; Büchi, L.; Haefele, S.M.; Bastiaans, L.; Runo, S.; Rodenburg, J. Combining host plant defence with targeted nutrition: Key to durable control of hemiparasitic Striga in cereals in sub–Saharan Africa? New Phytol. 2021, 230, 2164–2178. [Google Scholar] [CrossRef]
- Hammond-Kosack, K.; Jones, J. Responses to Plant Pathogens. In Biochemistry and Molecular Biology of Plants; Buchanan, B., Gruissem, W., Jones, R.L., Eds.; Wiley: Rockville, MD, USA, 2000; pp. 1102–1156. [Google Scholar]
- Stevens, E.J.; Bates, K.A.; King, K.C. Host microbiota can facilitate pathogen infection. PLoS Pathog. 2021, 17, e1009514. [Google Scholar] [CrossRef]
- Casadevall, A.; Pirofski, L.-A. Host-Pathogen Interactions: Basic Concepts of Microbial Commensalism, Colonization, Infection, and Disease. Infect. Immun. 2000, 68, 6511–6518. [Google Scholar] [CrossRef] [PubMed]
- De Wit, P.J.G.M. How plants recognize pathogens and defend themselves. Cell. Mol. Life Sci. 2007, 64, 2726–2732. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.-M.; Ibrahim, R.K. Tricin—A potential multifunctional nutraceutical. Phytochem. Rev. 2010, 9, 413–424. [Google Scholar] [CrossRef]
- Wang, J.; Wei, X.; Qin, X.; Tian, X.; Liao, L.; Li, K.; Zhou, X.; Yang, X.; Wang, F.; Zhang, T.; et al. Antiviral Merosesquiterpenoids Produced by the Antarctic Fungus Aspergillus ochraceopetaliformis SCSIO 05702. J. Nat. Prod. 2016, 79, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Mattio, L.; Catinella, G.; Iriti, M.; Vallone, L. Inhibitory activity of stilbenes against filamentous fungi. Ital. J. Food Saf. 2021, 10, 8461. [Google Scholar] [CrossRef]
- Dvorakova, M.; Landa, P. Anti-inflammatory activity of natural stilbenoids: A review. Pharmacol. Res. 2017, 124, 126–145. [Google Scholar] [CrossRef]
- Samanta, A.; Das, G.; Das, S.K. Roles of Flavonoids in Plants. Int. J. Pharm. Sci. Technol. 2011, 6, 12–35. [Google Scholar]
- Bro, R.; Smilde, A.K. Principal component analysis. Anal. Methods 2014, 6, 2812–2831. [Google Scholar] [CrossRef]
- Barker, M.; Rayens, W. Partial least squares for discrimination. J. Chemom. 2003, 17, 166–173. [Google Scholar] [CrossRef]
- Roque, J.V.; Cardoso, W.; Peternelli, L.A.; Teófilo, R.F. Comprehensive new approaches for variable selection using ordered predictors selection. Anal. Chim. Acta 2019, 1075, 57–70. [Google Scholar] [CrossRef]
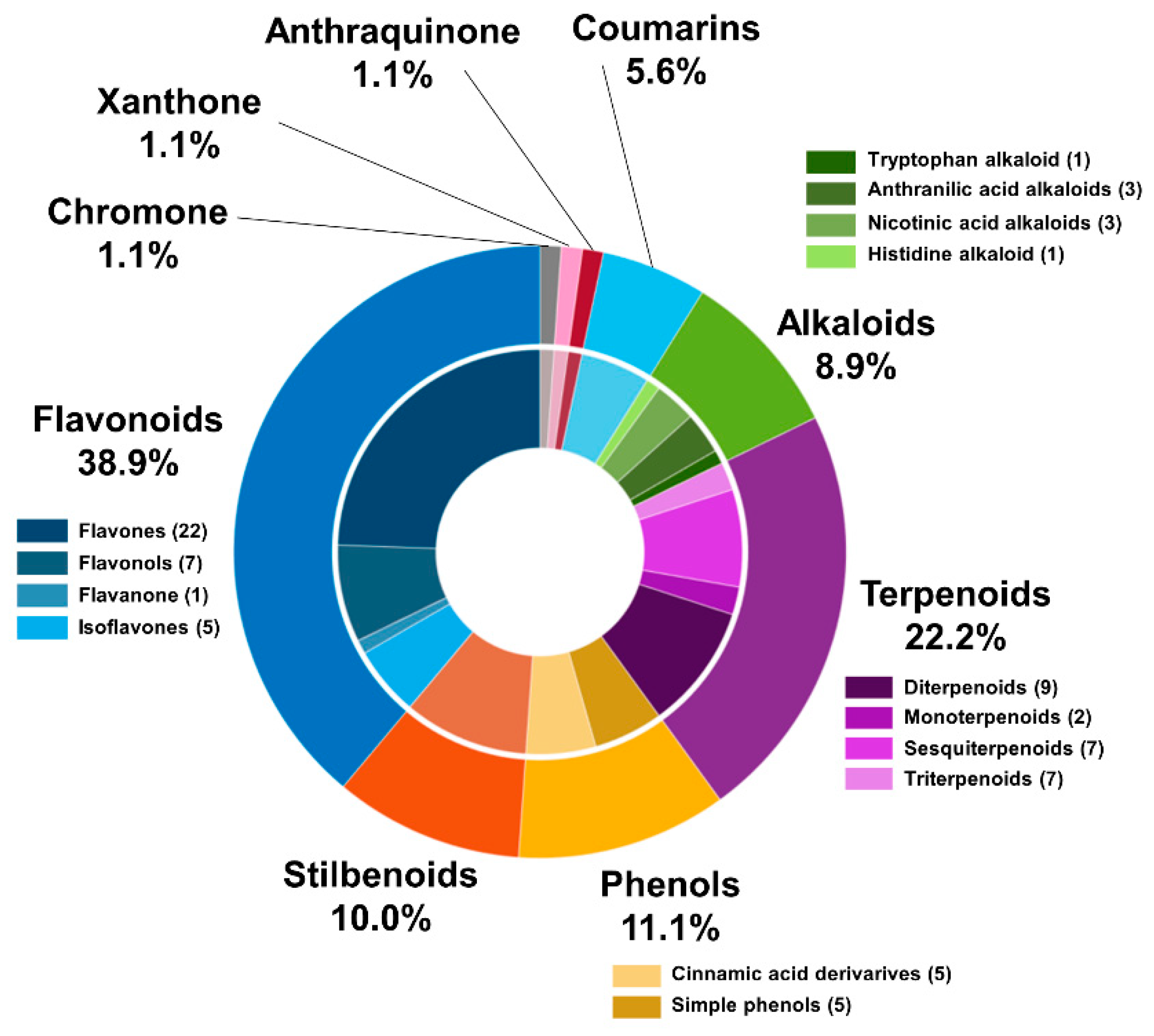
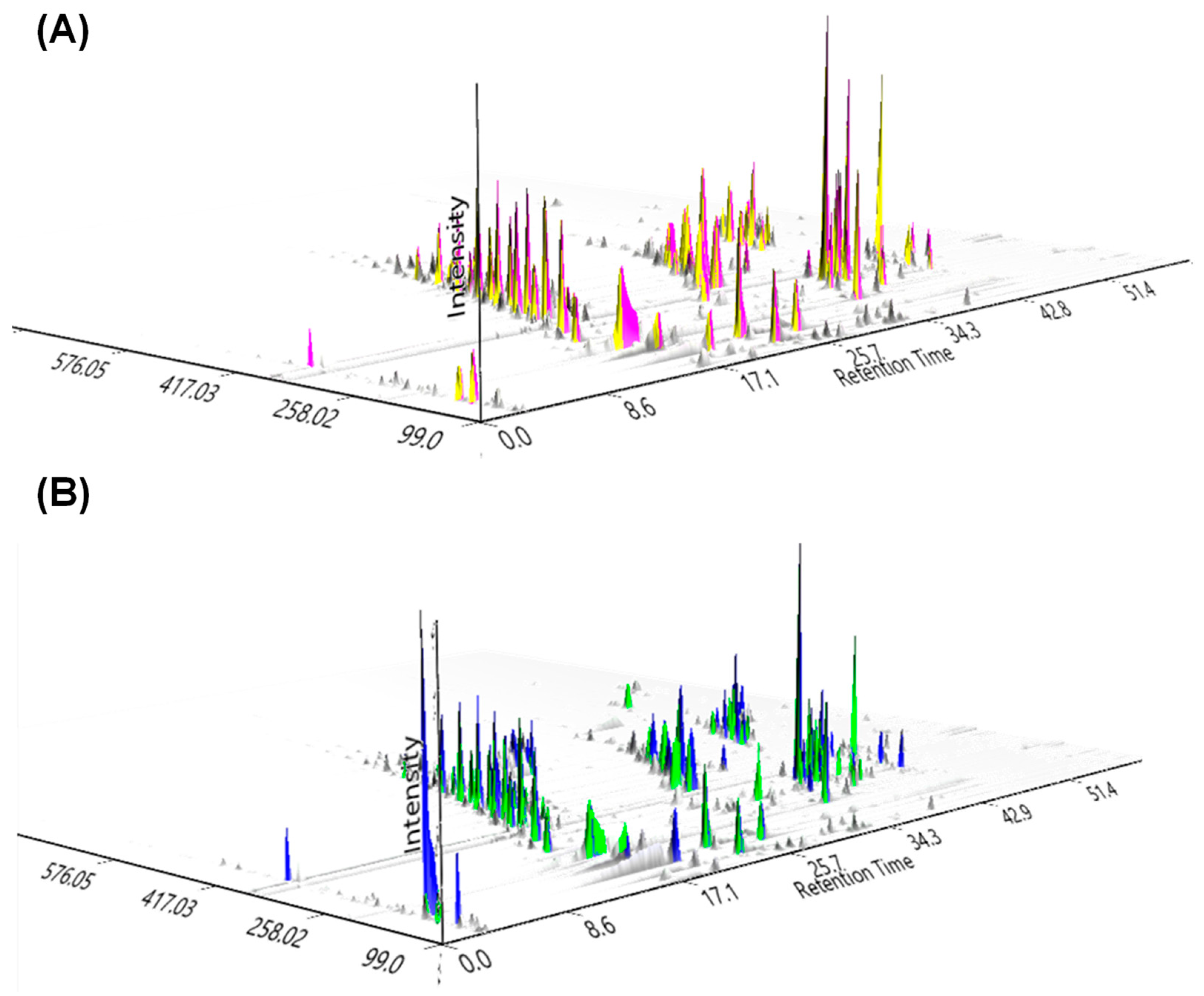
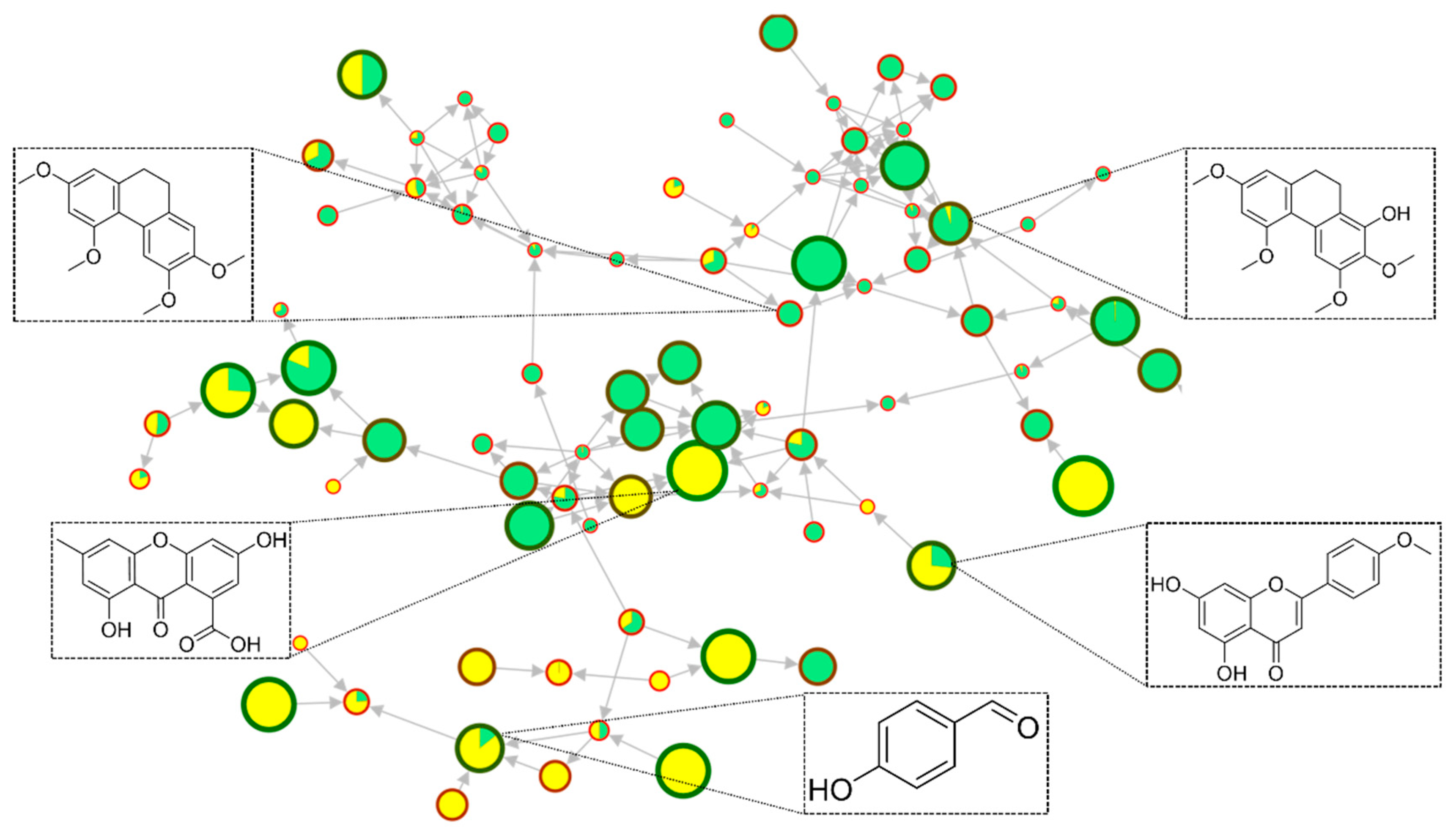
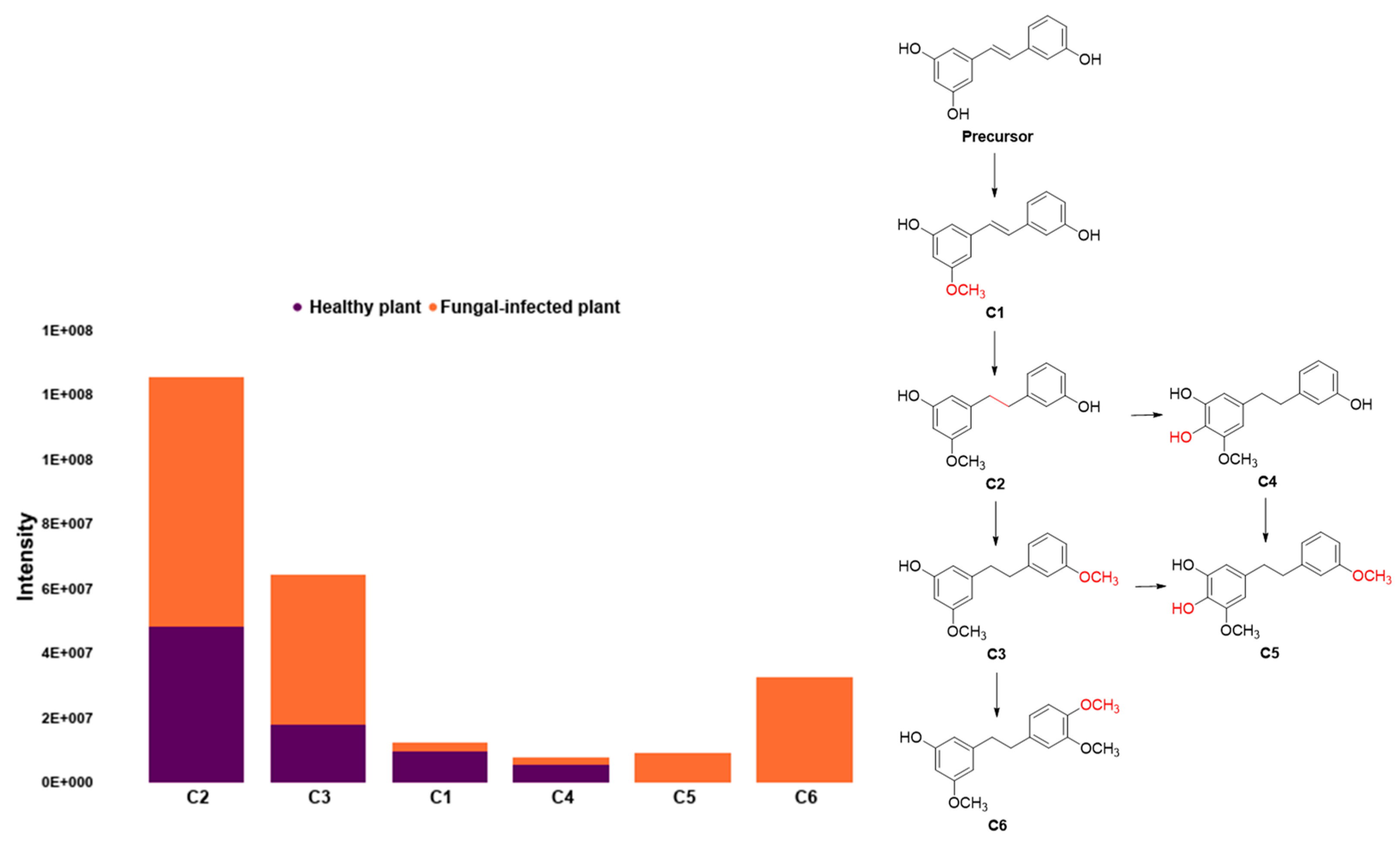
| Genus | RT (Min) | Exact Mass | Molecular Formula | Metabolite Name | Chemical Class |
|---|---|---|---|---|---|
| Vanda | 1.72 | 122.037 | C7H6O2 | 4-Hydroxybenzaldehyde | Phenolic acid |
| Vanda | 1.77 | 124.052 | C7H8O2 | 3-hydroxybenzyl alcohol | Benzenoid |
| Vanda | 24.22 | 134.073 | C9H10O | Cinnamic alcohol | Phenylpropanoid |
| Vanda/Cattleya | 2.02 | 150.068 | C9H10O2 | 4-methoxy-3-methylbenzaldehyde | Benzenoid |
| Vanda | 28.78 | 162.068 | C10H10O2 | 4-Methylcinnamic acid | Phenylpropanoid |
| Vanda | 1.72 | 164.047 | C9H8O3 | p-coumaric acid | Phenylpropanoid |
| Cattleya | 12.53 | 168.042 | C8H8O4 | Benzoic acid, 2,4-dihydroxy-, methyl ester | Phenolic acid |
| Vanda | 20.78 | 196.110 | C11H16O3 | Loliolide | Terpenoid |
| Vanda | 29.26 | 210.089 | C11H14O4 | Sinapyl alcohol | Phenylpropanoid |
| Vanda/Cattleya | 16.80 | 224.068 | C11H12O5 | Sinapic acid | Phenylpropanoid |
| Vanda/Cattleya | 22.06 | 242.094 | C15H14O3 | Thunalbene | Stilbenoid |
| Vanda/Cattleya | 29.32 | 244.109 | C15H16O3 | 3-[2-(3-hydroxyphenyl)ethyl]-5-methoxyphenol | Stilbenoid |
| Cattleya | 31.36 | 254.058 | C15H10O4 | Daidzein | Isoflavonoid |
| Vanda | 27.28 | 256.074 | C15H12O4 | Liquiritigenin | Flavonoid |
| Vanda/Cattleya | 33.93 | 258.125 | C16H18O3 | 3′-O-Methylbatatasin III | Stilbenoid |
| Vanda/Cattleya | 260.104 | C15H16O4 | Tristin | Stilbenoid | |
| Vanda/Cattleya | 30.11 | 260.105 | C15H16O4 | Dendrosinene B | Stilbenoid |
| Vanda | 33.56 | 266.094 | C17H14O3 | 4′-Methoxy-6-methylflavone | Flavonoid |
| Vanda/Cattleya | 39.50 | 272.177 | C18H24O2 | Galaxolidone | Terpenoid |
| Vanda/Cattleya | 28.65 | 274.121 | C16H18O4 | Gigantol | Stilbenoid |
| Vanda/Cattleya | 32.23 | 274.121 | C16H18O4 | 3-Methoxy-5-[2-(3-methoxyphenyl)ethyl]-1,2-benzenediol | Stilbenoid |
| Cattleya | 31.93 | 284.068 | C16H12O5 | Acacetin | Flavonoid |
| Vanda/Cattleya | 31.68 | 289.143 | C17H20O4 | 3-O-Methylgigantol | Stilbenoid |
| Cattleya | 34.35 | 298.084 | C17H14O5 | Afrormosin | Isoflavonoid |
| Cattleya | 10.01 | 300.120 | C14H20O7 | Salidroside | Phenol |
| Vanda | 36.41 | 300.136 | C18H20O4 | 2,3,5,7-tetramethoxy-9,10-dihydrophenanthrene | Stilbenoid |
| Vanda/Cattleya | 31.44 | 318.146 | C18H22O5 | Erianin | Stilbenoid |
| Vanda | 31.22 | 330.147 | C19H22O5 | 3-(4-hydroxy-3-methoxyphenyl)propyl 3-(4-hydroxyphenyl)propanoate | Phenol |
| Cattleya | 26.91 | 348.209 | C24H28O2 | Bexarotene | Terpenoid |
| Vanda/Cattleya | 22.81 | 374.230 | C19H34O7 | (2R,3S,4S,5R,6R)-2-(hydroxymethyl)-6-[4-(4-hydroxy-2,6,6-trimethylcyclohexen-1-yl)butan-2-yloxy]oxane-3,4,5-triol | Terpenoid |
| Vanda/Cattleya | 17.70 | 389.217 | C19H32O8 | (2R)-4-[(1S)-1-Hydroxy-2,6,6-trimethyl-4-oxo-2-cyclohexen-1-yl]-2-butanyl beta-D-glucopyranoside | Terpenoid |
| Vanda/Cattleya | 35.31 | 390.204 | C22H30O6 | 7b,9-Dihydroxy-3-(hydroxymethyl)-1,1,6,8-tetramethyl-5-oxo-1,1a,1b,4,4a,5,7a,7b,8,9-decahydro-9aH-cyclopropa [3,4]benzo [1,2-e]azulen-9a-yl acetate | Terpenoid |
| Vanda/Cattleya | 21.65 | 420.142 | C21H24O9 | Rhapontin | Stilbenoid |
| Vanda | 16.08 | 432.105 | C21H20O10 | Isovitexin | Flavonoid |
| Vanda/Cattleya | 12.84 | 432.163 | C19H28O11 | Darendoside A | Phenylethanoid |
| Cattleya | 44.93 | 442.381 | C30H50O2 | Allobetulinol | Terpenoid |
| Vanda | 16.61 | 448.100 | C21H20O11 | Homoorientin | Flavonoid |
| Vanda | 22.65 | 464.095 | C21H20O12 | Hyperoside | Flavonoid |
| Vanda | 24.66 | 478.111 | C22H22O12 | Isorhamnetin 3-galactoside | Flavonoid |
| Cattleya | 29.47 | 492.127 | C23H24O12 | Tricin 5-glucoside | Flavonoid |
| Vanda | 1.80 | 564.148 | C26H28O14 | Isoschaftoside | Flavonoid |
| Cattleya | 21.31 | 564.148 | C26H28O14 | Isovitexin 2′’-O-arabinoside | Flavonoid |
| Cattleya | 18.13 | 582.231 | C28H38O13 | 2-[[5-(4-hydroxy-3,5-dimethoxyphenyl)-6,7-bis(hydroxymethyl)-1,3-dimethoxy-5,6,7,8-tetrahydronaphthalen-2-yl]oxy]-6-(hydroxymethyl)oxane-3,4,5-triol | Lignan |
| Vanda/Cattleya | 15.80 | 594.158 | C27H30O15 | Saponarin | Flavonoid |
| Vanda | 22.84 | 610.153 | C27H30O16 | Rutin | Flavonoid |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lima, G.S.; Lima, N.M.; Roque, J.V.; de Aguiar, D.V.A.; Oliveira, J.V.A.; dos Santos, G.F.; Chaves, A.R.; Vaz, B.G. LC-HRMS/MS-Based Metabolomics Approaches Applied to the Detection of Antifungal Compounds and a Metabolic Dynamic Assessment of Orchidaceae. Molecules 2022, 27, 7937. https://doi.org/10.3390/molecules27227937
Lima GS, Lima NM, Roque JV, de Aguiar DVA, Oliveira JVA, dos Santos GF, Chaves AR, Vaz BG. LC-HRMS/MS-Based Metabolomics Approaches Applied to the Detection of Antifungal Compounds and a Metabolic Dynamic Assessment of Orchidaceae. Molecules. 2022; 27(22):7937. https://doi.org/10.3390/molecules27227937
Chicago/Turabian StyleLima, Gesiane S., Nerilson M. Lima, Jussara V. Roque, Deborah V. A. de Aguiar, João V. A. Oliveira, Gabriel F. dos Santos, Andrea R. Chaves, and Boniek G. Vaz. 2022. "LC-HRMS/MS-Based Metabolomics Approaches Applied to the Detection of Antifungal Compounds and a Metabolic Dynamic Assessment of Orchidaceae" Molecules 27, no. 22: 7937. https://doi.org/10.3390/molecules27227937
APA StyleLima, G. S., Lima, N. M., Roque, J. V., de Aguiar, D. V. A., Oliveira, J. V. A., dos Santos, G. F., Chaves, A. R., & Vaz, B. G. (2022). LC-HRMS/MS-Based Metabolomics Approaches Applied to the Detection of Antifungal Compounds and a Metabolic Dynamic Assessment of Orchidaceae. Molecules, 27(22), 7937. https://doi.org/10.3390/molecules27227937







