Bile Acids: Physiological Activity and Perspectives of Using in Clinical and Laboratory Diagnostics
Abstract
1. Introduction
2. The Structure and Metabolism of Bile Acids
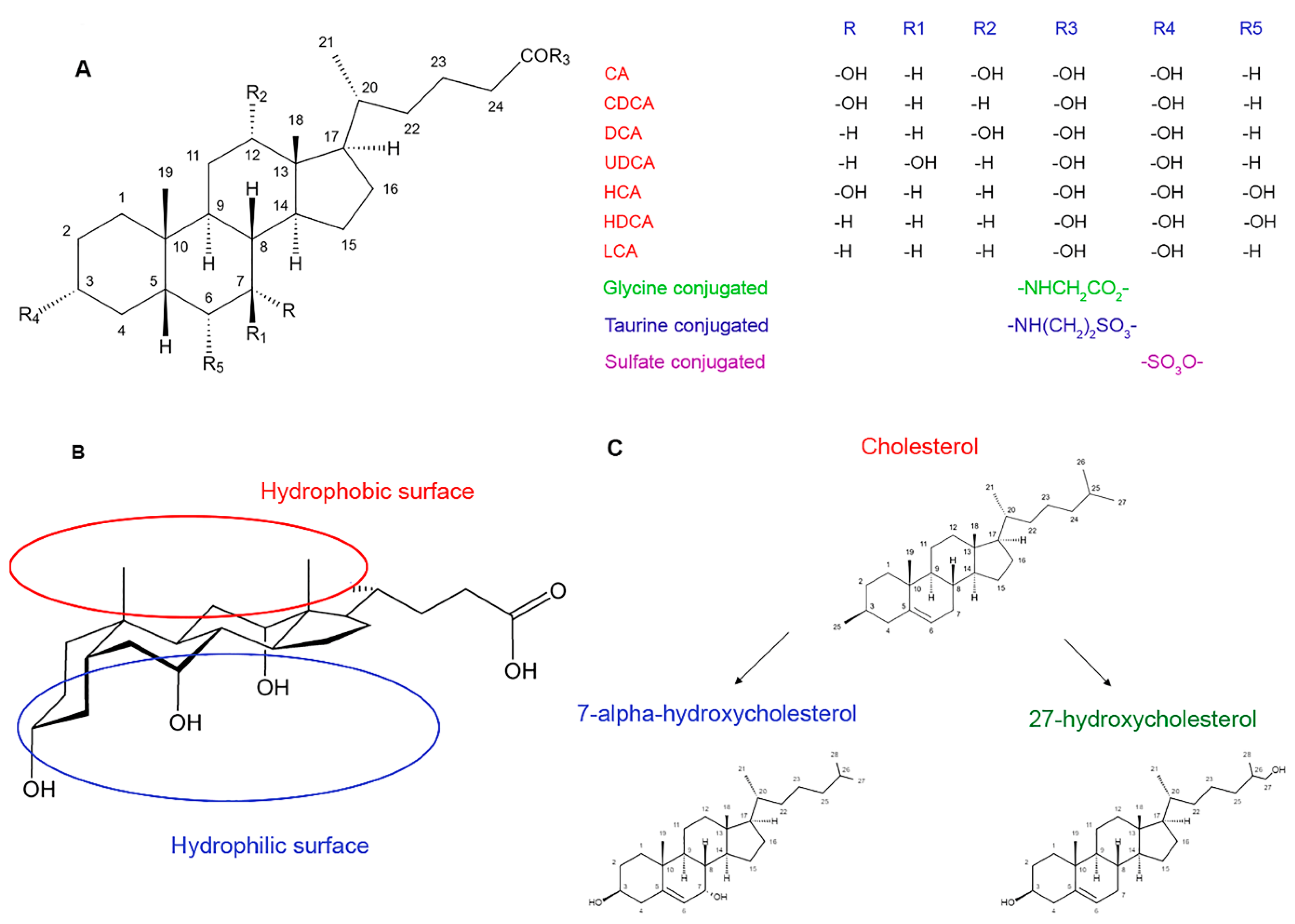
2.1. The Structure and Physicochemical Properties
2.2. Biosynthesis and Metabolism
2.3. Regulation of Biosynthesis
3. The Role of Bile Acids in Human Physiology
3.1. Digestion
3.2. Signaling Pathways
3.3. Bile Acids and Gut Microbiota
4. Bile Acids in Pathology
4.1. Dysbiosis
4.2. Crohn’s Disease
4.3. Overweight and Obesity
4.4. Cancer Progression
5. Approaches to the Analysis of Bile Acids
5.1. Possibilities of High-Performance Liquid Chromatography in Detection of Metabolites
5.2. Challenges
6. The Metabolomics of Bile Acids
7. The Perspectives and Limitations in Medicine
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Daviss, B. Growing Pains for Metabolomics. Scientist 2005, 19, 25–28. [Google Scholar]
- Miggiels, P.; Wouters, B.; van Westen, G.J.P.; Dubbelman, A.-C.; Hankemeier, T. Novel Technologies for Metabolomics: More for Less. TrAC Trends Anal. Chem. 2019, 120, 115323. [Google Scholar] [CrossRef]
- Witkiewicz, Z.; Neffe, S. Chromatographic Analysis of Chemical Warfare Agents and Their Metabolites in Biological Samples. TrAC Trends Anal. Chem. 2020, 130, 115960. [Google Scholar] [CrossRef]
- Lokhov, P.G.; Balashova, E.E.; Trifonova, O.P.; Maslov, D.L.; Archakov, A.I. A Decade of Russian Metabolomics: The History of Development and Achievements. Biochem. (Mosc.) Suppl. Ser. B Biomed. Chem. 2021, 15, 1–15. [Google Scholar] [CrossRef]
- Xie, A.J.; Mai, C.T.; Zhu, Y.Z.; Liu, X.C.; Xie, Y. Bile Acids as Regulatory Molecules and Potential Targets in Metabolic Diseases. Life Sci. 2021, 287. [Google Scholar] [CrossRef]
- Chiang, J.Y.L. Bile Acid Metabolism and Signaling. Compr. Physiol. 2013, 3, 1191–1212. [Google Scholar] [CrossRef]
- Hofmann, A.F.; Hagey, L.R. Bile Acids: Chemistry, Pathochemistry, Biology, Pathobiology, and Therapeutics. Cell. Mol. Life Sci. 2008, 65, 2461–2483. [Google Scholar] [CrossRef] [PubMed]
- Humbert, L.; Maubert, M.A.; Wolf, C.; Duboc, H.; Mahé, M.; Farabos, D.; Seksik, P.; Mallet, J.M.; Trugnan, G.; Masliah, J.; et al. Bile Acid Profiling in Human Biological Samples: Comparison of Extraction Procedures and Application to Normal and Cholestatic Patients. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2012, 899, 135–145. [Google Scholar] [CrossRef]
- Ridlon, J.M.; Kang, D.J.; Hylemon, P.B.; Bajaj, J.S. Bile Acids and the Gut Microbiome. Curr. Opin. Gastroenterol. 2014, 30, 332–338. [Google Scholar] [CrossRef]
- Shulpekova, Y.; Shirokova, E.; Zharkova, M.; Tkachenko, P.; Tikhonov, I.; Stepanov, A.; Sinitsyna, A.; Izotov, A.; Butkova, T.; Shulpekova, N.; et al. A Recent Ten-Year Perspective: Bile Acid Metabolism and Signaling. Molecules 2022, 27, 1983. [Google Scholar] [CrossRef]
- di Gregorio, M.C.; Cautela, J.; Galantini, L. Physiology and Physical Chemistry of Bile Acids. Int. J. Mol. Sci. 2021, 22, 1780. [Google Scholar] [CrossRef] [PubMed]
- Kuhajda, K.; Kandrac, J.; Kevresan, S.; Mikov, M.; Fawcett, J.P. Structure and Origin of Bile Acids: An Overview. Eur. J. Drug Metab. Pharmacokinet. 2006, 31, 135–143. [Google Scholar] [CrossRef]
- Chiang, J.Y.L.; Ferrell, J.M. Bile Acid Metabolism in Liver Pathobiology. Gene Expr. 2018, 18, 71–87. [Google Scholar] [CrossRef]
- Kim, I.; Ahn, S.H.; Inagaki, T.; Choi, M.; Ito, S.; Guo, G.L.; Kliewer, S.A.; Gonzalez, F.J. Differential Regulation of Bile Acid Homeostasis by the Farnesoid X Receptor in Liver and Intestine. J. Lipid Res. 2007, 48, 2664–2672. [Google Scholar] [CrossRef]
- Russell, D.W. Fifty Years of Advances in Bile Acid Synthesis and Metabolism. J. Lipid Res. 2009, 50, S120–S125. [Google Scholar] [CrossRef] [PubMed]
- Sticova, E.; Jirsa, M.; Pawłowska, J. New Insights in Genetic Cholestasis: From Molecular Mechanisms to Clinical Implications. Can. J. Gastroenterol. Hepatol. 2018, 2018. [Google Scholar] [CrossRef]
- Dawson, P.A.; Karpen, S.J. Intestinal Transport and Metabolism of Bile Acids. J. Lipid Res. 2015, 56, 1085–1099. [Google Scholar] [CrossRef]
- Bathena, S.P.R.; Mukherjee, S.; Olivera, M.; Alnouti, Y. The Profile of Bile Acids and Their Sulfate Metabolites in Human Urine and Serum. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2013, 942–943, 53–62. [Google Scholar] [CrossRef]
- Wang, S.; Sheng, F.; Zou, L.; Xiao, J.; Li, P. Hyperoside Attenuates Non-Alcoholic Fatty Liver Disease in Rats via Cholesterol Metabolism and Bile Acid Metabolism. J. Adv. Res. 2021, 34, 109–122. [Google Scholar] [CrossRef] [PubMed]
- Jani, M.; Beéry, E.; Heslop, T.; Tóth, B.; Jagota, B.; Kis, E.; Kevin Park, B.; Krajcsi, P.; Weaver, R.J. Kinetic Characterization of Bile Salt Transport by Human NTCP (SLC10A1). Toxicol. In Vitro 2018, 46, 189–193. [Google Scholar] [CrossRef]
- Zwicker, B.L.; Agellon, L.B. Transport and Biological Activities of Bile Acids. Int. J. Biochem. Cell Biol. 2013, 45, 1389–1398. [Google Scholar] [CrossRef] [PubMed]
- Ferrebee, C.B.; Li, J.; Haywood, J.; Pachura, K.; Robinson, B.S.; Hinrichs, B.H.; Jones, R.M.; Rao, A.; Dawson, P.A. Organic Solute Transporter α-β Protects Ileal Enterocytes From Bile Acid–Induced Injury. Cell Mol. Gastroenterol. Hepatol. 2018, 5, 499–522. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Chiang, J.Y.L. Bile Acids as Metabolic Regulators. Curr. Opin. Gastroenterol. 2015, 31, 159–165. [Google Scholar] [CrossRef]
- Staley, C.; Weingarden, A.R.; Khoruts, A.; Sadowsky, M.J. Interaction of Gut Microbiota with Bile Acid Metabolism and Its Influence on Disease States. Appl. Microbiol. Biotechnol. 2016, 101, 47–64. [Google Scholar] [CrossRef]
- Funabashi, M.; Grove, T.L.; Wang, M.; Varma, Y.; McFadden, M.E.; Brown, L.C.; Guo, C.; Higginbottom, S.; Almo, S.C.; Fischbach, M.A. A Metabolic Pathway for Bile Acid Dehydroxylation by the Gut Microbiome. Nature 2020, 582, 566–570. [Google Scholar] [CrossRef]
- de Aguiar Vallim, T.Q.; Tarling, E.J.; Edwards, P.A. Pleiotropic Roles of Bile Acids in Metabolism. Cell Metab. 2013, 17, 657–669. [Google Scholar] [CrossRef]
- Meier-Abt, F.; Mokrab, Y.; Mizuguchi, K. Organic Anion Transporting Polypeptides of the OATP/SLCO Superfamily: Identification of New Members in Nonmammalian Species, Comparative Modeling and a Potential Transport Mode. J. Membr. Biol. 2006, 208, 213–227. [Google Scholar] [CrossRef] [PubMed]
- Wilson, A.; Almousa, A.; Teft, W.A.; Kim, R.B. Attenuation of Bile Acid-Mediated FXR and PXR Activation in Patients with Crohn’s Disease. Sci. Rep. 2020, 10, 1–11. [Google Scholar] [CrossRef]
- Goodwin, B.; Jones, S.A.; Price, R.R.; Watson, M.A.; McKee, D.D.; Moore, L.B.; Galardi, C.; Wilson, J.G.; Lewis, M.C.; Roth, M.E.; et al. A Regulatory Cascade of the Nuclear Receptors FXR, SHP-1, and LRH-1 Represses Bile Acid Biosynthesis. Mol. Cell 2000, 6, 517–526. [Google Scholar] [CrossRef]
- Macierzanka, A.; Torcello-Gómez, A.; Jungnickel, C.; Maldonado-Valderrama, J. Bile Salts in Digestion and Transport of Lipids. Adv. Colloid. Interface Sci. 2019, 274, 102045. [Google Scholar] [CrossRef]
- Naso, J.N.; Bellesi, F.A.; Pizones Ruiz-Henestrosa, V.M.; Pilosof, A.M.R. Studies on the Interactions between Bile Salts and Food Emulsifiers under in Vitro Duodenal Digestion Conditions to Evaluate Their Bile Salt Binding Potential. Colloids Surf B Biointerfaces 2019, 174, 493–500. [Google Scholar] [CrossRef] [PubMed]
- Naumann, S.; Schweiggert-Weisz, U.; Eglmeier, J.; Haller, D.; Eisner, P. In Vitro Interactions of Dietary Fibre Enriched Food Ingredients with Primary and Secondary Bile Acids. Nutrients 2019, 11, 1424. [Google Scholar] [CrossRef]
- Shokry, D.S.; Waters, L.J.; Parkes, G.M.B.; Mitchell, J.C.; Snowden, M.J. Formation of a Bile Salt-Drug Hydrogel to Predict Human Intestinal Absorption. J. Pharm. Sci. 2019, 108, 279–287. [Google Scholar] [CrossRef]
- Guo, C.; Xie, C.; Ding, P.; Qin, G.; Mo, W.; Cao, X.; Zheng, S. Quantification of Glycocholic Acid in Human Serum by Stable Isotope Dilution Ultra Performance Liquid Chromatography Electrospray Ionization Tandem Mass Spectrometry. J. Chromatogr. B 2018, 1072, 315–319. [Google Scholar] [CrossRef]
- Dikkers, A.; Tietge, U.J.F. Biliary Cholesterol Secretion: More than a Simple ABC. World J. Gastroenterol. WJG 2010, 16, 5936. [Google Scholar] [CrossRef] [PubMed]
- Roscam Abbing, R.L.P.; Slijepcevic, D.; Donkers, J.M.; Havinga, R.; Duijst, S.; Paulusma, C.C.; Kuiper, J.; Kuipers, F.; Groen, A.K.; Oude Elferink, R.P.J.; et al. Blocking Sodium-Taurocholate Cotransporting Polypeptide Stimulates Biliary Cholesterol and Phospholipid Secretion in Mice. Hepatology 2020, 71, 247–258. [Google Scholar] [CrossRef] [PubMed]
- Chiang, J.Y.L.; Li, T. Regulation of Bile Acid and Cholesterol Metabolism by PPARs. PPAR Res. 2009. [CrossRef]
- Sannasiddappa, T.H.; Lund, P.A.; Clarke, S.R. In Vitro Antibacterial Activity of Unconjugated and Conjugated Bile Salts on Staphylococcus Aureus. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef]
- Chang, K.-O.; George, D.W. Bile Acids Promote the Expression of Hepatitis C Virus in Replicon-Harboring Cells. J. Virol. 2007, 81, 9633. [Google Scholar] [CrossRef]
- Graf, D.; Haselow, K.; Münks, I.; Bode, J.G.; Häussinger, D. Inhibition of Interferon-α-Induced Signaling by Hyperosmolarity and Hydrophobic Bile Acids. Biol. Chem. 2010, 391, 1175–1187. [Google Scholar] [CrossRef]
- Steiner, C.; von Eckardstein, A.; Rentsch, K.M. Quantification of the 15 Major Human Bile Acids and Their Precursor 7α-Hydroxy-4-Cholesten-3-One in Serum by Liquid Chromatography–Tandem Mass Spectrometry. J. Chromatogr. B 2010, 878, 2870–2880. [Google Scholar] [CrossRef]
- Amaral, J.D.; Viana, R.J.S.; Ramalho, R.M.; Steer, C.J.; Rodrigues, C.M.P. Bile Acids: Regulation of Apoptosis by Ursodeoxycholic Acid. J. Lipid Res. 2009, 50, 1721–1734. [Google Scholar] [CrossRef] [PubMed]
- Fiorucci, S.; Biagioli, M.; Zampella, A.; Distrutti, E. Bile Acids Activated Receptors Regulate Innate Immunity. Front. Immunol. 2018, 9, 1853. [Google Scholar] [CrossRef] [PubMed]
- Lajczak-McGinley, N.K.; Porru, E.; Fallon, C.M.; Smyth, J.; Curley, C.; McCarron, P.A.; Tambuwala, M.M.; Roda, A.; Keely, S.J. The Secondary Bile Acids, Ursodeoxycholic Acid and Lithocholic Acid, Protect against Intestinal Inflammation by Inhibition of Epithelial Apoptosis. Physiol. Rep. 2020, 8. [Google Scholar] [CrossRef] [PubMed]
- BAPS—Overview: Bile Acid Profile, Serum. Available online: https://www.mayocliniclabs.com/test-catalog/overview/62538#Clinical-and-Interpretive (accessed on 17 June 2022).
- DeKeyser, J.G.; Omiecinski, C.J. Constitutive Androstane Receptor. In Comprehensive Toxicology, 2nd ed.; Elsevier: Amsterdam, Poland, 2010; Volume 2–14, pp. 169–181. [Google Scholar] [CrossRef]
- Garcia, M.; Thirouard, L.; Sedès, L.; Monrose, M.; Holota, H.; Caira, F.; Volle, D.H.; Beaudoin, C. Nuclear Receptor Metabolism of Bile Acids and Xenobiotics: A Coordinated Detoxification System with Impact on Health and Diseases. Int. J. Mol. Sci. 2018, 19, 3630. [Google Scholar] [CrossRef]
- Oliviero, F.; Lukowicz, C.; Boussadia, B.; Forner-Piquer, I.; Pascussi, J.M.; Marchi, N.; Mselli-Lakhal, L. Constitutive Androstane Receptor: A Peripheral and a Neurovascular Stress or Environmental Sensor. Cells 2020, 9, 2426. [Google Scholar] [CrossRef]
- Chen, Y.X.; Xie, G.C.; Pan, D.; Du, Y.R.; Pang, L.L.; Song, J.D.; Duan, Z.J.; Hu, B.R. Three-Dimensional Culture of Human Airway Epithelium in Matrigel for Evaluation of Human Rhinovirus C and Bocavirus Infections. Biomed. Environ. Sci. 2018, 31, 136–145. [Google Scholar] [CrossRef]
- Klindt, C.; Reich, M.; Hellwig, B.; Stindt, J.; Rahnenführer, J.; Hengstler, J.G.; Köhrer, K.; Schoonjans, K.; Häussinger, D.; Keitel, V. The G Protein-Coupled Bile Acid Receptor TGR5 (Gpbar1) Modulates Endothelin-1 Signaling in Liver. Cells 2019, 8, 1467. [Google Scholar] [CrossRef]
- Chen, J.L.; Wang, L.; Li, R.; Jiao, Y.F.; Yu, W.F. High Expression of Endothelial Progenitor Cell-Induced Angiogenic Markers Is Associated with Bile Acid Levels in HCC. Oncol. Lett. 2020, 20, 2729–2738. [Google Scholar] [CrossRef]
- Hanafi, N.I.; Mohamed, A.S.; Kadir, S.H.S.A.; Othman, M.H.D. Overview of Bile Acids Signaling and Perspective on the Signal of Ursodeoxycholic Acid, the Most Hydrophilic Bile Acid, in the Heart. Biomolecules 2018, 8, 159. [Google Scholar] [CrossRef]
- Ticho, A.L.; Malhotra, P.; Dudeja, P.K.; Gill, R.K.; Alrefai, W.A. Bile Acid Receptors and Gastrointestinal Functions. Liver Res. 2019, 3, 31–39. [Google Scholar] [CrossRef]
- Fiorucci, S.; Zampella, A.; Cirino, G.; Bucci, M.; Distrutti, E. Decoding the Vasoregulatory Activities of Bile Acid-Activated Receptors in Systemic and Portal Circulation: Role of Gaseous Mediators. Am. J. Physiol. Heart Circ. Physiol. 2017, 312, H21–H32. [Google Scholar] [CrossRef] [PubMed]
- Lyu, J.; Li, H.; Yin, D.; Zhao, M.; Sun, Q.; Guo, M. Analysis of Eight Bile Acids in Urine of Gastric Cancer Patients Based on Covalent Organic Framework Enrichment Coupled with Liquid Chromatography-Tandem Mass Spectrometry. J. Chromatogr. A 2021, 1653. [Google Scholar] [CrossRef]
- Chiang, J.Y.L. Bile Acid Regulation of Gene Expression: Roles of Nuclear Hormone Receptors. Endocr. Rev. 2002, 23, 443–463. [Google Scholar] [CrossRef] [PubMed]
- Schulman, I.G. Liver X Receptors Link Lipid Metabolism and Inflammation. FEBS Lett. 2017, 591, 2978–2991. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.Q.H.; Tazuma, S.; Cohen, D.E.; Carey, M.C. Feeding Natural Hydrophilic Bile Acids Inhibits Intestinal Cholesterol Absorption: Studies in the Gallstone-Susceptible Mouse. Am. J. Physiol. Gastrointest. Liver Physiol. 2003, 285. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, I.; Lu, R.; Zhang, Y.; Zhang, J.; Dai, Y.; Xia, Y.; Sun, J. Vitamin D Receptor Promotes Healthy Microbial Metabolites and Microbiome. Sci. Rep. 2020, 10, 1–18. [Google Scholar] [CrossRef]
- Makishima, M.; Lu, T.T.; Xie, W.; Whitfield, G.K.; Domoto, H.; Evans, R.M.; Haussler, M.R.; Mangelsdorf, D.J. Vitamin D Receptor as an Intestinal Bile Acid Sensor. Science (1979) 2002, 296, 1313–1316. [Google Scholar] [CrossRef]
- Ocvirk, S.; O’Keefe, S.J.D. Dietary Fat, Bile Acid Metabolism and Colorectal Cancer. Semin. Cancer Biol. 2021, 73, 347–355. [Google Scholar] [CrossRef]
- Biagioli, M.; Fiorucci, S. Bile Acid Activated Receptors: Integrating Immune and Metabolic Regulation in Non-Alcoholic Fatty Liver Disease. Liver Res. 2021, 5, 119–141. [Google Scholar] [CrossRef]
- Dutta, M.; Lim, J.J.; Cui, J.Y. Pregnane X Receptor and the Gut-Liver Axis: A Recent Update. Drug Metab. Dispos. 2022, 50, 478–491. [Google Scholar] [CrossRef]
- Fan, S.; Liu, C.; Jiang, Y.; Gao, Y.; Chen, Y.; Fu, K.; Yao, X.; Huang, M.; Bi, H. Lignans from Schisandra Sphenanthera Protect against Lithocholic Acid-Induced Cholestasis by Pregnane X Receptor Activation in Mice. J. Ethnopharmacol. 2019, 245, 112103. [Google Scholar] [CrossRef]
- Hassani-Nezhad-Gashti, F.; Kummu, O.; Karpale, M.; Rysä, J.; Hakkola, J. Nutritional Status Modifies Pregnane X Receptor Regulated Transcriptome. Sci. Rep. 2019, 9, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Reich, N.W.; Bell, N.; Finn, P.D.; Rodriguez, D.; Kohler, J.; Kozuka, K.; He, L.; Spencer, A.G.; Charmot, D.; et al. Design of Gut-Restricted Thiazolidine Agonists of G Protein-Coupled Bile Acid Receptor 1 (GPBAR1, TGR5). J. Med. Chem. 2018, 61, 7589–7613. [Google Scholar] [CrossRef] [PubMed]
- di Leva, F.S.; Festa, C.; Carino, A.; de Marino, S.; Marchianò, S.; di Marino, D.; Finamore, C.; Monti, M.C.; Zampella, A.; Fiorucci, S.; et al. Discovery of ((1,2,4-Oxadiazol-5-Yl)Pyrrolidin-3-Yl)Ureidyl Derivatives as Selective Non-Steroidal Agonists of the G-Protein Coupled Bile Acid Receptor-1. Sci. Rep. 2019, 9, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Pathak, P.; Xie, C.; Nichols, R.G.; Ferrell, J.M.; Boehme, S.; Krausz, K.W.; Patterson, A.D.; Gonzalez, F.J.; Chiang, J.Y.L. Intestine Farnesoid X Receptor Agonist and the Gut Microbiota Activate G-Protein Bile Acid Receptor-1 Signaling to Improve Metabolism. Hepatology 2018, 68, 1574–1588. [Google Scholar] [CrossRef]
- Hou, L.; Yang, L.; Chang, N.; Zhao, X.; Zhou, X.; Dong, C.; Liu, F.; Yang, L.; Li, L. Macrophage Sphingosine 1-Phosphate Receptor 2 Blockade Attenuates Liver Inflammation and Fibrogenesis Triggered by NLRP3 Inflammasome. Front. Immunol. 2020, 11, 1149. [Google Scholar] [CrossRef]
- Liu, R.; Li, X.; Hylemon, P.B.; Zhou, H. Conjugated Bile Acids Promote Invasive Growth of Esophageal Adenocarcinoma Cells and Cancer Stem Cell Expansion via Sphingosine 1-Phosphate Receptor 2–Mediated Yes-Associated Protein Activation. Am. J. Pathol. 2018, 188, 2042–2058. [Google Scholar] [CrossRef]
- Yang, T.; Wang, X.; Yuan, Z.; Miao, Y.; Wu, Z.; Chai, Y.; Yu, Q.; Wang, H.; Sun, L.; Huang, X.; et al. Sphingosine 1-Phosphate Receptor-1 Specific Agonist SEW2871 Ameliorates ANIT-Induced Dysregulation of Bile Acid Homeostasis in Mice Plasma and Liver. Toxicol. Lett. 2020, 331, 242–253. [Google Scholar] [CrossRef]
- Zhao, S.; Gong, Z.; Du, X.; Tian, C.; Wang, L.; Zhou, J.; Xu, C.; Chen, Y.; Cai, W.; Wu, J. Deoxycholic Acid-Mediated Sphingosine-1-Phosphate Receptor 2 Signaling Exacerbates DSS-Induced Colitis through Promoting Cathepsin b Release. J. Immunol. Res. 2018, 2018. [Google Scholar] [CrossRef]
- Ibrahim, E.; Diakonov, I.; Arunthavarajah, D.; Swift, T.; Goodwin, M.; McIlvride, S.; Nikolova, V.; Williamson, C.; Gorelik, J. Bile Acids and Their Respective Conjugates Elicit Different Responses in Neonatal Cardiomyocytes: Role of Gi Protein, Muscarinic Receptors and TGR5. Sci. Rep. 2018, 8, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Kiriyama, Y.; Nochi, H. The Biosynthesis, Signaling, and Neurological Functions of Bile Acids. Biomolecules 2019, 9, 232. [Google Scholar] [CrossRef] [PubMed]
- Kovacevic, B.; Jones, M.; Ionescu, C.; Walker, D.; Wagle, S.; Chester, J.; Foster, T.; Brown, D.; Mikov, M.; Mooranian, A.; et al. The Emerging Role of Bile Acids as Critical Components in Nanotechnology and Bioengineering: Pharmacology, Formulation Optimizers and Hydrogel-Biomaterial Applications. Biomaterials 2022, 283, 121459. [Google Scholar] [CrossRef] [PubMed]
- Kundu, S.; Bansal, S.; Muthukumarasamy, K.M.; Sachidanandan, C.; Motiani, R.K.; Bajaj, A. Deciphering the Role of Hydrophobic and Hydrophilic Bile Acids in Angiogenesis Using in Vitro and in Vivo Model Systems. Medchemcomm 2017, 8, 2248–2257. [Google Scholar] [CrossRef] [PubMed]
- White, J.H. Vitamin D Deficiency and the Pathogenesis of Crohn’s Disease. J. Steroid. Biochem. Mol. Biol. 2018, 175, 23–28. [Google Scholar] [CrossRef]
- Zelcer, N.; Reid, G.; Wielinga, P.; Kuil, A.; van der Heijden, I.; Schuetz, J.D.; Borst, P. Steroid and Bile Acid Conjugates Are Substrates of Human Multidrug-Resistance Protein (MRP) 4 (ATP-Binding Cassette C4). Biochemical. J. 2003, 371, 361–367. [Google Scholar] [CrossRef]
- Rius, M.; Hummel-Eisenbeiss, J.; Hofmann, A.F.; Keppler, D. Substrate Specificity of Human ABCC4 (MRP4)-Mediated Cotransport of Bile Acids and Reduced Glutathione. Am. J. Physiol. Gastrointest. Liver Physiol. 2006, 290, 640–647. [Google Scholar] [CrossRef]
- Monteiro-Cardoso, V.F.; Corlianò, M.; Singaraja, R.R. Bile Acids: A Communication Channel in the Gut-Brain Axis. NeuroMolecular Med. 2020, 23, 99–117. [Google Scholar] [CrossRef]
- Wang, W.; Kim, M.T.; Sedykh, A.; Zhu, H. Developing Enhanced Blood–Brain Barrier Permeability Models: Integrating External Bio-Assay Data in QSAR Modeling. Pharm. Res. 2015, 32, 3055–3065. [Google Scholar] [CrossRef]
- Tian, Y.; Gui, W.; Koo, I.; Smith, P.B.; Allman, E.L.; Nichols, R.G.; Rimal, B.; Cai, J.; Liu, Q.; Patterson, A.D. The Microbiome Modulating Activity of Bile Acids. Gut Microbes 2020, 11, 979–996. [Google Scholar] [CrossRef]
- Garrido, A.; Kim, E.; Teijeiro, A.; Sánchez Sánchez, P.; Gallo, R.; Nair, A.; Matamala Montoya, M.; Perna, C.; Vicent, G.P.; Muñoz, J.; et al. Histone Acetylation of Bile Acid Transporter Genes Plays a Critical Role in Cirrhosis. J. Hepatol. 2022, 76, 850–861. [Google Scholar] [CrossRef] [PubMed]
- Kakiyama, G.; Pandak, W.M.; Gillevet, P.M.; Hylemon, P.B.; Heuman, D.M.; Daita, K.; Takei, H.; Muto, A.; Nittono, H.; Ridlon, J.M.; et al. Modulation of the Fecal Bile Acid Profile by Gut Microbiota in Cirrhosis. J. Hepatol. 2013, 58, 949–955. [Google Scholar] [CrossRef]
- Hofmann, A.F.; Eckmann, L. How Bile Acids Confer Gut Mucosal Protection against Bacteria. Proc. Natl. Acad. Sci. USA 2006, 103, 4333–4334. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.D.; Myers, C.J.; Harris, S.C.; Kakiyama, G.; Lee, I.K.; Yun, B.S.; Matsuzaki, K.; Furukawa, M.; Min, H.K.; Bajaj, J.S.; et al. Bile Acid 7α-Dehydroxylating Gut Bacteria Secrete Antibiotics That Inhibit Clostridium Difficile: Role of Secondary Bile Acids. Cell Chem. Biol. 2019, 26, 27–34.e4. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Han, M.; Heinrich, B.; Fu, Q.; Zhang, Q.; Sandhu, M.; Agdashian, D.; Terabe, M.; Berzofsky, J.A.; Fako, V.; et al. Gut Microbiome–Mediated Bile Acid Metabolism Regulates Liver Cancer via NKT Cells. Science (1979) 2018, 360. [Google Scholar] [CrossRef] [PubMed]
- di Ciaula, A.; Wang, D.Q.H.; Molina, E.M.; Baccetto, R.L.; Calamita, G.; Palmieri, V.O.; Portincasa, P. Bile Acids and Cancer: Direct and Environmental-Dependent Effects. Ann. Hepatol. 2017, 16, S87–S105. [Google Scholar] [CrossRef]
- Zhao, B.; Liu, L.; Mao, J.; Liu, K.; Fan, W.; Liu, J.; Zhang, Z.; Li, Q. Sinomenine Hydrochloride Attenuates the Proliferation, Migration, Invasiveness, Angiogenesis and Epithelial-Mesenchymal Transition of Clear-Cell Renal Cell Carcinoma Cells via Targeting Smad in Vitro. Biomed. Pharmacother. 2017, 96, 1036–1044. [Google Scholar] [CrossRef]
- Blanchet, M.; Brunel, J.M. Bile Acid Derivatives: From Old Molecules to a New Potent Therapeutic Use: An Overview. Curr. Med. Chem. 2018, 25, 3613–3636. [Google Scholar] [CrossRef]
- Brossard, D.; el Kihel, L.; Clément, M.; Sebbahi, W.; Khalid, M.; Roussakis, C.; Rault, S. Synthesis of Bile Acid Derivatives and in Vitro Cytotoxic Activity with Pro-Apoptotic Process on Multiple Myeloma (KMS-11), Glioblastoma Multiforme (GBM), and Colonic Carcinoma (HCT-116) Human Cell Lines. Eur. J. Med. Chem. 2010, 45, 2912–2918. [Google Scholar] [CrossRef]
- Park, K.; Kim, Y.S.; Lee, G.Y.; Nam, J.O.; Lee, S.K.; Park, R.W.; Kim, S.Y.; Kim, I.S.; Byun, Y. Antiangiogenic Effect of Bile Acid Acylated Heparin Derivative. Pharm. Res. 2007, 24, 176–185. [Google Scholar] [CrossRef]
- Park, K.; Ki Lee, S.; Hyun Son, D.; Ah Park, S.; Kim, K.; Won Chang, H.; Jeong, E.J.; Park, R.W.; Kim, I.S.; Chan Kwon, I.; et al. The Attenuation of Experimental Lung Metastasis by a Bile Acid Acylated-Heparin Derivative. Biomaterials 2007, 28, 2667–2676. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.; Singh, A.; Kundu, S.; Bansal, S.; Bajaj, A. Deciphering the Role of Charge, Hydration, and Hydrophobicity for Cytotoxic Activities and Membrane Interactions of Bile Acid Based Facial Amphiphiles. Biochim. Biophys. Acta (BBA)—Biomembr. 2013, 1828, 1926–1937. [Google Scholar] [CrossRef] [PubMed]
- Ďurč, P.; Dosedělová, V.; Foret, F.; Dolina, J.; Konečný, Š.; Himmelsbach, M.; Buchberger, W.; Kubáň, P. Analysis of Major Bile Acids in Saliva Samples of Patients with Barrett’s Esophagus Using High-Performance Liquid Chromatography-Electrospray Ionization-Mass Spectrometry. J. Chromatogr. A 2020, 1625. [Google Scholar] [CrossRef] [PubMed]
- Quante, M.; Bhagat, G.; Abrams, J.A.; Marache, F.; Good, P.; Lee, M.D.; Lee, Y.; Friedman, R.; Asfaha, S.; Dubeykovskaya, Z.; et al. Bile Acid and Inflammation Activate Gastric Cardia Stem Cells in a Mouse Model of Barrett-Like Metaplasia. Cancer Cell 2012, 21, 36–51. [Google Scholar] [CrossRef] [PubMed]
- Alnouti, Y. Bile Acid Sulfation: A Pathway of Bile Acid Elimination and Detoxification. Toxicological. Sci. 2009, 108, 225–246. [Google Scholar] [CrossRef]
- Yang, T.; Shu, T.; Liu, G.; Mei, H.; Zhu, X.; Huang, X.; Zhang, L.; Jiang, Z. Quantitative Profiling of 19 Bile Acids in Rat Plasma, Liver, Bile and Different Intestinal Section Contents to Investigate Bile Acid Homeostasis and the Application of Temporal Variation of Endogenous Bile Acids. J. Steroid Biochem. Mol. Biol. 2017, 172, 69–78. [Google Scholar] [CrossRef]
- Huang, R.; Chen, Z.; He, L.; He, N.; Xi, Z.; Li, Z.; Deng, Y.; Zeng, X. Mass Spectrometry-Assisted Gel-Based Proteomics in Cancer Biomarker Discovery: Approaches and Application. Theranostics 2017, 7, 3559–3572. [Google Scholar] [CrossRef]
- Heřmánková, E.; Žák, A.; Poláková, L.; Hobzová, R.; Hromádka, R.; Širc, J. Polymeric Bile Acid Sequestrants: Review of Design, in Vitro Binding Activities, and Hypocholesterolemic Effects. Eur. J. Med. Chem. 2018, 144, 300–317. [Google Scholar] [CrossRef]
- Patel, V.; Ray, D.; Bahadur, A.; Ma, J.; Aswal, V.K.; Bahadur, P. Pluronic®-Bile Salt Mixed Micelles. Colloids Surf B Biointerfaces 2018, 166, 119–126. [Google Scholar] [CrossRef]
- Khan, T.J.; Hasan, M.N.; Azhar, E.I.; Yasir, M. Association of Gut Dysbiosis with Intestinal Metabolites in Response to Antibiotic Treatment. Hum. Microb. J. 2019, 11, 100054. [Google Scholar] [CrossRef]
- Liu, L.; Yang, M.; Dong, W.; Liu, T.; Song, X.; Gu, Y.; Wang, S.; Liu, Y.; Abla, Z.; Qiao, X.; et al. Undefined Gut Dysbiosis and Abnormal Bile Acid Metabolism in Colitis-Associated Cancer. Gastroenterol. Res. Pract. 2021, 2021. [Google Scholar] [CrossRef]
- Li, R.; Andreu-Sánchez, S.; Kuipers, F.; Fu, J. Gut Microbiome and Bile Acids in Obesity-Related Diseases. Best Pract. Res. Clin. Endocrinol. Metab. 2021, 35, 101493. [Google Scholar] [CrossRef]
- Xu, M.; Cen, M.; Shen, Y.; Zhu, Y.; Cheng, F.; Tang, L.; Hu, W.; Dai, N. Deoxycholic Acid-Induced Gut Dysbiosis Disrupts Bile Acid Enterohepatic Circulation and Promotes Intestinal Inflammation. Dig. Dis. Sci. 2021, 66, 568–576. [Google Scholar] [CrossRef] [PubMed]
- Franco, P.; Porru, E.; Fiori, J.; Gioiello, A.; Cerra, B.; Roda, G.; Caliceti, C.; Simoni, P.; Roda, A. Identification and Quantification of Oxo-Bile Acids in Human Faeces with Liquid Chromatography–Mass Spectrometry: A Potent Tool for Human Gut Acidic Sterolbiome Studies. J. Chromatogr. A 2019, 1585, 70–81. [Google Scholar] [CrossRef]
- Fiorucci, S.; Carino, A.; Baldoni, M.; Santucci, L.; Costanzi, E.; Graziosi, L.; Distrutti, E.; Biagioli, M. Bile Acid Signaling in Inflammatory Bowel Diseases. Dig. Dis. Sci. 2020, 66, 674–693. [Google Scholar] [CrossRef]
- Tatsugami, M.; Ito, M.; Tanaka, S.; Yoshihara, M.; Matsui, H.; Haruma, K.; Chayama, K. Bile Acid Promotes Intestinal Metaplasia and Gastric Carcinogenesis. Cancer Epidemiol. Biomark. Prev. 2012, 21, 2101–2107. [Google Scholar] [CrossRef]
- Ciocan, D.; Sebastian Voican, C.; Wrzosek, L.; Hugot, C.; Rainteau, D.; Humbert, L.; Cassard, A.-M.M.; Voican, C.S.; Wrzosek, L.; Hugot, C.; et al. Bile Acid Homeostasis and Intestinal Dysbiosis in Alcoholic Hepatitis. Aliment. Pharmacol. Ther. 2018, 48, 961–974. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Killinger, B.A.; Ensink, E.; Beddows, I.; Yilmaz, A.; Lubben, N.; Lamp, J.; Schilthuis, M.; Vega, I.E.; Woltjer, R.; et al. Gut Microbiota Dysbiosis Is Associated with Elevated Bile Acids in Parkinson’s Disease. Metabolites 2021, 11, 29. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Liu, S.; Wang, M.; Hu, H.; Yin, J.; Liu, C.; Huang, Y. Undefined Gut Microbiota Dysbiosis Associated with Bile Acid Metabolism in Neonatal Cholestasis Disease. Sci. Rep. 2020, 10, 7686. [Google Scholar] [CrossRef]
- Ushiroda, C.; Naito, Y.; Takagi, T.; Uchiyama, K.; Mizushima, K.; Higashimura, Y.; Yasukawa, Z.; Okubo, T.; Inoue, R.; Honda, A.; et al. Green Tea Polyphenol (Epigallocatechin-3-Gallate) Improves Gut Dysbiosis and Serum Bile Acids Dysregulation in High-Fat Diet-Fed Mice. J. Clin. Biochem. Nutr. 2019, 65, 34–46. [Google Scholar] [CrossRef]
- Jin, D.; Wu, S.; Zhang, Y.G.; Lu, R.; Xia, Y.; Dong, H.; Sun, J. Lack of Vitamin D Receptor Causes Dysbiosis and Changes the Functions of the Murine Intestinal Microbiome. Clin. Ther. 2015, 37, 996–1009.e7. [Google Scholar] [CrossRef] [PubMed]
- Shang, M.; Sun, J. Vitamin D/VDR, Probiotics, and Gastrointestinal Diseases. Curr. Med. Chem. 2016, 24, 876–887. [Google Scholar] [CrossRef]
- Parséus, A.; Sommer, N.; Sommer, F.; Caesar, R.; Molinaro, A.; Stahlman, M.; Greiner, T.U.; Perkins, R.; Bäckhed, F. Microbiota-Induced Obesity Requires Farnesoid X Receptor. Gut 2017, 66, 429–437. [Google Scholar] [CrossRef] [PubMed]
- Bisanz, J.E.; Upadhyay, V.; Turnbaugh, J.A.; Ly, K.; Turnbaugh, P.J. Meta-Analysis Reveals Reproducible Gut Microbiome Alterations in Response to a High-Fat Diet. Cell Host. Microbe. 2019, 26, 265–272.e4. [Google Scholar] [CrossRef] [PubMed]
- Sinha, S.R.; Haileselassie, Y.; Nguyen, L.P.; Tropini, C.; Wang, M.; Becker, L.S.; Sim, D.; Jarr, K.; Spear, E.T.; Singh, G.; et al. Dysbiosis-Induced Secondary Bile Acid Deficiency Promotes Intestinal Inflammation. Cell Host. Microbe. 2020, 27, 659–670.e5. [Google Scholar] [CrossRef]
- Liu, T.C.; Kern, J.T.; Jain, U.; Sonnek, N.M.; Xiong, S.; Simpson, K.F.; VanDussen, K.L.; Winkler, E.S.; Haritunians, T.; Malique, A.; et al. Western Diet Induces Paneth Cell Defects through Microbiome Alterations and Farnesoid X Receptor and Type I Interferon Activation. Cell Host. Microbe. 2021, 29, 988–1001.e6. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.T.; Krutzik, S.R.; Modlin, R.L. Therapeutic Implications of the TLR and VDR Partnership. Trends. Mol. Med. 2007, 13, 117–124. [Google Scholar] [CrossRef]
- Verway, M.; Bouttier, M.; Wang, T.T.; Carrier, M.; Calderon, M.; An, B.S.; Devemy, E.; McIntosh, F.; Divangahi, M.; Behr, M.A.; et al. Vitamin D Induces Interleukin-1β Expression: Paracrine Macrophage Epithelial Signaling Controls M. Tuberculosis Infection. PLoS Pathog. 2013, 9, e1003407. [Google Scholar] [CrossRef]
- Chávez-Talavera, O.; Haas, J.; Grzych, G.; Tailleux, A.; Staels, B. Bile Acid Alterations in Nonalcoholic Fatty Liver Disease, Obesity, Insulin Resistance and Type 2 Diabetes: What Do the Human Studies Tell? Curr. Opin. Lipidol. 2019, 30, 244–254. [Google Scholar] [CrossRef]
- Lin, H.; An, Y.; Tang, H.; Wang, Y. Alterations of Bile Acids and Gut Microbiota in Obesity Induced by High Fat Diet in Rat Model. J. Agric. Food Chem. 2019, 67, 3624–3632. [Google Scholar] [CrossRef]
- Sipe, L.M.; Chaib, M.; Pingili, A.K.; Pierre, J.F.; Makowski, L. Microbiome, Bile Acids, and Obesity: How Microbially Modified Metabolites Shape Anti-Tumor Immunity. Immunol. Rev. 2020, 295, 220–239. [Google Scholar] [CrossRef]
- Sun, L.; Cai, J.; Gonzalez, F.J. The Role of Farnesoid X Receptor in Metabolic Diseases, and Gastrointestinal and Liver Cancer. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 335–347. [Google Scholar] [CrossRef]
- Wu, Q.; Sun, L.; Hu, X.; Wang, X.; Xu, F.; Chen, B.; Liang, X.; Xia, J.; Wang, P.; Aibara, D.; et al. Suppressing the Intestinal Farnesoid X Receptor/Sphingomyelin Phosphodiesterase 3 Axis Decreases Atherosclerosis. J. Clin. Invest. 2021, 131, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z. Variable Selection with Stepwise and Best Subset Approaches. Ann. Transl. Med. 2016, 4, 136. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Wei, Y.; Xiong, A.; Li, Y.; Guan, H.; Wang, Q.; Miao, Q.; Bian, Z.; Xiao, X.; Lian, M.; et al. Comprehensive Analysis of Serum and Fecal Bile Acid Profiles and Interaction with Gut Microbiota in Primary Biliary Cholangitis. Clin. Rev. Allergy Immunol. 2019, 58, 25–38. [Google Scholar] [CrossRef] [PubMed]
- Fujita, K.; Iguchi, Y.; Une, M.; Watanabe, S. Ursodeoxycholic Acid Suppresses Lipogenesis in Mouse Liver: Possible Role of the Decrease in β-Muricholic Acid, a Farnesoid X Receptor Antagonist. Lipids 2017, 52, 335–344. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-García, A.; Sahebkar, A.; Simental-Mendía, M.; Simental-Mendía, L.E. Effect of Ursodeoxycholic Acid on Glycemic Markers: A Systematic Review and Meta-Analysis of Clinical Trials. Pharmacol. Res. 2018, 135, 144–149. [Google Scholar] [CrossRef]
- Ocvirk, S.; Wilson, A.S.; Posma, J.M.; Li, J.v.; Koller, K.R.; Day, G.M.; Flanagan, C.A.; Otto, J.E.; Sacco, P.E.; Sacco, F.D.; et al. A Prospective Cohort Analysis of Gut Microbial Co-Metabolism in Alaska Native and Rural African People at High and Low Risk of Colorectal Cancer. Am. J. Clin. Nutr. 2020, 111, 406–419. [Google Scholar] [CrossRef]
- Fabian, T.; Leung, A. Epidemiology of Barrett’s Esophagus and Esophageal Carcinoma. Surg. Clin. N. Am. 2021, 101, 381–389. [Google Scholar] [CrossRef]
- Kauer, W.K.H.; Peters, J.H.; DeMeester, T.R.; Feussner, H.; Ireland, A.P.; Stein, H.J.; Siewert, R.J. Composition and Concentration of Bile Acid Reflux into the Esophagus of Patients with Gastroesophageal Reflux Disease. Surgery 1997, 122, 874–881. [Google Scholar] [CrossRef]
- Straub, D.; Oude Elferink, R.P.J.; Jansen, P.L.M.; Bergman, J.J.G.H.M.; Parikh, K.; Krishnadath, K.K. Glyco-Conjugated Bile Acids Drive the Initial Metaplastic Gland Formation from Multi-Layered Glands through Crypt-Fission in a Murine Model. PLoS ONE 2019, 14, e0220050. [Google Scholar] [CrossRef]
- Li, X.Y.; Zhu, S.H.; Yang, F.; Hu, G.X.; Yuan, L. An Ultra-Performance Liquid Chromatography-Tandem Mass Spectrometry Method for the Determination of Obeticholic Acid in Rat Plasma and Its Application in Preclinical Pharmacokinetic Studies. J. Chromatogr. B 2019, 1121, 82–88. [Google Scholar] [CrossRef]
- Barros, R.; Freund, J.N.; David, L.; Almeida, R. Gastric Intestinal Metaplasia Revisited: Function and Regulation of CDX2. Trends. Mol. Med. 2012, 18, 555–563. [Google Scholar] [CrossRef]
- Katsidzira, L.; Ocvirk, S.; Wilson, A.; Li, J.; Mahachi, C.B.; Soni, D.; DeLany, J.; Nicholson, J.K.; Zoetendal, E.G.; O’Keefe, S.J.D. Differences in Fecal Gut Microbiota, Short-Chain Fatty Acids and Bile Acids Link Colorectal Cancer Risk to Dietary Changes Associated with Urbanization Among Zimbabweans. Nutr. Cancer 2019, 71, 1313–1324. [Google Scholar] [CrossRef]
- Sheng, L.; Jena, P.K.; Hu, Y.; Liu, H.X.; Nagar, N.; Kalanetra, K.M.; French, S.W.; French, S.W.; Mills, D.A.; Wan, Y.J.Y. Hepatic Inflammation Caused by Dysregulated Bile Acid Synthesis Is Reversible by Butyrate Supplementation. J Pathol 2017, 243, 431–441. [Google Scholar] [CrossRef] [PubMed]
- Cheng, K.; Metry, M.; Felton, J.; Shang, A.C.; Drachenberg, C.B.; Xu, S.; Zhan, M.; Schumacher, J.; Guo, G.L.; Polli, J.E.; et al. Diminished Gallbladder Filling, Increased Fecal Bile Acids, and Promotion of Colon Epithelial Cell Proliferation and Neoplasia in Fibroblast Growth Factor 15-Deficient Mice. Oncotarget 2018, 9, 25572–25585. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.; Chen, J.; Drachenberg, C.B.; Raufman, J.P.; Xie, G. Farnesoid X Receptor Represses Matrix Metalloproteinase 7 Expression, Revealing This Regulatory Axis as a Promising Therapeutic Target in Colon Cancer. J. Biol. Chem. 2019, 294, 8529–8542. [Google Scholar] [CrossRef] [PubMed]
- Hwang, H.H.; Jeong, H.J.; Yun, S.; Byun, Y.; Okano, T.; Kim, S.W.; Lee, D.Y. Anticancer Effect of Heparin–Taurocholate Conjugate on Orthotopically Induced Exocrine and Endocrine Pancreatic Cancer. Cancers 2021, 13, 5775. [Google Scholar] [CrossRef]
- Birse, C.E.; Lagier, R.J.; FitzHugh, W.; Pass, H.I.; Rom, W.N.; Edell, E.S.; Bungum, A.O.; Maldonado, F.; Jett, J.R.; Mesri, M.; et al. Blood-Based Lung Cancer Biomarkers Identified through Proteomic Discovery in Cancer Tissues, Cell Lines and Conditioned Medium. Clin. Proteomics. 2015, 12. [Google Scholar] [CrossRef]
- Flores-Morales, A.; Iglesias-Gato, D. Quantitative Mass Spectrometry-Based Proteomic Profiling for Precision Medicine in Prostate Cancer. Front. Oncol. 2017, 7, 1–8. [Google Scholar] [CrossRef]
- van den Broek, I.; Sparidans, R.W.; Schellens, J.H.M.; Beijnen, J.H. Quantitative Assay for Six Potential Breast Cancer Biomarker Peptides in Human Serum by Liquid Chromatography Coupled to Tandem Mass Spectrometry. J. Chromatogr. B 2010, 878, 590–602. [Google Scholar] [CrossRef] [PubMed]
- Atak, A.; Khurana, S.; Gollapalli, K.; Reddy, P.J.; Levy, R.; Ben-Salmon, S.; Hollander, D.; Donyo, M.; Heit, A.; Hotz-Wagenblatt, A.; et al. Quantitative Mass Spectrometry Analysis Reveals a Panel of Nine Proteins as Diagnostic Markers for Colon Adenocarcinomas. Oncotarget 2018, 9, 13530–13544. [Google Scholar] [CrossRef] [PubMed]
- al Kadhi, O.; Melchini, A.; Mithen, R.; Saha, S. Development of a LC-MS/MS Method for the Simultaneous Detection of Tricarboxylic Acid Cycle Intermediates in a Range of Biological Matrices. J. Anal. Methods Chem. 2017, 2017, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Ansari, D.; Aronsson, L.; Sasor, A.; Welinder, C.; Rezeli, M.; Marko-Varga, G.; Andersson, R. The Role of Quantitative Mass Spectrometry in the Discovery of Pancreatic Cancer Biomarkers for Translational Science. J. Transl. Med. 2014, 12, 87. [Google Scholar] [CrossRef] [PubMed]
- Beretov, J.; Wasinger, V.C.; Millar, E.K.A.; Schwartz, P.; Graham, P.H.; Li, Y. Proteomic Analysis of Urine to Identify Breast Cancer Biomarker Candidates Using a Label-Free LC-MS/MS Approach. PLoS ONE 2015, 10, e0141876. [Google Scholar] [CrossRef]
- Goto, R.; Nakamura, Y.; Takami, T.; Sanke, T.; Tozuka, Z. Quantitative LC-MS/MS Analysis of Proteins Involved in Metastasis of Breast Cancer. PLoS ONE 2015, 10, e0130760. [Google Scholar] [CrossRef]
- Zeng, X.; Hood, B.L.; Zhao, T.; Conrads, T.P.; Sun, M.; Gopalakrishnan, V.; Grover, H.; Day, R.S.; Weissfeld, J.L.; Wilson, D.O.; et al. Lung Cancer Serum Biomarker Discovery Using Label-Free Liquid Chromatography-Tandem Mass Spectrometry. J. Thorac. Oncol. 2011, 6, 725–734. [Google Scholar] [CrossRef]
- Larkin, S.E.T.; Johnston, H.E.; Jackson, T.R.; Jamieson, D.G.; Roumeliotis, T.I.; Mockridge, C.I.; Michael, A.; Manousopoulou, A.; Papachristou, E.K.; Brown, M.D.; et al. Detection of Candidate Biomarkers of Prostate Cancer Progression in Serum: A Depletion-Free 3D LC/MS Quantitative Proteomics Pilot Study. Br. J. Cancer 2016, 115, 1078–1086. [Google Scholar] [CrossRef]
- McGlone, E.R.; Bloom, S.R. Bile Acids and the Metabolic Syndrome. Ann. Clin. Biochem. 2019, 56, 326–337. [Google Scholar] [CrossRef]
- Cao, Y.; Li, W.; Gong, X.; Niu, X.; Zheng, J.; Yu, J.; Li, J.; Tu, P.; Song, Y. Widely Quasi-Quantitative Analysis Enables Temporal Bile Acids-Targeted Metabolomics in Rat after Oral Administration of Ursodeoxycholic Acid. Anal. Chim. Acta. 2022, 1212, 339885. [Google Scholar] [CrossRef]
- Gika, H.; Virgiliou, C.; Theodoridis, G.; Plumb, R.S.; Wilson, I.D. Untargeted LC/MS-Based Metabolic Phenotyping (Metabonomics/Metabolomics): The State of the Art. J. Chromatogr. B 2019, 1117, 136–147. [Google Scholar] [CrossRef] [PubMed]
- Barupal, D.K.; Fan, S.; Fiehn, O. Integrating Bioinformatics Approaches for a Comprehensive Interpretation of Metabolomics Datasets. Curr. Opin. Biotechnol. 2018, 54, 1–9. [Google Scholar] [CrossRef] [PubMed]
- van den Berg, R.A.; Hoefsloot, H.C.J.; Westerhuis, J.A.; Smilde, A.K.; van der Werf, M.J. Centering, Scaling, and Transformations: Improving the Biological Information Content of Metabolomics Data. BMC Genomics 2006, 7, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Human Metabolome Database: Other Databases. Available online: https://hmdb.ca/w/databases (accessed on 2 August 2022).
- Charkoftaki, G.; Tan, W.Y.; Berrios-Carcamo, P.; Orlicky, D.J.; Golla, J.P.; Garcia-Milian, R.; Aalizadeh, R.; Thomaidis, N.S.; Thompson, D.C.; Vasiliou, V. Liver Metabolomics Identifies Bile Acid Profile Changes at Early Stages of Alcoholic Liver Disease in Mice. Chem. Biol. Interact. 2022, 360, 109931. [Google Scholar] [CrossRef] [PubMed]
- Luo, C.; Zhang, X.; He, Y.; Chen, H.; Liu, M.; Wang, H.; Tang, L.; Tu, G.; Ding, M. A Pseudo-Targeted Metabolomics Study Based on Serum Bile Acids Profiling for the Differential Diagnosis of Benign and Malignant Breast Lesions. Steroids 2021, 175, 108914. [Google Scholar] [CrossRef]
- Tian, M.; Yan, J.; Zhang, H.; Wei, Y.; Zhang, M.; Rao, Z.; Zhang, M.; Wang, H.; Wang, Y.; Li, X. Screening and Validation of Biomarkers for Cadmium-Induced Liver Injury Based on Targeted Bile Acid Metabolomics. Environ. Pollut. 2022, 300, 118837. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Cheng, J.; Luo, M.; Yang, S.; Xing, Q.; Cheng, J.; Lv, J.; Yu, C.; Sun, L.; Shi, D.; et al. Targeted Metabolomics Analysis of Bile Acids and Cell Biology Studies Reveal the Critical Role of Glycodeoxycholic Acid in Buffalo Follicular Atresia. J. Steroid. Biochem. Mol. Biol. 2022, 221, 106115. [Google Scholar] [CrossRef] [PubMed]
- Dewaele, D.; Annaert, P.; Hoeben, E. LC-MS/MS Analysis of Bile Acids in In Vitro Samples. Methods Mol. Biol. 2019, 1981, 15–23. [Google Scholar] [CrossRef]
- Liu, C.; Zhu, Y.; Lu, Z.; Guo, W.; Tumen, B.; He, Y.; Chen, C.; Hu, S.; Xu, K.; Wang, Y.; et al. Cadmium Induces Acute Liver Injury by Inhibiting Nrf2 and the Role of NF-ΚB, NLRP3, and MAPKs Signaling Pathway. Int. J. Environ. Res. Public Health 2020, 17, 138. [Google Scholar] [CrossRef]
- Ren, C.; Ren, L.; Yan, J.; Bai, Z.; Zhang, L.; Zhang, H.; Xie, Y.; Li, X. Cadmium Causes Hepatopathy by Changing the Status of DNA Methylation in the Metabolic Pathway. Toxicol. Lett. 2021, 340, 101–113. [Google Scholar] [CrossRef]
- Zhang, A.; Sun, H.; Wang, P.; Han, Y.; Wang, X. Recent and Potential Developments of Biofluid Analyses in Metabolomics. J. Proteomics. 2012, 75, 1079–1088. [Google Scholar] [CrossRef]
- Bujak, R.; Struck-Lewicka, W.; Markuszewski, M.J.; Kaliszan, R. Metabolomics for Laboratory Diagnostics. J. Pharm. Biomed. Anal. 2015, 113, 108–120. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Wang, Y.; Wang, X.; Ren, Q.; Han, S.; Ding, L.; Li, Z.; Zhou, X.; Li, W.; Zhang, L. Comparative Salivary Proteomics Analysis of Children with and without Dental Caries Using the ITRAQ/MRM Approach. J. Transl. Med. 2018, 16, 11. [Google Scholar] [CrossRef]
- Chauhan, S.; Chauhan, S.; D’Cruz, R.; Faruqi, S.; Singh, K.K.; Varma, S.; Singh, M.; Karthik, V. Chemical Warfare Agents. Environ. Toxicol. Pharmacol. 2008, 26, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhang, H.; Si, Y.; Du, Y.; Wu, J.; Li, J. High-Coverage Lipidomics Analysis Reveals Biomarkers for Diagnosis of Acute Exacerbation of Chronic Obstructive Pulmonary Disease. J. Chromatogr. B 2022, 123278. [Google Scholar] [CrossRef] [PubMed]
- Minno, A.D.; Gelzo, M.; Caterino, M.; Costanzo, M.; Ruoppolo, M.; Castaldo, G. Challenges in Metabolomics-Based Tests, Biomarkers Revealed by Metabolomic Analysis, and the Promise of the Application of Metabolomics in Precision Medicine. Int. J. Mol. Sci. 2022, 23, 5213. [Google Scholar] [CrossRef]

| Type of Receptor | Receptor | Function | Localization | Ref. |
|---|---|---|---|---|
| Nuclear | Farnesoid X receptor | Regulation of enterohepatic circulation Suppression of the synthesis of BAs Suppression of the uptake of BAs Increasing of BAs export of bile Suppression of the absorption of BAs by ileal epithelium and cholangiocytes Vasodilation activity | Ileal epithelium Hepatocytes Cholangiocytes Endothelium of sinusoids Renal epithelium Adrenal cortex Cells of innate and adaptive immunity | [28,43,54,55] |
| Nuclear | Nuclear receptor subfamily 1 group H member 3 | Regulation of the remodeling of the phospholipids of the endoplasmic reticulum Suppression of the stress of the endoplasmic reticulum Reduction of the absorption of cholesterol in the intestine Increases the synthesis of BAs through increasing of CYP7A1 activity Promotion of the transport of cholesterol from peripheral tissues to liver Activation of sterol response element-binding protein-1c | Hepatocytes Enterocytes Renal epithelium Adipose tissue Skeletal muscles Cells of innate and adaptive immunity | [56,57,58] |
| Nuclear | Vitamin D receptor | Modulation of the intestinal microbiota composition Regulation of secondary BAs production Potential impact on the risk of developing colorectal cancer | Ileum Endocrine glands Skin Cells of innate and adaptive immunity | [59,60,61] |
| Nuclear | Constitutive activated receptor | Suppression of CYP7A expression and BAs synthesis Activation of phase II enzymes for the detoxification of xenobiotics Activation of transporters (MRP, MDR, and OATP) Suppression of gluconeogenesis, development of steatosis, and decrease in thyroxine activity | Hepatocytes Renal tubular epithelium | [46,47,48,62] |
| Nuclear | Pregnane X receptor | Suppression of CYP7A expression and BAs synthesis Activation of phase II enzymes for the detoxification of xenobiotics Activation of transporters (MRP, MDR, and OATP) Suppression of gluconeogenesis, development of steatosis, and decrease in thyroxine activity CYP3A43 activation Suppression of the hepatocytes and intestinal inflammatory cascade Suppression of CYP7A1 | Hepatocytes Intestinal epithelium | [28,63,64,65] |
| Membrane | G protein–coupled bile acid receptor 1, Takeda G-protein receptor 5 | Systemic effects of BAs Regulation of intestinal motility and metabolism Relaxation of the gallbladder (together with FGF19) Vasodilating action Regulation of the proliferation of non-ciliated cholangiocytes Possible development of cholangiocellular cancer | Ileal epithelium Cholangiocytes epithelium Smooth muscle cells Endothelium Adipose tissue Cells of innate and adaptive immunity | [50,66,67,68] |
| Membrane | Sphingosine-1-phosphate receptor 2 | Increasing of enzymes of lipid (SREBP1c, FAS, LDLR, FXRα, and PPARγ) and glucose metabolism (ERK1/2) Regulation of the differentiation of endothelial cells Promotion of the growth and metastasis of cholangiocarcinoma | Hepatocytes Intestinal epithelium Endothelium Vascular smooth muscle cells Myocardium Fibroblasts | [69,70,71,72] |
| Membrane | Muscarinic receptors M2, M3 | Stimulation of intestinal motility, negative chronotropic action. Probable promotion of colon cancer growth | Intestinal smooth muscle cells Exocrine glands Myocardium | [52,53,73,74] |
| Membrane | Vascular endothelial growth factor | Prevention of bile duct injury, possibly fibrosis. New vessel formation. | Cell lines of stomach and colon cancer | [51,75,76] |
| No. | Compound Name | SRM 1 (Q1/Q3) |
|---|---|---|
| 1 | Glycoursodeoxycholic acid-3-sulfate (GUDCA-3S) | 528.3/528.3 |
| 2 | Ursodeoxycholic acid-3-sulfate (UDCA-3S) | 471.2/471.2 |
| 3 | Tauroursodeoxycholic acid (TUDCA) | 498.2/432.2 |
| 4 | Glycoursodeoxycholic acid (GUDCA) | 448.2/404.2 |
| 5 | Cholic acid-3-sulfate (CA-3S) | 487.2/97.0 |
| 6 | Glycolithocholic acid-3-sulfate (GLCA-3S) | 512.2/74.0 |
| 7 | Тауринхиoдезoксихoлевая кислoта (THDCA) | 498.2/80.0 |
| 8 | Taurochenodeoxycholic acid (TCA) | 514.2/496.2 |
| 9 | Glycocholic acid (GCA) | 464.2/402.2 |
| 10 | Chenodeoxycholic acid-3-sulfate (CDCA-3S) | 471.4/97.0 |
| 11 | Deoxycholic acid-3-sulfate (DCA-3S) | 471.2/97.0 |
| 12 | Ursodeoxycholic acid (UDCA) | 391.2/373.2 |
| 13 | Hyocholic acid (HCA) | 407.2/345.2 |
| 14 | Taurochenodeoxycholic acid (TCDCA) | 498.2/80.0 |
| 15 | Glycochenodeoxycholic acid (GCDCA) | 448.2/74.0 |
| 16 | Chenodeoxycholic acid (HDCA) | 391.4/391.4 |
| 17 | Taurodeoxycholic acid (TDCA) | 498.2/355.2 |
| 18 | Lithocholic acid-3-sulfate (LCA-3S) | 455.4/97.0 |
| 19 | Glycodeoxycholic acid (GDCA) | 448.4/386.2 |
| 20 | Cholic acid (CA) | 407.2/345.2 |
| 21 | Taurolithocholic acid (TLCA) | 482.2/416.2 |
| 22 | Glycolithocholic acid (GLCA) | 432.2/74.0 |
| 23 | Chenodeoxycholic acid (CDCA) | 391.2/373.2 |
| 24 | Deoxycholic acid (DCA) | 391.2/345.2 |
| 25 | Lithocholic acid (LCA) | 375.3/356.2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shansky, Y.; Bespyatykh, J. Bile Acids: Physiological Activity and Perspectives of Using in Clinical and Laboratory Diagnostics. Molecules 2022, 27, 7830. https://doi.org/10.3390/molecules27227830
Shansky Y, Bespyatykh J. Bile Acids: Physiological Activity and Perspectives of Using in Clinical and Laboratory Diagnostics. Molecules. 2022; 27(22):7830. https://doi.org/10.3390/molecules27227830
Chicago/Turabian StyleShansky, Yaroslav, and Julia Bespyatykh. 2022. "Bile Acids: Physiological Activity and Perspectives of Using in Clinical and Laboratory Diagnostics" Molecules 27, no. 22: 7830. https://doi.org/10.3390/molecules27227830
APA StyleShansky, Y., & Bespyatykh, J. (2022). Bile Acids: Physiological Activity and Perspectives of Using in Clinical and Laboratory Diagnostics. Molecules, 27(22), 7830. https://doi.org/10.3390/molecules27227830









