Multi-Bioactivity of Protein Digests and Peptides from Oat (Avena sativa L.) Kernels in the Prevention of the Cardiometabolic Syndrome
Abstract
1. Introduction
2. Results and Discussion
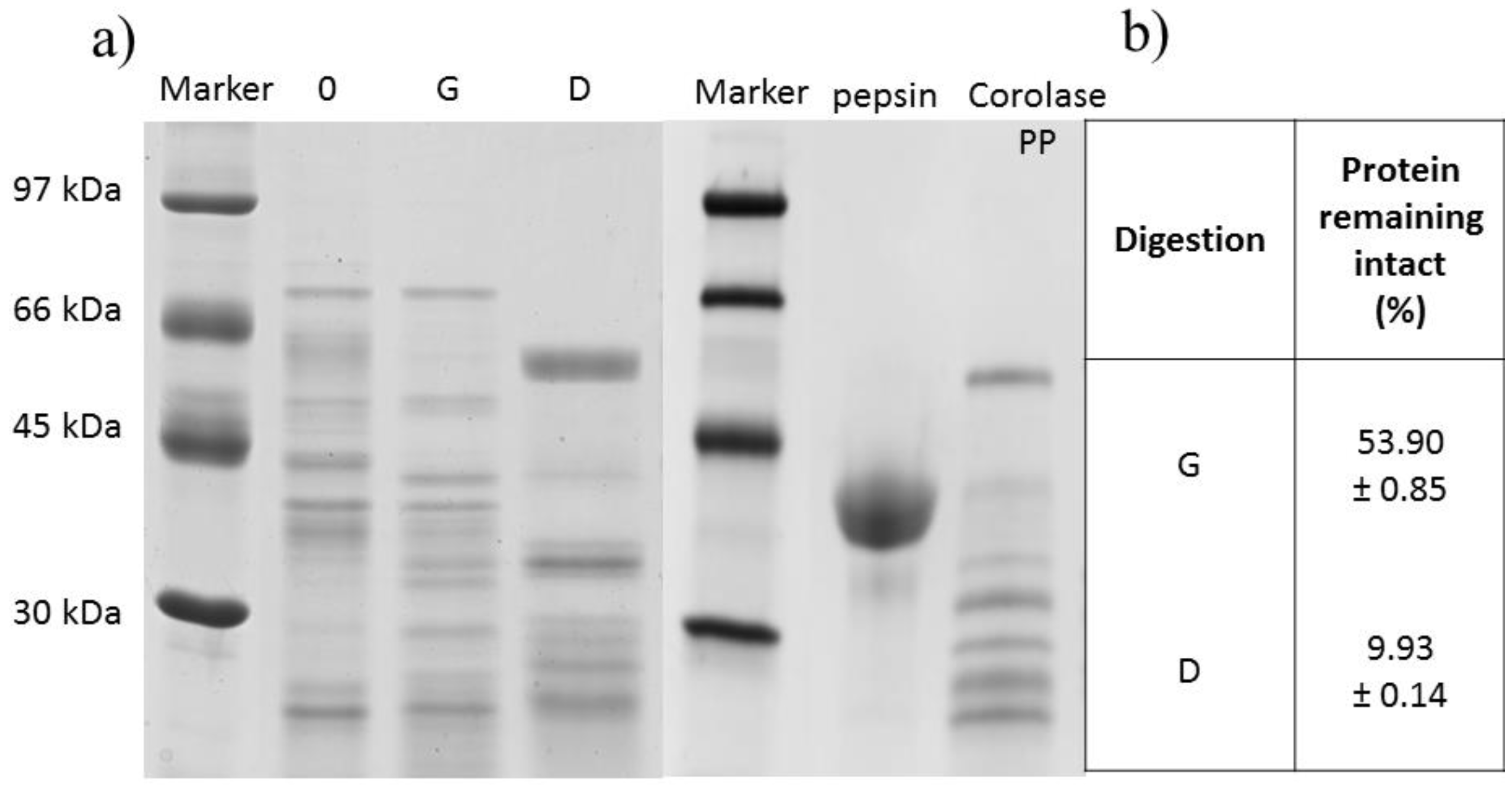
2.1. Oat Kernel Proteins In Vitro Human-Like Gastrointestinal Digestion
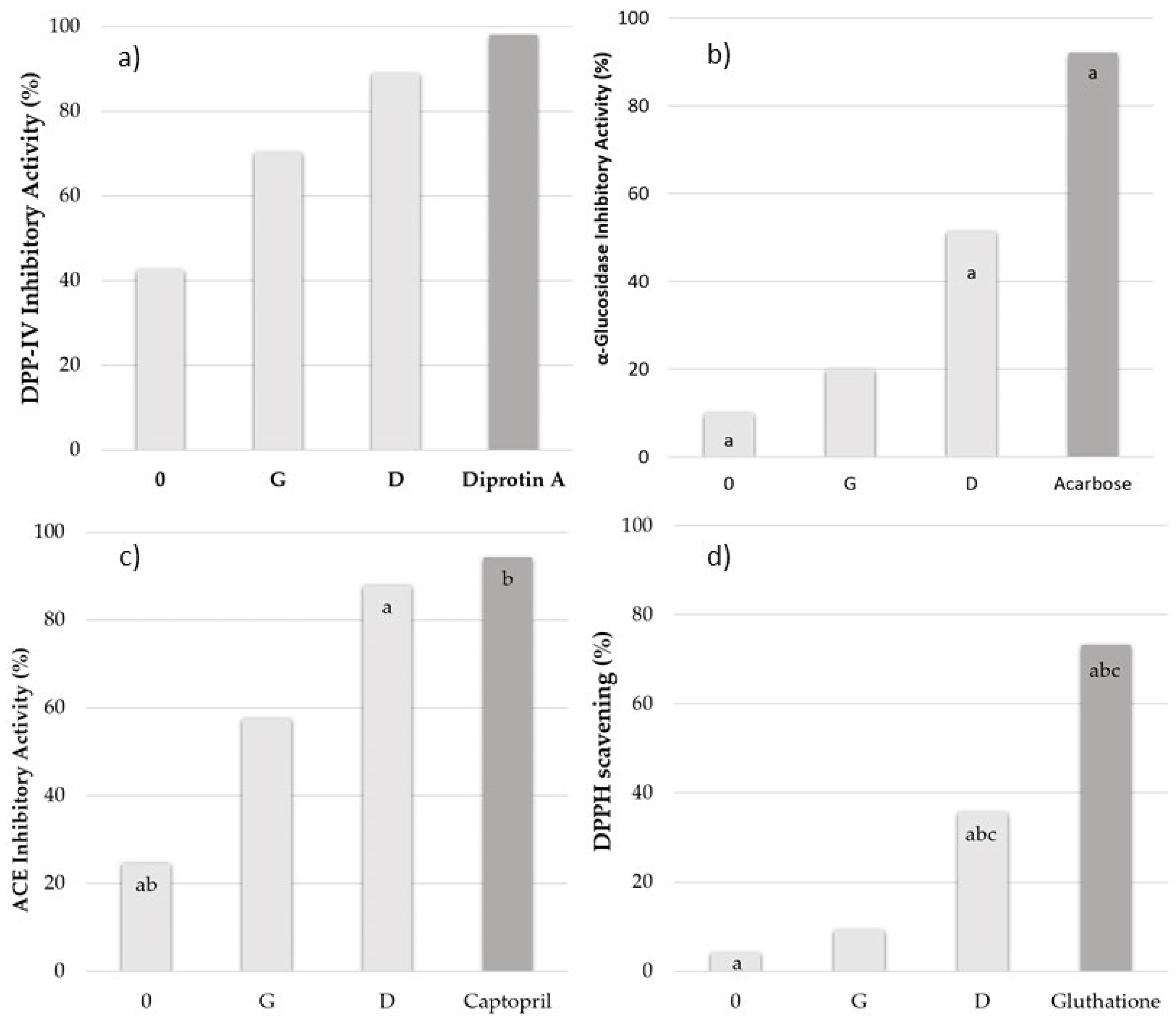
2.2. Dipeptidyl Peptidase IV Inhibitory Activity
2.3. α-Glucosidase Inhibitory Activity
2.4. Angiotensin I-Converting Enzyme Inhibitory Activity
2.5. Antioxidative Activity
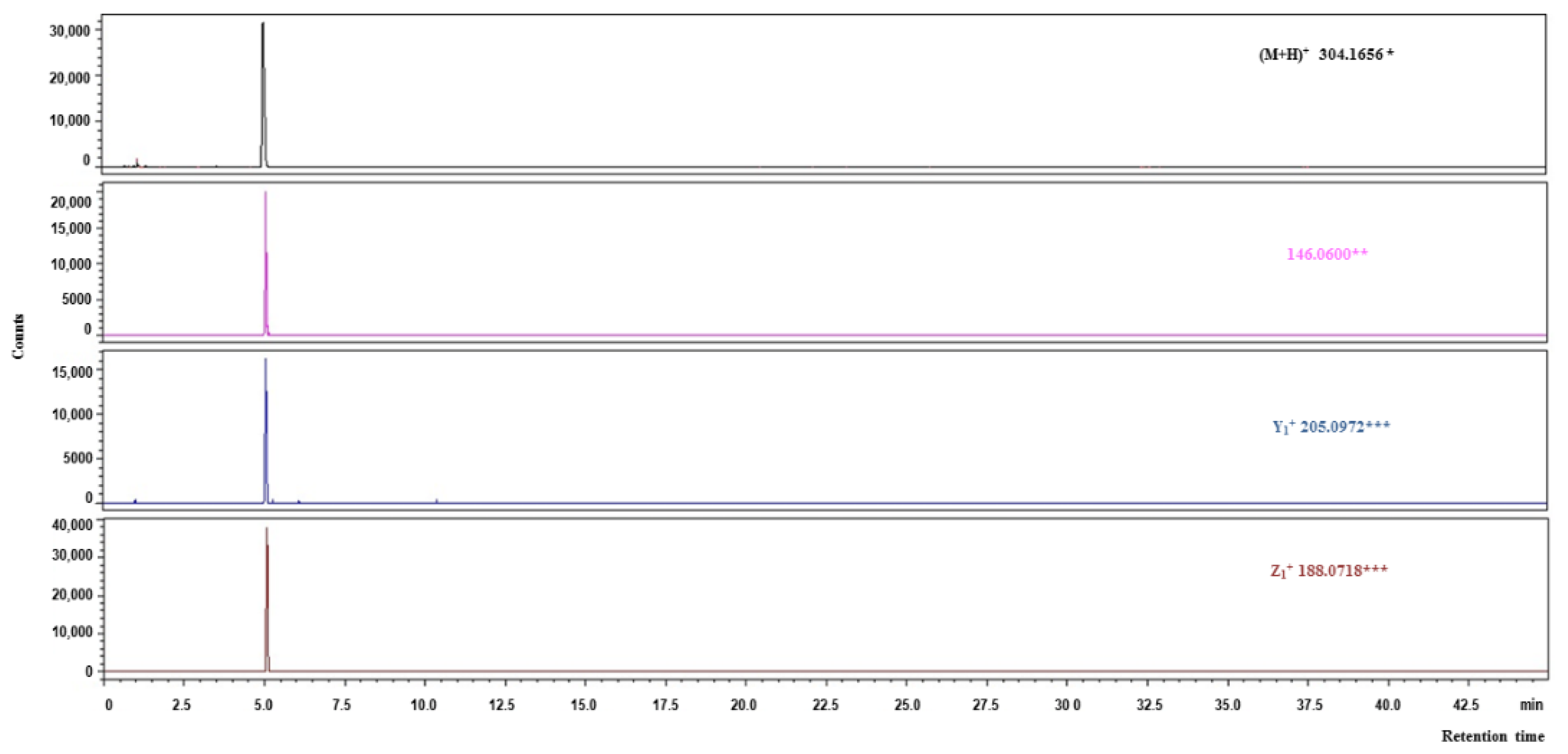
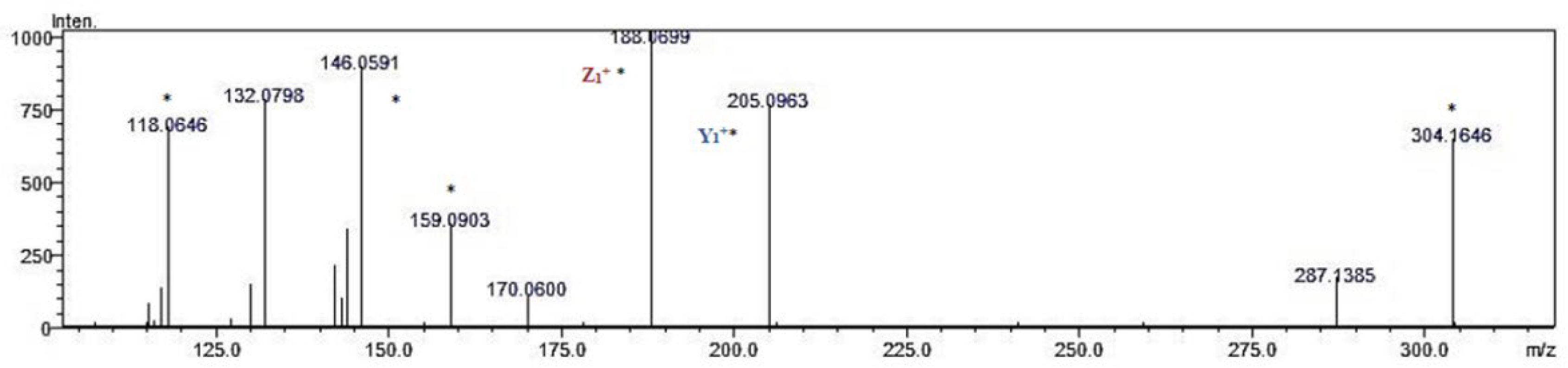
2.6. Identification of Cardiometabolic Syndrome-Preventive Peptides
2.7. Transepithelial Transport of Peptides
3. Materials and Methods
3.1. Chemicals and Reagents
3.2. Materials
3.3. In Silico Digestion
3.3.1. Dataset Construction
3.3.2. In Silico Evaluation of Bioactive Peptides Potential
3.4. Extraction of Oat Kernel Proteins
3.5. In Vitro Human-Like Gastrointestinal Digestion Using the INFOGEST Static Model
3.6. Protein/Peptide Content
3.7. Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis (SDS-PAGE)
3.8. Enzyme Inhibition Assays
3.9. DPPH• Scavenging Activity Assay
3.10. Identification of Peptides Using Reversed-Phase Liquid Chromatography Coupled with Mass Spectrometry with Quadrupole Time of Flight Analyzer (LC-Q-TOF-MS/MS)
3.11. Caco-2 Transportation Experiments
3.11.1. Caco-2 Cells Culture
3.11.2. Transport Studies
3.12. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Mora, L.; Aristoy, M.-C.; Toldrá, F. Bioactive peptides. Reference Module in Food Science. In Encyclopedia of Food Chemistry; Elsevier: Amsterdam, The Netherlands, 2019; pp. 381–389. [Google Scholar]
- Tu, M.; Cheng, S.; Lu, W.; Du, M. Advancement and prospects of bioinformatics analysis for studying bioactive peptides from food-derived protein: Sequence, structure, and functions. TrAC—Trends Anal. Chem. 2018, 105, 7–17. [Google Scholar] [CrossRef]
- Zambrowicz, A.; Timmer, M.; Polanowski, A.; Lubec, G.; Trziszka, T. Manufacturing of peptides exhibiting biological activity. Amino Acids 2013, 44, 315–320. [Google Scholar] [CrossRef] [PubMed]
- Minkiewicz, P.; Dziuba, J.; Darewicz, M.; Iwaniak, A.; Dziuba, M.; Nałęcz, D. Food peptidomics. Food Technol. Biotechnol. 2008, 46, 1–10. [Google Scholar]
- Saklayen, M.G. The epidemic of the metabolic syndrome. Curr. Hypertens. Rep. 2018, 20, 12. [Google Scholar] [CrossRef] [PubMed]
- Lemieux, I.; Després, J.P. Metabolic syndrome: Past, present and future. Nutrients 2020, 12, 3501. [Google Scholar] [CrossRef] [PubMed]
- Kelli, H.M.; Kassas, I.; Lattouf, O.M. Cardio Metabolic Syndrome: A Global Epidemic. J. Diabetes Metab. 2015, 6, 1000513. [Google Scholar] [CrossRef]
- Iwaniak, A.; Mogut, D. Metabolic syndrome-preventive peptides derived from milk proteins and their presence in cheeses: A review. Appl. Sci. 2020, 10, 2772. [Google Scholar] [CrossRef]
- Garcia-Mora, P.; Peñas, E.; Frias, J.; Martínez-Villaluenga, C. Savinase, the most suitable enzyme for releasing peptides from lentil (Lens culinaris var. Castellana) protein concentrates with multifunctional properties. J. Agric. Food Chem. 2014, 62, 4166–4174. [Google Scholar] [CrossRef]
- Ji, W.; Zhang, C.; Ji, H. Two Novel Bioactive Peptides from Antarctic Krill with Dual Angiotensin Converting Enzyme and Dipeptidyl Peptidase IV Inhibitory Activities. J. Food Sci. 2017, 82, 1742–1749. [Google Scholar] [CrossRef]
- Zambrowicz, A.; Pokora, M.; Setner, B.; Dąbrowska, A.; Szołtysik, M.; Babij, K.; Szewczuk, Z.; Trziszka, T.; Lubec, G.; Chrzanowska, J. Multifunctional peptides derived from an egg yolk protein hydrolysate: Isolation and characterization. Amino Acids 2015, 47, 369–380. [Google Scholar] [CrossRef]
- Gangopadhyay, N.; Hossain, M.B.; Rai, D.K.; Brunton, N.P. A review of extraction and analysis of bioactives in oat and barley and scope for use of novel food processing technologies. Molecules 2015, 20, 10884–10909. [Google Scholar] [CrossRef]
- Iwaniak, A.; Darewicz, M.; Minkiewicz, P. Peptides Derived from Foods as Supportive Diet Components in the Prevention of Metabolic Syndrome. Compr. Rev. Food Sci. Food Saf. 2018, 17, 63–81. [Google Scholar] [CrossRef]
- Nongonierma, A.B.; Fitzgerald, R.J. An in silico model to predict the potential of dietary proteins as sources of dipeptidyl peptidase IV (DPP-IV) inhibitory peptides. Food Chem. 2014, 165, 489–498. [Google Scholar] [CrossRef]
- Bleakley, S.; Hayes, M.; O’ Shea, N.; Gallagher, E.; Lafarga, T. Predicted release and analysis of novel ACE-I, renin, and DPP-IV inhibitory peptides from common oat (Avena sativa) protein hydrolysates using in Silico analysis. Foods 2017, 6, 108. [Google Scholar] [CrossRef]
- Cheung, I.W.Y.; Nakayama, S.; Hsu, M.N.K.; Samaranayaka, A.G.P.; Li-Chan, E.C.Y. Angiotensim-I converting enzyme inhibitory activity of hydrolysates from oat (Avena sativa) proteins by in silico and in vitro analyses. J. Agric. Food Chem. 2009, 57, 9234–9242. [Google Scholar] [CrossRef]
- Wang, F.; Yu, G.; Zhang, Y.; Zhang, B.; Fan, J. Dipeptidyl Peptidase IV Inhibitory Peptides Derived from Oat (Avena sativa L.), Buckwheat (Fagopyrum esculentum), and Highland Barley (Hordeum vulgare trifurcatum (L.) Trofim) Proteins. J. Agric. Food Chem. 2015, 63, 9543–9549. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, J.; Liu, Y.; Sun, B. Peptides derived from oats improve insulin sensitivity and lower blood glucose in streptozotocin-induced diabetic mice. J. Biomed. Sci. 2015, 4, 1–7. [Google Scholar] [CrossRef]
- Dugardin, C.; Cudennec, B.; Tourret, M.; Caron, J.; Guérin-Deremaux, L.; Behra-Miellet, J.; Lefranc-Millot, C.; Ravallec, R. Explorative screening of bioactivities generated by plant-based proteins after in vitro static gastrointestinal digestion. Nutrients 2020, 12, 3746. [Google Scholar] [CrossRef]
- Versantvoort, C.H.M.; Oomen, A.G.; Van De Kamp, E.; Rompelberg, C.J.M.; Sips, J.A.M. Applicability of an in vitro digestion model in assessing the bioaccessibility of mycotoxins from food. Food Chem. Toxicol. 2005, 43, 31–40. [Google Scholar] [CrossRef]
- Ben Halima, N.; Saad, R.B.; Khemakhem, B.; Fendri, I.; Abdelkafi, S. Oat (Avena sativa L.): Oil and Nutriment Compounds Valorization for Potential Use in Industrial Applications. J. Oleo Sci. 2015, 64, 915–932. [Google Scholar] [CrossRef]
- Sanchón, J.; Fernández-Tomé, S.; Miralles, B.; Hernández-Ledesma, B.; Tomé, D.; Gaudichon, C.; Recio, I. Protein degradation and peptide release from milk proteins in human jejunum. Comparison with in vitro gastrointestinal simulation. Food Chem. 2018, 239, 486–494. [Google Scholar] [CrossRef] [PubMed]
- Sousa, R.; Portmann, R.; Dubois, S.; Recio, I.; Egger, L. Protein digestion of different protein sources using the INFOGEST static digestion model. Food Res. Int. 2020, 130, 108996. [Google Scholar] [CrossRef] [PubMed]
- Schalk, K.; Lexhaller, B.; Koehler, P.; Scherf, K.A. Isolation and characterization of gluten protein types from wheat, rye, barley and oats for use as reference materials. PLoS ONE 2017, 12, e0172819. [Google Scholar] [CrossRef]
- Borawska, J.; Darewicz, M.; Vegarud, G.E.; Iwaniak, A.; Minkiewicz, P. Ex vivo digestion of carp muscle tissue—ACE inhibitory and antioxidant activities of the obtained hydrolysates. Food Funct. 2015, 6, 210–217. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.A.; Ekeberg, D.; Rukke, E.O.; Vegarud, G.E. Ex vivo digestion of proteins and fat in buffalo milk. Int. Dairy J. 2016, 52, 82–91. [Google Scholar] [CrossRef]
- Liu, R.; Cheng, J.; Wu, H. Discovery of food-derived dipeptidyl peptidase IV inhibitory peptides: A review. Int. J. Mol. Sci. 2019, 20, 463. [Google Scholar] [CrossRef]
- Nongonierma, A.B.; Hennemann, M.; Paolella, S.; Fitzgerald, R.J. Generation of wheat gluten hydrolysates with dipeptidyl peptidase IV (DPP-IV) inhibitory properties. Food Funct. 2017, 8, 2249–2257. [Google Scholar] [CrossRef]
- Nongonierma, A.B.; Le Maux, S.; Dubrulle, C.; Barre, C.; FitzGerald, R.J. Quinoa (Chenopodium quinoa Willd.) protein hydrolysates with in vitro dipeptidyl peptidase IV (DPP-IV) inhibitory and antioxidant properties. J. Cereal Sci. 2015, 65, 112–118. [Google Scholar] [CrossRef]
- González-Montoya, M.; Hernández-Ledesma, B.; Mora-Escobedo, R.; Martínez-Villaluenga, C. Bioactive Peptides from Germinated Soybean with Anti-Diabetic Potential by Inhibition of Dipeptidyl Peptidase-IV, α-Amylase, and α-Glucosidase Enzymes. Int. J. Mol. Sci. 2018, 19, 2883. [Google Scholar] [CrossRef]
- Lammi, C.; Arnoldi, A.; Aiello, G. Soybean Peptides Exert Multifunctional Bioactivity Modulating 3-Hydroxy-3-Methylglutaryl-CoA Reductase and Dipeptidyl Peptidase-IV Targets in Vitro. J. Agric. Food Chem. 2019, 67, 4824–4830. [Google Scholar] [CrossRef]
- Li, Y.; Aiello, G.; Bollati, C.; Bartolomei, M.; Arnoldi, A.; Lammi, C. Phycobiliproteins from Arthrospira platensis (spirulina): A new source of peptides with dipeptidyl peptidase-IV inhibitory activity. Nutrients 2020, 12, 794. [Google Scholar] [CrossRef]
- Mojica, L.; Luna-Vital, D.A.; González de Mejía, E. Characterization of peptides from common bean protein isolates and their potential to inhibit markers of type-2 diabetes, hypertension and oxidative stress. J. Sci. Food Agric. 2017, 97, 2401–2410. [Google Scholar] [CrossRef] [PubMed]
- Mojica, L.; De Mejía, E.G. Optimization of enzymatic production of anti-diabetic peptides from black bean (Phaseolus vulgaris L.) proteins, their characterization and biological potential. Food Funct. 2016, 7, 713–727. [Google Scholar] [CrossRef] [PubMed]
- Oseguera-Toledo, M.E.; Gonzalez de Mejia, E.; Amaya-Llano, S.L. Hard-to-cook bean (Phaseolus vulgaris L.) proteins hydrolyzed by alcalase and bromelain produced bioactive peptide fractions that inhibit targets of type-2 diabetes and oxidative stress. Food Res. Int. 2015, 76, 839–851. [Google Scholar] [CrossRef] [PubMed]
- Iwaniak, A.; Minkiewicz, P.; Darewicz, M. Food-originating ACE inhibitors, including antihypertensive peptides, as preventive food components in blood pressure reduction. Compr. Rev. Food Sci. Food Saf. 2014, 13, 114–134. [Google Scholar] [CrossRef]
- Jakubczyk, A.; Baraniak, B. Activities and sequences of the angiotensin I-converting enzyme (ACE) inhibitory peptides obtained from the digested lentil (Lens culinaris) globulins. Int. J. Food Sci. Technol. 2013, 48, 2363–2369. [Google Scholar] [CrossRef]
- Durak, A.; Baraniak, B.; Jakubczyk, A.; Świeca, M. Biologically active peptides obtained by enzymatic hydrolysis of Adzuki bean seeds. Food Chem. 2013, 141, 2177–2187. [Google Scholar] [CrossRef]
- Darewicz, M.; Borawska, J.; Vegarud, G.E.; Minkiewicz, P.; Iwaniak, A. Angiotensin I-converting enzyme (ACE) inhibitory activity and ace inhibitory peptides of salmon (Salmo salar) protein hydrolysates obtained by human and porcine gastrointestinal enzymes. Int. J. Mol. Sci. 2014, 15, 14077–14101. [Google Scholar] [CrossRef]
- Akıllıoğlu, H.G.; Karakaya, S. Effects of heat treatment and in vitro digestion on the Angiotensin converting enzyme inhibitory activity of some legume species. Eur. Food Res. Technol. 2009, 229, 915–921. [Google Scholar] [CrossRef]
- Chen, Z.; Bertin, R.; Froldi, G. EC50 estimation of antioxidant activity in DPPH* assay using several statistical programs. Food Chem. 2013, 138, 414–420. [Google Scholar] [CrossRef]
- Esteve, C.; Marina, M.L.; García, M.C. Novel strategy for the revalorization of olive (Olea europaea) residues based on the extraction of bioactive peptides. Food Chem. 2015, 167, 272–280. [Google Scholar] [CrossRef]
- Ngoh, Y.Y.; Gan, C.Y. Enzyme-assisted extraction and identification of antioxidative and α-amylase inhibitory peptides from Pinto beans (Phaseolus vulgaris cv. Pinto). Food Chem. 2016, 190, 331–337. [Google Scholar] [CrossRef]
- Park, S.Y.; Lee, J.S.; Baek, H.H.; Lee, H.G. Purification and characterization of antioxidant peptides from soy protein hydrolysate. J. Food Biochem. 2010, 34, 120–132. [Google Scholar] [CrossRef]
- Ashaolu, T.J. Antioxidative peptides derived from plants for human nutrition: Their production, mechanisms and applications. Eur. Food Res. Technol. 2020, 246, 853–865. [Google Scholar] [CrossRef]
- Darewicz, M.; Dziuba, J.; Dziuba, M. Functional properties and biological activities of bovine casein proteins and peptides. Polish J. Food Nutr. Sci. 2006, 15, 79–86. [Google Scholar]
- Garcia-Vaquero, M.; Mora, L.; Hayes, M. In vitro and in silico approaches to generating and identifying angiotensin-converting enzyme I inhibitory peptides from green macroalga ulva lactuca. Mar. Drugs 2019, 17, 204. [Google Scholar] [CrossRef]
- Mooney, C.; Haslam, N.J.; Pollastri, G.; Shields, D.C. Towards the Improved Discovery and Design of Functional Peptides: Common Features of Diverse Classes Permit Generalized Prediction of Bioactivity. PLoS ONE 2012, 7, e45012. [Google Scholar] [CrossRef]
- Mudgil, P.; Kamal, H.; Yuen, G.C.; Maqsood, S. Characterization and identification of novel antidiabetic and anti-obesity peptides from camel milk protein hydrolysates. Food Chem. 2018, 259, 46–54. [Google Scholar] [CrossRef]
- Lafarga, T.; Rai, D.K.; O’connor, P.; Hayes, M. Generation of bioactive hydrolysates and peptides from bovine hemoglobin with in vitro renin, angiotensin-I-converting enzyme and dipeptidyl peptidase-IV inhibitory activities. J. Food Biochem. 2016, 40, 673–685. [Google Scholar] [CrossRef]
- Acquah, C.; Chan, Y.W.; Pan, S.; Agyei, D.; Udenigwe, C.C. Structure-informed separation of bioactive peptides. J. Food Biochem. 2019, 43, e12765. [Google Scholar] [CrossRef]
- Hrynkiewicz, M.; Iwaniak, A.; Bucholska, J.; Minkiewicz, P.; Darewicz, M. Structure–activity prediction of ACE inhibitory/bitter dipeptides—A chemometric approach based on stepwise regression. Molecules 2019, 24, 950. [Google Scholar] [CrossRef]
- Hsu, K.-C.; Tung, Y.-S.; Huang, S.-L.; Jao, C.-L. Dipeptidyl Peptidase-IV Inhibitory Activity of Peptides in Porcine Skin Gelatin Hydrolysates. In Bioactive Food Peptides in Health and Disease; IntechOpen: London, UK, 2013; pp. 205–218. ISBN 978-953-51-0964-8. [Google Scholar]
- Hernández-Ledesma, B.; Del Mar Contreras, M.; Recio, I. Antihypertensive peptides: Production, bioavailability and incorporation into foods. In Proceedings of the Advances in Colloid and Interface Science; Elsevier: Amsterdam, The Netherlands, 2011; Volume 165, pp. 23–35. [Google Scholar]
- Borawska, J.; Darewicz, M.; Vegarud, G.E.; Minkiewicz, P. Antioxidant properties of carp (Cyprinus carpio L.) protein ex vivo and in vitro hydrolysates. Food Chem. 2016, 194, 770–779. [Google Scholar] [CrossRef] [PubMed]
- Roblet, C.; Amiot, J.; Lavigne, C.; Marette, A.; Lessard, M.; Jean, J.; Ramassamy, C.; Moresoli, C.; Bazinet, L. Screening of in vitro bioactivities of a soy protein hydrolysate separated by hollow fiber and spiral-wound ultrafiltration membranes. Food Res. Int. 2012, 46, 237–249. [Google Scholar] [CrossRef]
- Sarmadi, B.H.; Ismail, A. Antioxidative peptides from food proteins: A review. Peptides 2010, 31, 1949–1956. [Google Scholar] [CrossRef] [PubMed]
- Medzihradszky, K.F.; Chalkley, R.J. Lessons in de novo peptide sequencing by tandem mass spectrometry. Mass Spectrom. Rev. 2015, 34, 43–63. [Google Scholar] [CrossRef]
- Guijas, C.; Montenegro-Burke, J.R.; Domingo-Almenara, X.; Palermo, A.; Warth, B.; Hermann, G.; Koellensperger, G.; Huan, T.; Uritboonthai, W.; Aisporna, A.E.; et al. METLIN: A Technology Platform for Identifying Knowns and Unknowns. Anal. Chem. 2018, 90, 3156–3164. [Google Scholar] [CrossRef]
- Roepstorff, P.; Fohlman, J. Letter to the editors. Biol. Mass Spectrom. 1984, 11, 601. [Google Scholar] [CrossRef]
- Taga, Y.; Hayashida, O.; Kusubata, M.; Ogawa-Goto, K.; Hattori, S. Production of a novel wheat gluten hydrolysate containing dipeptidyl peptidase-IV inhibitory tripeptides using ginger protease. Biosci. Biotechnol. Biochem. 2017, 81, 1823–1828. [Google Scholar] [CrossRef]
- Minkiewicz, P.; Darewicz, M.; Iwaniak, A.; Sokołowska, J.; Starowicz, P.; Bucholska, J.; Hrynkiewicz, M. Common amino acid subsequences in a universal proteome-relevance for food science. Int. J. Mol. Sci. 2015, 16, 20748–20773. [Google Scholar] [CrossRef]
- Iwaniak, A.; Hrynkiewicz, M.; Minkiewicz, P.; Bucholska, J.; Darewicz, M. Soybean (Glycine max) protein hydrolysates as sources of peptide bitter-tasting indicators: An analysis based on hybrid and fragmentomic approaches. Appl. Sci. 2020, 10, 2514. [Google Scholar] [CrossRef]
- Bucholska, J.; Minkiewicz, P. The use of peptide markers of carp and herring allergens as an example of detection of sequenced and non-sequenced proteins. Food Technol. Biotechnol. 2016, 54, 266–274. [Google Scholar] [CrossRef]
- Giromini, C.; Cheli, F.; Rebucci, R.; Baldi, A. Dairy proteins and bioactive peptides: Modeling digestion and the intestinal barrier. J. Dairy Sci. 2019, 102, 929–942. [Google Scholar] [CrossRef]
- Tonolo, F.; Sandre, M.; Ferro, S.; Folda, A.; Scalcon, V.; Scutari, G.; Feller, E.; Marin, O.; Bindoli, A.; Rigobello, M.P. Milk-derived bioactive peptides protect against oxidative stress in a Caco-2 cell model. Food Funct. 2018, 9, 1245–1253. [Google Scholar] [CrossRef]
- Lammi, C.; Aiello, G.; Vistoli, G.; Zanoni, C.; Arnoldi, A.; Sambuy, Y.; Ferruzza, S.; Ranaldi, G. A multidisciplinary investigation on the bioavailability and activity of peptides from lupin protein. J. Funct. Foods 2016, 24, 297–306. [Google Scholar] [CrossRef]
- Ding, L.; Wang, L.; Zhang, T.; Yu, Z.; Liu, J. Hydrolysis and transepithelial transport of two corn gluten derived bioactive peptides in human Caco-2 cell monolayers. Food Res. Int. 2018, 106, 475–480. [Google Scholar] [CrossRef]
- Lin, Q.; Xu, Q.; Bai, J.; Wu, W.; Hong, H.; Wu, J. Transport of soybean protein-derived antihypertensive peptide LSW across Caco-2 monolayers. J. Funct. Foods 2017, 39, 96–102. [Google Scholar] [CrossRef]
- Rohm, F.; Daniel, H.; Spanier, B. Transport Versus Hydrolysis: Reassessing Intestinal Assimilation of Di- and Tripeptides by LC–MS/MS Analysis. Mol. Nutr. Food Res. 2019, 63, 1900263. [Google Scholar] [CrossRef]
- Udenigwe, C.C.; Aluko, R.E. Food protein-derived bioactive peptides: Production, processing, and potential health benefits. J. Food Sci. 2012, 77, R11–R24. [Google Scholar] [CrossRef]
- Lacroix, I.M.E.; Chen, X.M.; Kitts, D.D.; Li-Chan, E.C.Y. Investigation into the bioavailability of milk protein-derived peptides with dipeptidyl-peptidase IV inhibitory activity using Caco-2 cell monolayers. Food Funct. 2017, 8, 701–709. [Google Scholar] [CrossRef]
- Miner-Williams, W.M.; Stevens, B.R.; Moughan, P.J. Are intact peptides absorbed from the healthy gut in the adult human? Nutr. Res. Rev. 2014, 27, 308–329. [Google Scholar] [CrossRef]
- Horner, K.; Drummond, E.; Brennan, L. Bioavailability of milk protein-derived bioactive peptides: A glycaemic management perspective. Nutr. Res. Rev. 2016, 29, 91–101. [Google Scholar] [CrossRef] [PubMed]
- The UniProt Consortium UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; Mcgettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- Minkiewicz, P.; Iwaniak, A.; Darewicz, M. BIOPEP-UWM database of bioactive peptides: Current opportunities. Int. J. Mol. Sci. 2019, 20, 5978. [Google Scholar] [CrossRef] [PubMed]
- Kyte, J.; Doolittle, R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982, 157, 105–132. [Google Scholar] [CrossRef]
- Gupta, S.; Kapoor, P.; Chaudhary, K.; Gautam, A.; Kumar, R.; Raghava, G.P.S. In Silico Approach for Predicting Toxicity of Peptides and Proteins. PLoS ONE 2013, 8, e73957. [Google Scholar] [CrossRef]
- Dimitrov, I.; Bangov, I.; Flower, D.R.; Doytchinova, I. AllerTOP v.2—A server for in silico prediction of allergens. J. Mol. Model. 2014, 20, 2278. [Google Scholar] [CrossRef]
- Trabuco, L.G.; Lise, S.; Petsalaki, E.; Russell, R.B. PepSite: Prediction of peptide-binding sites from protein surfaces. Nucleic Acids Res. 2012, 40, W423–W427. [Google Scholar] [CrossRef]
- PDBe-KB Consortium PDBe-KB: Collaboratively defining the biological The Protein Data Bank in Europe—Knowledge Base. Nucleic Acids Res. 2022, 50, D534–D542. [CrossRef]
- Ma, C.Y. Preparation, Composition and Functional Properties of Oat Protein Isolates. Can. Inst. Food Sci. Technol. J. 1983, 16, 201–205. [Google Scholar] [CrossRef]
- Minekus, M.; Alminger, M.; Alvito, P.; Ballance, S.; Bohn, T.; Bourlieu, C.; Carrière, F.; Boutrou, R.; Corredig, M.; Dupont, D.; et al. A standardised static in vitro digestion method suitable for food—an international consensus. Food Funct. 2014, 5, 1113–1124. [Google Scholar] [CrossRef]
- Lowry, O.H.; Rosebrough, N.J.; Farr, A.L.; Randall, R.J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Nongonierma, A.B.; Paolella, S.; Mudgil, P.; Maqsood, S.; FitzGerald, R.J. Identification of novel dipeptidyl peptidase IV (DPP-IV) inhibitory peptides in camel milk protein hydrolysates. Food Chem. 2018, 244, 340–348. [Google Scholar] [CrossRef]
- Zhang, C.; Zhang, Y.; Wang, Z.; Chen, S.; Luo, Y. Production and identification of antioxidant and angiotensin-converting enzyme inhibition and dipeptidyl peptidase IV inhibitory peptides from bighead carp (Hypophthalmichthys nobilis) muscle hydrolysate. J. Funct. Foods 2017, 35, 224–235. [Google Scholar] [CrossRef]
- Ben Ahmed, Z.; Yousfi, M.; Viaene, J.; Dejaegher, B.; Demeyer, K.; Mangelings, D.; Vander Heyden, Y. Potentially antidiabetic and antihypertensive compounds identified from Pistacia atlantica leaf extracts by LC fingerprinting. J. Pharm. Biomed. Anal. 2018, 149, 547–556. [Google Scholar] [CrossRef]
- Jimsheena, V.K.; Gowda, L.R. Colorimetric, high-throughput assay for screening angiotensin I-converting enzyme inhibitors. Anal. Chem. 2009, 81, 9388–9394. [Google Scholar] [CrossRef]
- Nakamura, Y.; Yamamoto, N.; Sakai, K.; Okubo, A.; Yamazaki, S.; Takano, T. Purification and characterization of angiotensin I-converting enzyme inhibitors from sour milk. J. Dairy Sci. 1995, 78, 777–783. [Google Scholar] [CrossRef]
- Savitzky, A.; Golay, M.J.E. Smoothing and Differentiation of Data by Simplified Least Squares Procedures. Anal. Chem. 1964, 36, 1627–1639. [Google Scholar] [CrossRef]
- Jarmołowska, B.; Teodorowicz, M.; Fiedorowicz, E.; Sienkiewicz-Szłapka, E.; Matysiewicz, M.; Kostyra, E. Glucose and calcium ions may modulate the efficiency of bovine β-casomorphin-7 permeability through a monolayer of Caco-2 cells. Peptides 2013, 49, 59–67. [Google Scholar] [CrossRef]
- Wang, L.; Ding, L.; Du, Z.; Yu, Z.; Liu, J. Hydrolysis and Transport of Egg White-Derived Peptides in Caco-2 Cell Monolayers and Everted Rat Sacs. J. Agric. Food Chem. 2019, 67, 4839–4848. [Google Scholar] [CrossRef] [PubMed]

| Sample | DPP-IV Inhibitory Activity | α-Glucosidase Inhibitory Activity | ACE Inhibitory Activity | DPPH Scavenging |
|---|---|---|---|---|
| IC50 (mg/mL) | μM Trolox/ mg Sample | |||
| 0 | 10.82 ± 0.04 a | 25.20 ± 0.06 ab | 34.52 ± 0.11 ab | 7.93 ± 0.07 a |
| G | 4.23 ± 0.05 a | 10.35 ± 0.03 | 8.89 ± 0.07 | 9.91 ± 0.15 b |
| D | 0.51 ± 0.07 a | 1.55 ± 0.04 a | 0.82 ± 0.10 a | 33.93 ± 0.26 abc |
| Positive control | 0.20 ± 0.04 a (Diprotin A) | 0.21 ± 0.17 b (Acarbose) | 0.20 ± 0.08 b (Captopril) | 290.00 ± 0.18 abc (Gluthatione) |
| No. | Amino Acid Sequence | Precursor Protein | Activity | PeptideRanker Score | Pepsite2 p-Value | Identication | BIOPEP-UWM ID | Kyte-Doolittle Hydrophobicity | Water Solubility | Toxicity | Potential Allergencity |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | MF | Other proteins | DPP-IV | 1.00 | 1.876 × 10−3 | No | 3385 | 4.7 | poor | No | No |
| ACE | 7.218 × 10−4 | 8827 | |||||||||
| 2 | MW | Other proteins | DPP-IV | 1.00 | 1.876 × 10−3 | No | 8690 | 1 | poor | No | No |
| 3 | GF | Globulins, other proteins | DPP-IV | 0.99 | 6.047 × 10−4 | Yes | 8782 | 2.4 | poor | No | No |
| ACE | 4.579 × 10−4 | 7591 | |||||||||
| 4 | GW | Other proteins | DPP-IV | 0.99 | 1.934 × 10−3 | No | 8787 | −1.3 | poor | No | No |
| ACE | 7.708 × 10−4 | 7579 | |||||||||
| 4 | PF | Globulins, other proteins | DPP-IV | 0.99 | 1.512 × 10−5 | No | 9505 | 1.2 | poor | No | No |
| 5 | PW | Other proteins | DPP-IV | 0.99 | 4.843 × 10−5 | Yes | 8865 | −2.5 | poor | No | No |
| AO | - | 8190 | |||||||||
| 6 | HF | Globulins, other proteins | DPP-IV | 0.95 | 3.062 × 10−3 | Yes | 8791 | −0.4 | poor | No | No |
| 8 | HW | Other proteins | DPP-IV | 0.95 | 5.719 × 10−3 | Yes | 8798 | −4.1 | poor | No | No |
| 9 | IF | Other proteins | ACE | 0.95 | 2.086 × 10−3 | Yes | 7593 | 7.3 | poor | No | No |
| 10 | SF | Globulins, other proteins | DPP-IV | 0.95 | 1.348 × 10−2 | Yes | 8891 | 2 | poor | No | No |
| ACE | 1.621 × 10−3 | 7685 | |||||||||
| 11 | IW | Other proteins | DPP-IV | 0.94 | 1.155 × 10−2 | No | 8807 | 3.6 | poor | No | No |
| ACE | 2.086 × 10−3 | 7544 | |||||||||
| 12 | NF | Globulins, other proteins | DPP-IV | 0.94 | 9.678 × 10−4 | No | 8842 | −0.7 | poor | No | No |
| 13 | SW | Globulins, other proteins | DPP-IV | 0.93 | 2.446 × 10−2 | No | 8896 | −1.7 | poor | No | No |
| 14 | ML | Globulins, other proteins | DPP-IV | 0.89 | 4.143 × 10−3 | Yes | 8832 | 5.7 | poor | No | No |
| 15 | PGL | Globulins, other proteins | ACE | 0.86 | 7.404 × 10−5 | No | 7507 | 1.8 | poor | No | No |
| 16 | MR | Other proteins | DPP-IV | 0.85 | 4.676 × 10−3 | No | 8836 | −2.6 | poor | No | No |
| 17 | MY | Other proteins | DPP-IV | 0.84 | 1.126 × 10−2 | No | 8838 | 0.6 | poor | No | No |
| ACE | 5.894 × 10−3 | 3388 | |||||||||
| AO | - | 8090 | |||||||||
| 18 | TF | Globulins, other proteins | DPP-IV | 0.83 | 8.881 × 10−4 | Yes | 8900 | 2.1 | poor | No | No |
| ACE | 1.143 × 10−3 | 8185 | |||||||||
| 19 | VF | Globulins, other proteins | DPP-IV | 0.82 | 2.740 × 10−3 | Yes | 8917 | 7 | poor | No | No |
| ACE | 3.016 × 10−3 | 3384 | |||||||||
| 20 | GL | Globulins, other proteins | DPP-IV | 0.81 | 3.881 × 10−3 | No | 8561 | 3.4 | poor | No | No |
| ACE | 3.881 × 10−3 | 7599 | |||||||||
| 21 | PL | Globulins, other proteins | DPP-IV | 0.81 | 4.843 × 10−5 | Yes | 8638 | 2.2 | poor | No | No |
| ACE | 2.612 × 10−5 | 7513 | |||||||||
| 22 | TW | Other proteins | DPP-IV | 0.81 | 2.839 × 10−3 | Yes | 8913 | −1.6 | poor | No | No |
| AO | - | ||||||||||
| 23 | VW | Other proteins | DPP-IV | 0.82 | 5.018 × 10−3 | Yes | 8928 | 7 | poor | No | No |
| ACE | 2.034 × 10−3 | 3486 | |||||||||
| AO | - | 8461 | |||||||||
| 24 | PR | Globulins, other proteins | DPP-IV | 0.80 | 2.964 × 10−5 | Yes | 9489 | −6.1 | poor | No | No |
| ACE | 6.661 × 10−6 | 3537 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Darewicz, M.; Pliszka, M.; Borawska-Dziadkiewicz, J.; Minkiewicz, P.; Iwaniak, A. Multi-Bioactivity of Protein Digests and Peptides from Oat (Avena sativa L.) Kernels in the Prevention of the Cardiometabolic Syndrome. Molecules 2022, 27, 7907. https://doi.org/10.3390/molecules27227907
Darewicz M, Pliszka M, Borawska-Dziadkiewicz J, Minkiewicz P, Iwaniak A. Multi-Bioactivity of Protein Digests and Peptides from Oat (Avena sativa L.) Kernels in the Prevention of the Cardiometabolic Syndrome. Molecules. 2022; 27(22):7907. https://doi.org/10.3390/molecules27227907
Chicago/Turabian StyleDarewicz, Małgorzata, Monika Pliszka, Justyna Borawska-Dziadkiewicz, Piotr Minkiewicz, and Anna Iwaniak. 2022. "Multi-Bioactivity of Protein Digests and Peptides from Oat (Avena sativa L.) Kernels in the Prevention of the Cardiometabolic Syndrome" Molecules 27, no. 22: 7907. https://doi.org/10.3390/molecules27227907
APA StyleDarewicz, M., Pliszka, M., Borawska-Dziadkiewicz, J., Minkiewicz, P., & Iwaniak, A. (2022). Multi-Bioactivity of Protein Digests and Peptides from Oat (Avena sativa L.) Kernels in the Prevention of the Cardiometabolic Syndrome. Molecules, 27(22), 7907. https://doi.org/10.3390/molecules27227907










