Post-Consumer Poly(ethylene terephthalate) (PET) Depolymerization by Yarrowia lipolytica: A Comparison between Hydrolysis Using Cell-Free Enzymatic Extracts and Microbial Submerged Cultivation
Abstract
1. Introduction
2. Results and Discussion
2.1. Hydrolysis Using Cell-Free Enzymatic Extracts
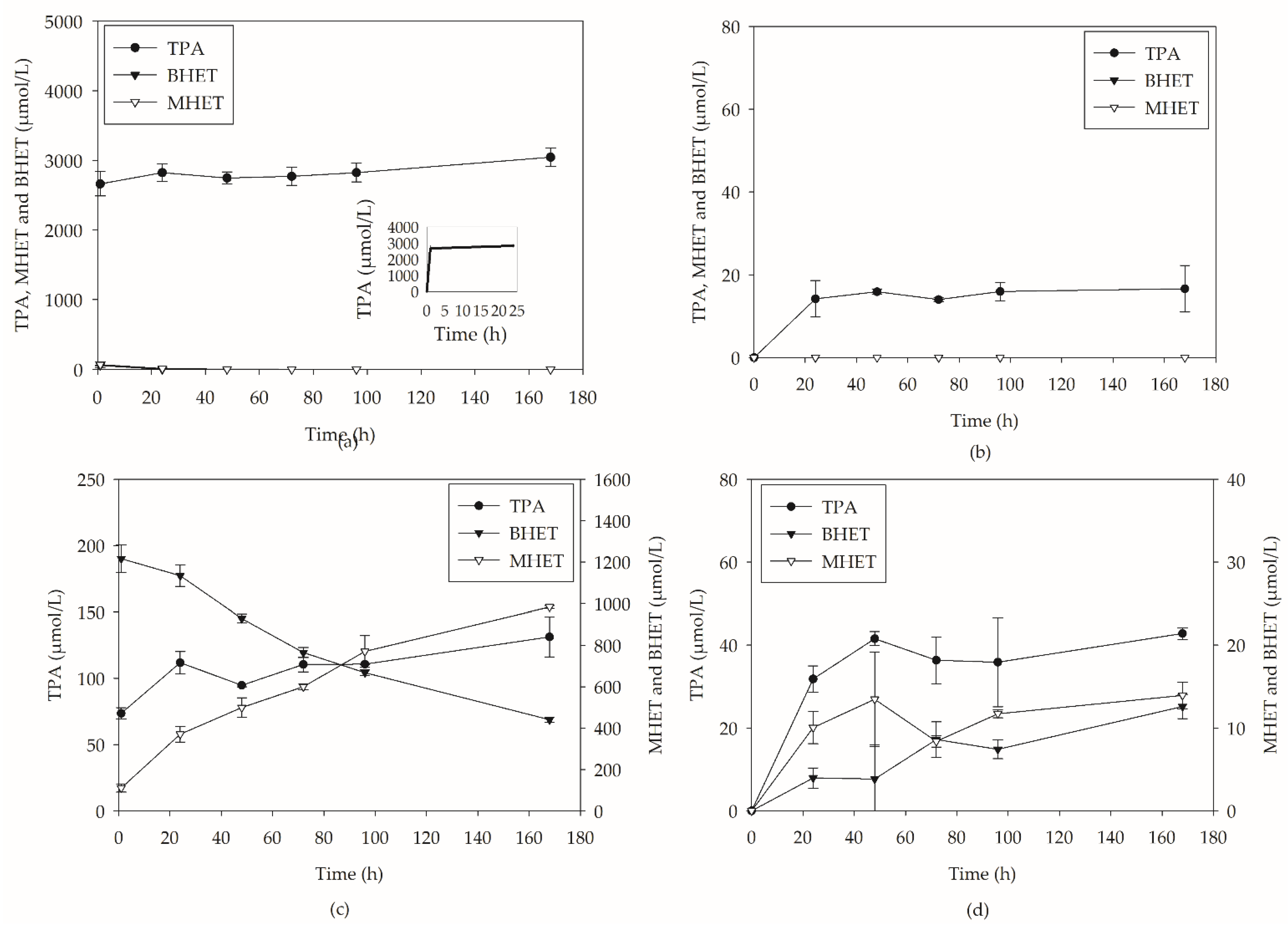
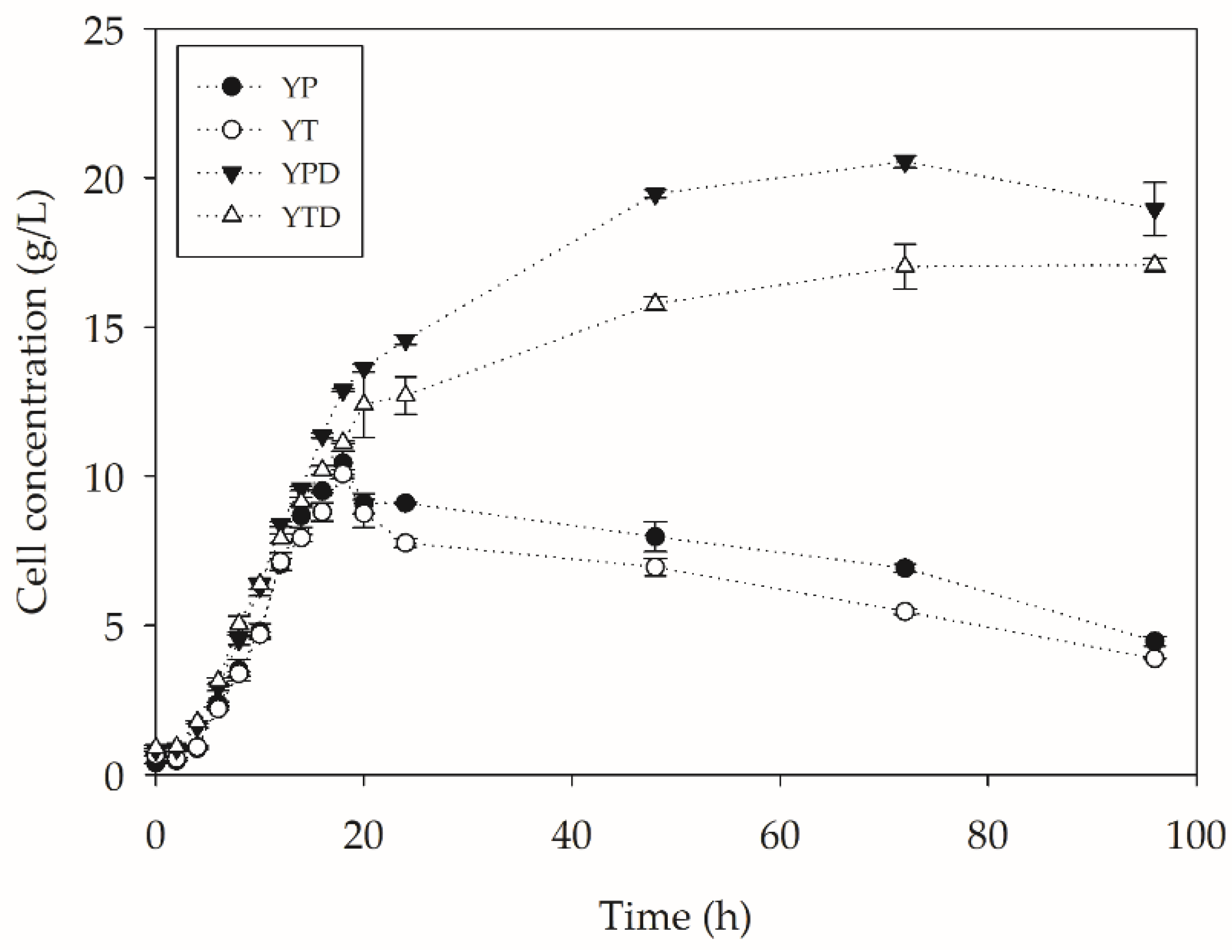
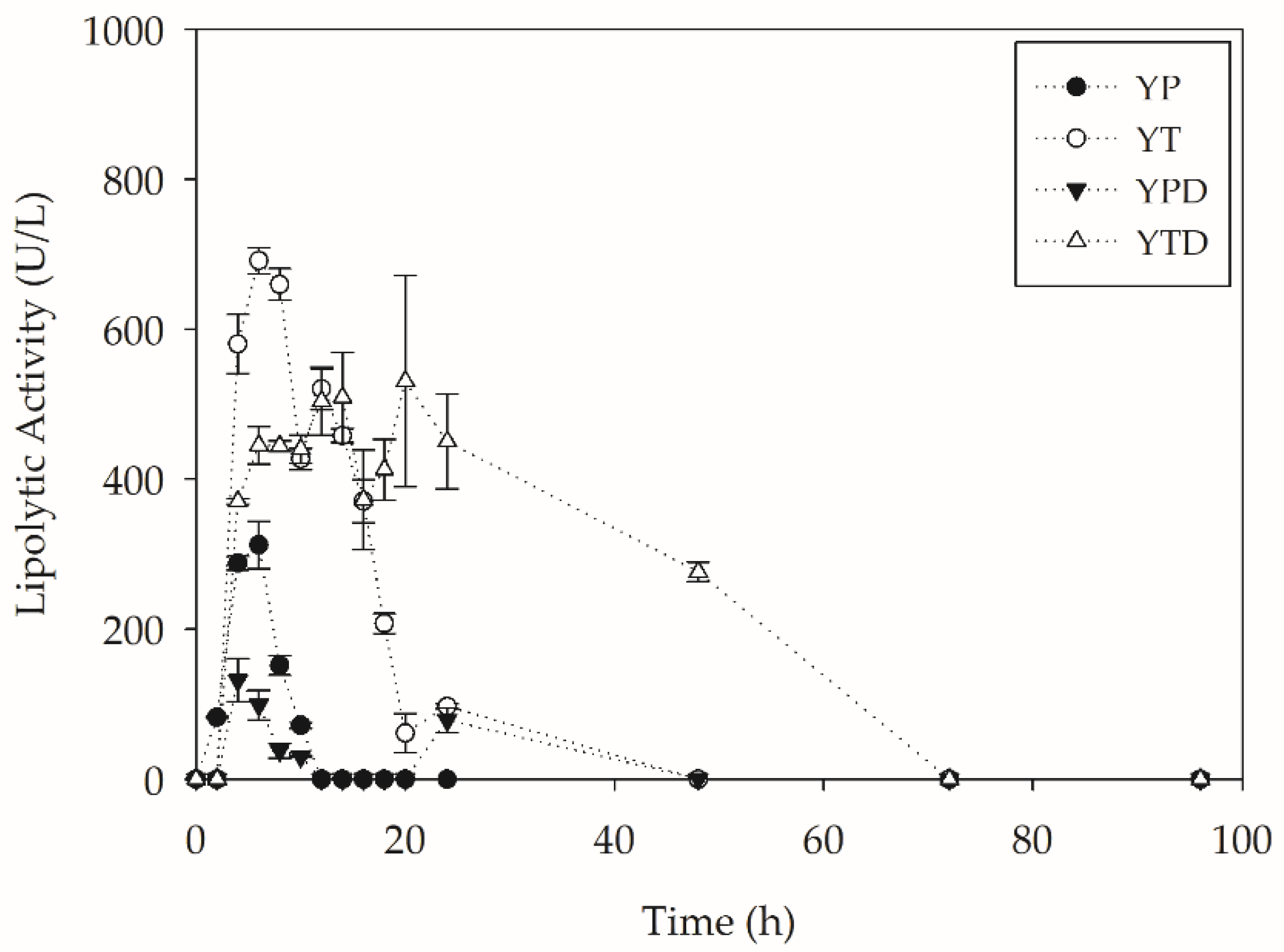
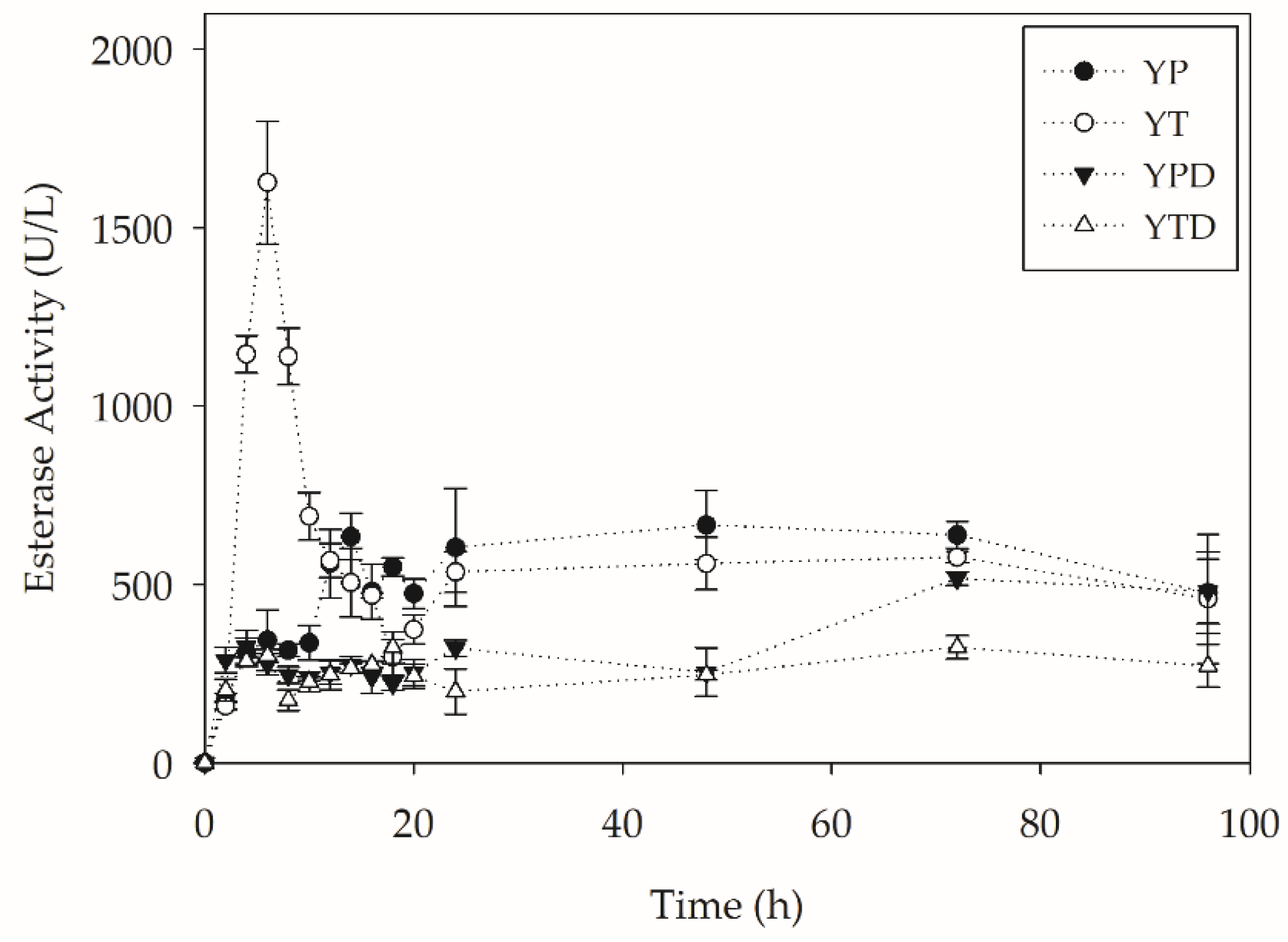
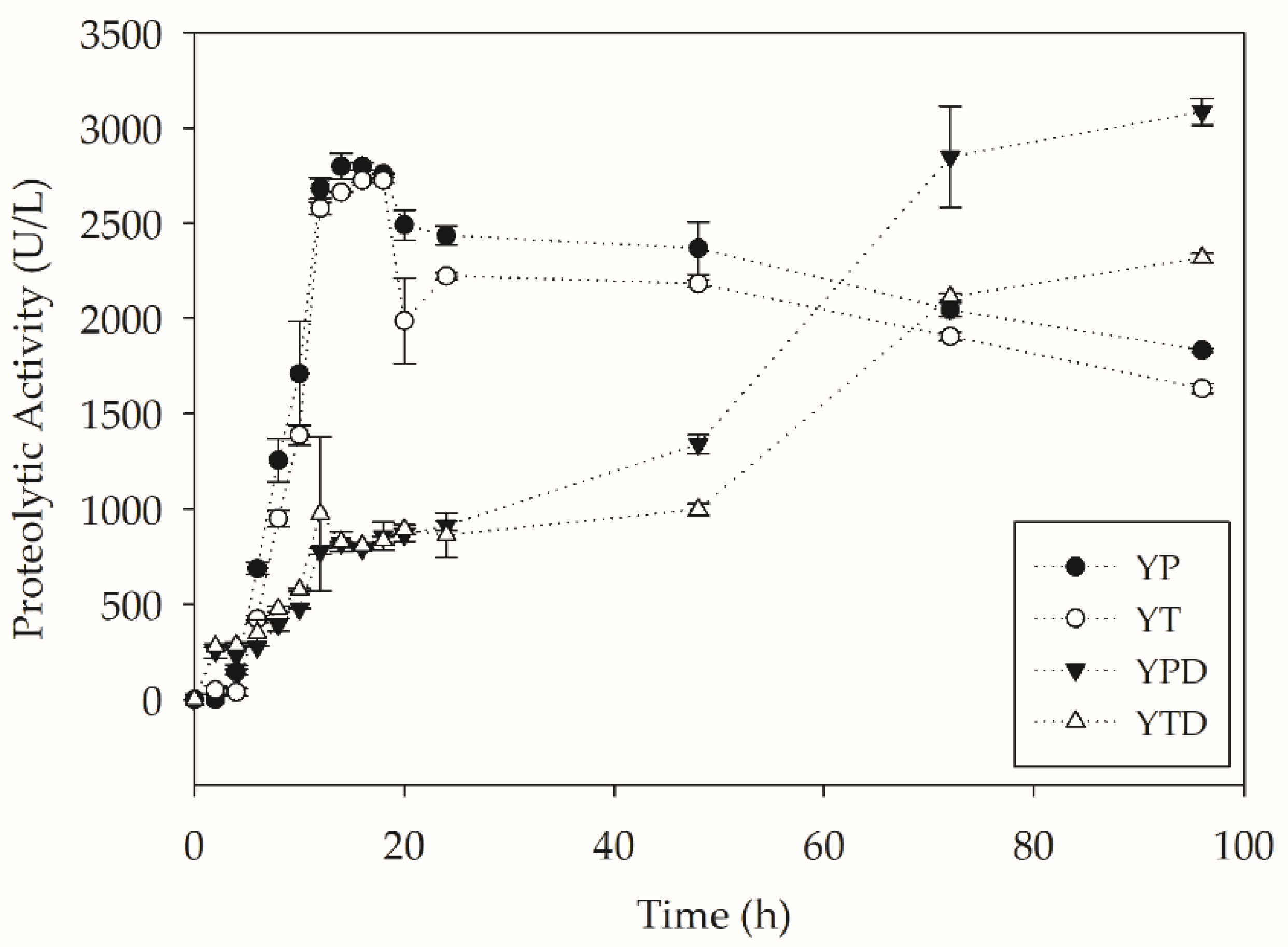
2.2. PC-PET Depolymerization in Submerged Cultivations
2.2.1. Depolymerization in Flasks
2.2.2. Depolymerization in Bioreactors
3. Materials and Methods
3.1. Materials
3.2. Microorganism
3.3. Solid-State Fermentation (SSF) and Hydrolysis Using Cell-Free Enzymatic Extracts
3.4. Submerged Cultivation in Flasks (SCF) and Submerged Cultivation in Bioreactors (SCB)
3.5. Analytical Methods
3.5.1. Biomass Quantification, pH and Growth Parameters
3.5.2. Enzymes Activities
3.5.3. PET Hydrolysis Products Quantification
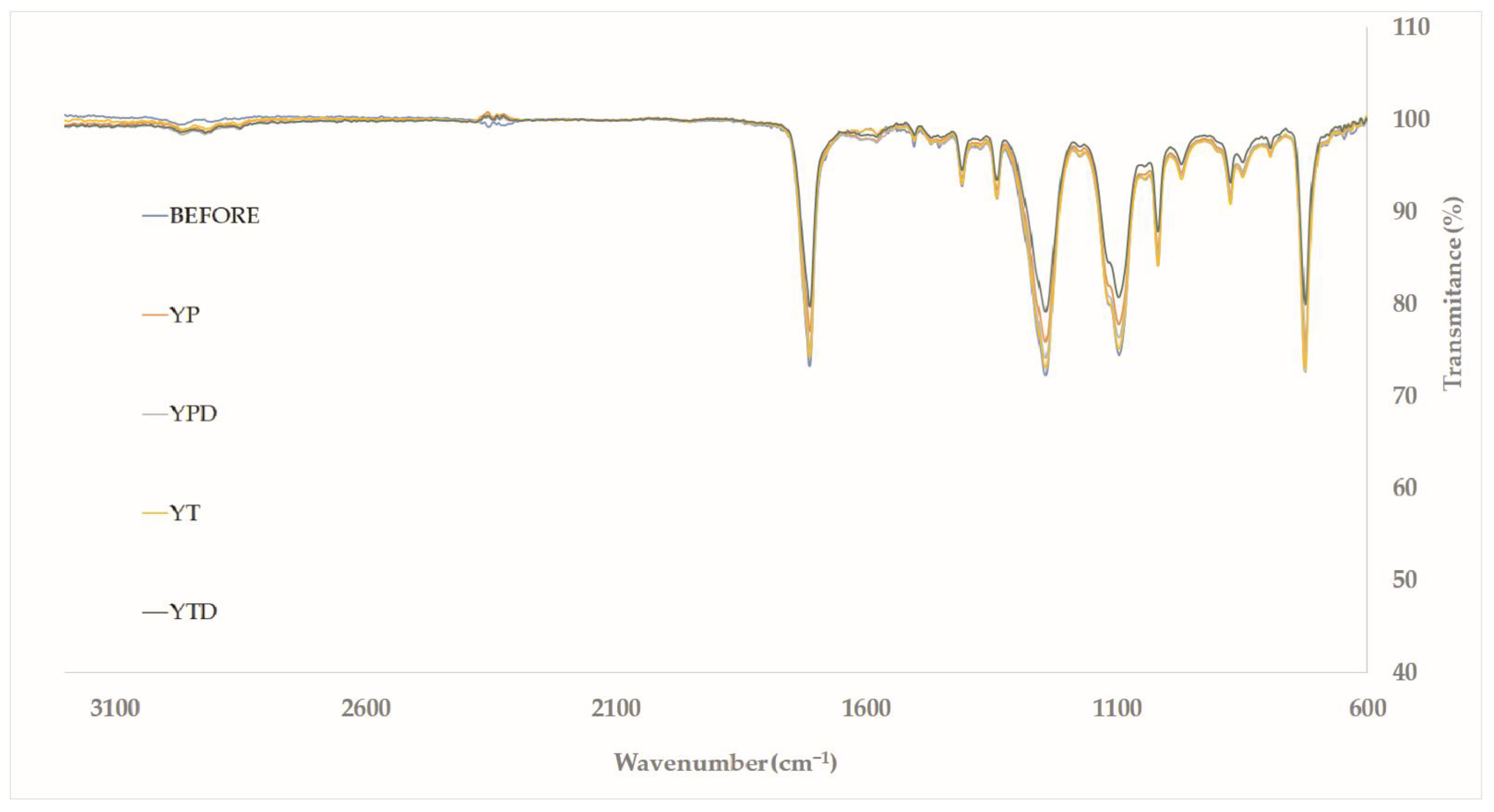
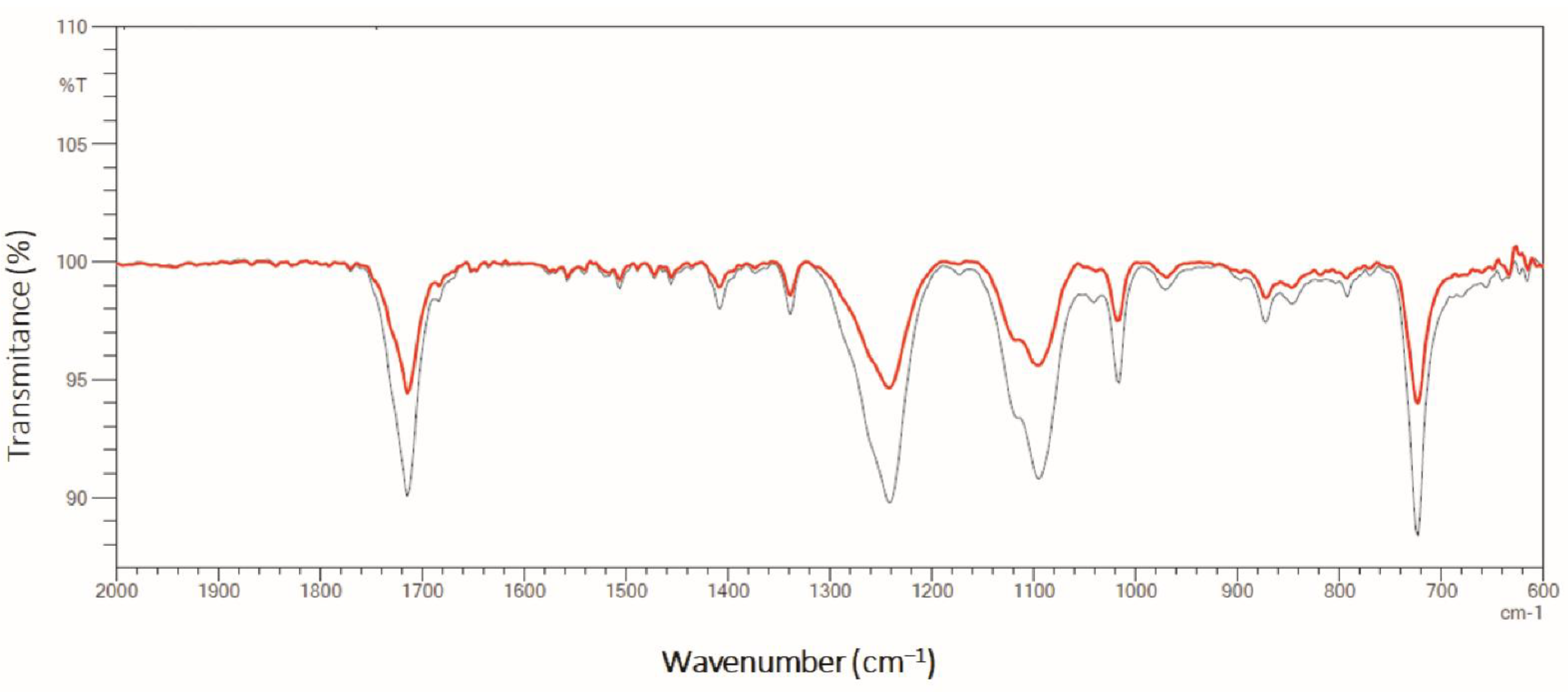
3.5.4. ATR-FTIR (Attenuated Total Reflectance—Fourier-Transform Infrared Spectroscopy)
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Geyer, R.; Jambeck, J.R.; Law, K.L. Production, Use, and Fate of All Plastics Ever Made. Sci. Adv. 2017, 3, e1700782. [Google Scholar] [CrossRef] [PubMed]
- Thushari, G.G.N.; Senevirathna, J.D.M. Plastic Pollution in the Marine Environment. Heliyon 2020, 6, e04709. [Google Scholar] [CrossRef] [PubMed]
- Shams, M.; Alam, I.; Mahbub, M.S. Plastic Pollution during COVID-19: Plastic Waste Directives and Its Long-Term Impact on the Environment. Environ. Adv. 2021, 5, 100119. [Google Scholar] [CrossRef] [PubMed]
- Phelps Bondaroff, T.; Cooke, S. Masks on the Beach: The Impact of COVID-19 on Marine Plastic Pollution. OceansAsia 2020, 1–79. [Google Scholar]
- Eriksen, M.; Lebreton, L.C.M.; Carson, H.S.; Thiel, M.; Moore, C.J.; Borerro, J.C.; Galgani, F.; Ryan, P.G.; Reisser, J. Plastic Pollution in the World’s Oceans: More than 5 Trillion Plastic Pieces Weighing over 250,000 Tons Afloat at Sea. PLoS ONE 2014, 9, e111913. [Google Scholar] [CrossRef]
- Solis, M.; Silveira, S. Technologies for Chemical Recycling of Household Plastics–A Technical Review and TRL Assessment. Waste Manag. 2020, 105, 128–138. [Google Scholar] [CrossRef]
- Wei, R.; Zimmermann, W. Microbial Enzymes for the Recycling of Recalcitrant Petroleum-Based Plastics: How Far Are We? Microb. Biotechnol. 2017, 10, 1308–1322. [Google Scholar] [CrossRef]
- Mohanan, N.; Montazer, Z.; Sharma, P.K.; Levin, D.B. Microbial and Enzymatic Degradation of Synthetic Plastics. Front. Microbiol. 2020, 11, 580709. [Google Scholar] [CrossRef]
- Vollmer, I.; Jenks, M.J.F.; Roelands, M.C.P.; White, R.J.; Van Harmelen, T.; De Wild, P.; Van Der Laan, G.P.; Meirer, F.; Keurentjes, J.T.F.; Weckhuysen, B.M. Beyond Mechanical Recycling: Giving New Life to Plastic Waste. Angew. Chemie Int. Ed. 2020, 59, 15402–15423. [Google Scholar] [CrossRef]
- Sales, J.C.S.; Santos, A.G.; de Castro, A.M.; Coelho, M.A.Z. A Critical View on the Technology Readiness Level (TRL) of Microbial Plastics Biodegradation. World J. Microbiol. Biotechnol. 2021, 37, 116. [Google Scholar] [CrossRef]
- Ronkvist, Å.M.; Xie, W.; Lu, W.; Gross, R.A. Cutinase-Catalyzed Hydrolysis of Poly(Ethylene Terephthalate). Macromolecules 2009, 42, 5128–5138. [Google Scholar] [CrossRef]
- Ribitsch, D.; Heumann, S.; Trotscha, E.; Acero, E.H.; Greimel, K.; Leber, R.; Birner-Gruenberger, R.; Deller, S.; Eiteljoerg, I.; Remler, P.; et al. Hydrolysis of Polyethyleneterephthalate by P-Nitrobenzylesterase from Bacillus Subtilis. Biotechnol. Prog. 2011, 27, 951–960. [Google Scholar] [CrossRef] [PubMed]
- de Castro, A.M.; Carniel, A.; Junior, J.N.; da Conceição Gomes, A.; Valoni, É. Screening of Commercial Enzymes for Poly(Ethylene Terephthalate) (PET) Hydrolysis and Synergy Studies on Different Substrate Sources. J. Ind. Microbiol. Biotechnol. 2017, 44, 835–844. [Google Scholar] [CrossRef]
- Barth, M.; Honak, A.; Oeser, T.; Wei, R.; Belisário-Ferrari, M.R.; Then, J.; Schmidt, J.; Zimmermann, W. A Dual Enzyme System Composed of a Polyester Hydrolase and a Carboxylesterase Enhances the Biocatalytic Degradation of Polyethylene Terephthalate Films. Biotechnol. J. 2016, 11, 1082–1087. [Google Scholar] [CrossRef] [PubMed]
- Müller, R.-J.J.; Schrader, H.; Profe, J.; Dresler, K.; Deckwer, W.-D.D. Enzymatic Degradation of Poly(Ethylene Terephthalate): Rapid Hydrolyse Using a Hydrolase from T. Fusca. Macromol. Rapid Commun. 2005, 26, 1400–1405. [Google Scholar] [CrossRef]
- Yoshida, S.; Hiraga, K.; Takehana, T.; Taniguchi, I.; Yamaji, H.; Maeda, Y.; Toyohara, K.; Miyamoto, K.; Kimura, Y.; Oda, K. A Bacterium That Degrades and Assimilates Poly(Ethylene Terephthalate). Science 2016, 351, 1196–1199. [Google Scholar] [CrossRef]
- da Costa, A.M.; de Oliveira Lopes, V.R.; Vidal, L.; Nicaud, J.-M.; de Castro, A.M.; Coelho, M.A.Z. Poly(Ethylene Terephthalate) (PET) Degradation by Yarrowia lipolytica: Investigations on Cell Growth, Enzyme Production and Monomers Consumption. Process Biochem. 2020, 95, 81–90. [Google Scholar] [CrossRef]
- Sales, J.C.S.; de Castro, A.M.; Ribeiro, B.D.; Coelho, M.A.Z. Improved Production of Biocatalysts by Yarrowia lipolytica Using Natural Sources of the Biopolyesters Cutin and Suberin, and Their Application in Hydrolysis of Poly (Ethylene Terephthalate) (PET). Bioprocess Biosyst. Eng. 2021, 44, 2277–2287. [Google Scholar] [CrossRef]
- Coelho, M.A.Z.; Amaral, P.F.F.; Belo, I. Yarrowia lipolytica: An Industrial Workhorse; Formatex Research Center: Badajoz, Spain, 2010; pp. 930–944. [Google Scholar]
- Wierzchowska, K.; Zieniuk, B.; Fabiszewska, A. Use of Non-Conventional Yeast Yarrowia lipolytica in Treatment or Upgradation of Hydrophobic Industry Wastes. Waste Biomass Valorization 2022, 13, 757–779. [Google Scholar] [CrossRef]
- Nascimento, F.V.D.; Lemes, A.C.; de Castro, A.M.; Secchi, A.R.; Coelho, M.A.Z. A Temporal Evolution Perspective of Lipase Production by Yarrowia lipolytica in Solid-State Fermentation. Processes 2022, 10, 381. [Google Scholar] [CrossRef]
- Pandey, A. Solid-State Fermentation. Biochem. Eng. J. 2003, 13, 81–84. [Google Scholar] [CrossRef]
- Ravindran, R.; Hassan, S.; Williams, G.; Jaiswal, A. A Review on Bioconversion of Agro-Industrial Wastes to Industrially Important Enzymes. Bioengineering 2018, 5, 93. [Google Scholar] [CrossRef] [PubMed]
- Carniel, A.; Valoni, É.; Nicomedes, J.; da Conceição Gomes, A.; de Castro, A.M. Lipase from Candida antarctica (CALB) and Cutinase from Humicola insolens Act Synergistically for PET Hydrolysis to Terephthalic Acid. Process Biochem. 2017, 59, 84–90. [Google Scholar] [CrossRef]
- Barth, M.; Oeser, T.; Wei, R.; Then, J.; Schmidt, J.; Zimmermann, W. Effect of Hydrolysis Products on the Enzymatic Degradation of Polyethylene Terephthalate Nanoparticles by a Polyester Hydrolase from Thermobifida Fusca. Biochem. Eng. J. 2015, 93, 222–228. [Google Scholar] [CrossRef]
- Bhutada, G.; Kavšček, M.; Ledesma-Amaro, R.; Thomas, S.; Rechberger, G.N.; Nicaud, J.M.; Natter, K. Sugar versus Fat: Elimination of Glycogen Storage Improves Lipid Accumulation in Yarrowia lipolytica. FEMS Yeast Res. 2017, 17, fox020. [Google Scholar] [CrossRef]
- Fickers, P.; Nicaud, J.M.; Destain, J.; Thonart, P. Involvement of Hexokinase Hxk1 in Glucose Catabolite Repression of LIP2 Encoding Extracellular Lipase in the Yeast Yarrowia lipolytica. Curr. Microbiol. 2005, 50, 133–137. [Google Scholar] [CrossRef]
- Fickers, P.; Nicaud, J.M.; Gaillardin, C.; Destain, J.; Thonart, P. Carbon and Nitrogen Sources Modulate Lipase Production in the Yeast Yarrowia lipolytica. J. Appl. Microbiol. 2004, 96, 742–749. [Google Scholar] [CrossRef]
- Fickers, P.; Fudalej, F.; Le Dall, M.T.; Casaregola, S.; Gaillardin, C.; Thonart, P.; Nicaud, J.M. Identification and Characterisation of LIP7 and LIP8 Genes Encoding Two Extracellular Triacylglycerol Lipases in the Yeast Yarrowia lipolytica. Fungal Genet. Biol. 2005, 42, 264–274. [Google Scholar] [CrossRef]
- Liu, W.-S.; Pan, X.-X.; Jia, B.; Zhao, H.-Y.; Xu, L.; Liu, Y.; Yan, Y.-J. Surface Display of Active Lipases Lip7 and Lip8 from Yarrowia lipolytica on Saccharomyces Cerevisiae. Appl. Microbiol. Biotechnol. 2010, 88, 885–891. [Google Scholar] [CrossRef]
- De Castro, A.M.; Carniel, A.; Stahelin, D.; Junior, L.S.C.; de Angeli Honorato, H.; de Menezes, S.M.C. High-Fold Improvement of Assorted Post-Consumer Poly(Ethylene Terephthalate) (PET) Packages Hydrolysis Using Humicola insolens Cutinase as a Single Biocatalyst. Process Biochem. 2019, 81, 85–91. [Google Scholar] [CrossRef]
- FICKERS, P.; BENETTI, P.; WACHE, Y.; MARTY, A.; MAUERSBERGER, S.; SMIT, M.; NICAUD, J. Hydrophobic Substrate Utilisation by the Yeast Yarrowia lipolytica, and Its Potential Applications. FEMS Yeast Res. 2005, 5, 527–543. [Google Scholar] [CrossRef] [PubMed]
- Ioakeimidis, C.; Fotopoulou, K.N.; Karapanagioti, H.K.; Geraga, M.; Zeri, C.; Papathanassiou, E.; Galgani, F.; Papatheodorou, G. The Degradation Potential of PET Bottles in the Marine Environment: An ATR-FTIR Based Approach. Sci. Rep. 2016, 6, 23501. [Google Scholar] [CrossRef]
- Sammon, C.; Yarwood, J.; Everall, N. An FT–IR Study of the Effect of Hydrolytic Degradation on the Structure of Thin PET Films. Polym. Degrad. Stab. 2000, 67, 149–158. [Google Scholar] [CrossRef]
- Botelho, A.; Penha, A.; Fraga, J.; Barros-Timmons, A.; Coelho, M.A.; Lehocky, M.; Štěpánková, K.; Amaral, P. Yarrowia lipolytica Adhesion and Immobilization onto Residual Plastics. Polymers 2020, 12, 649. [Google Scholar] [CrossRef] [PubMed]
- Amaral, P.F.F.; Lehocky, M.; Barros-Timmons, A.M.V.; Rocha-Leão, M.H.M.; Coelho, M.A.Z.; Coutinho, J.A.P. Cell Surface Characterization of Yarrowia lipolytica IMUFRJ 50682. Yeast 2006, 23, 867–877. [Google Scholar] [CrossRef] [PubMed]
- Amaral, P.F.F.; de Almeida, A.P.R.; Peixoto, T.; Rocha-Leão, M.H.M.; Coutinho, J.A.P.; Coelho, M.A.Z. Beneficial Effects of Enhanced Aeration Using Perfluorodecalin in Yarrowia lipolytica Cultures for Lipase Production. World J. Microbiol. Biotechnol. 2007, 23, 339–344. [Google Scholar] [CrossRef]
- Barth, G.; Gaillardin, C. Yarrowia lipolytica. In Nonconventional Yeasts in Biotechnology; Springer: Berlin/Heidelberg, Germany, 1996. [Google Scholar]
- Kadić, A.; Palmqvist, B.; Lidén, G. Effects of Agitation on Particle-Size Distribution and Enzymatic Hydrolysis of Pretreated Spruce and Giant Reed. Biotechnol. Biofuels 2014, 7, 77. [Google Scholar] [CrossRef]
- Chen, Z.; Hay, J.N.; Jenkins, M.J. FTIR Spectroscopic Analysis of Poly(Ethylene Terephthalate) on Crystallization. Eur. Polym. J. 2012, 48, 1586–1610. [Google Scholar] [CrossRef]
- Hagler, A.N.; Mendonça-Hagler, L.C. Yeasts from Marine and Estuarine Waters with Different Levels of Pollution in the State of Rio de Janeiro, Brazil. Appl. Environ. Microbiol. 1981, 41, 173–178. [Google Scholar] [CrossRef]
- Souza, C.E.C.; Farias, M.A.; Ribeiro, B.D.; Coelho, M.A.Z. Adding Value to Agro-Industrial Co-Products from Canola and Soybean Oil Extraction Through Lipase Production Using Yarrowia lipolytica in Solid-State Fermentation. Waste Biomass Valorization 2017, 8, 1163–1176. [Google Scholar] [CrossRef]
- Farias, M.A.; Valoni, E.A.; Castro, A.M.; Coelho, M.A.Z. Lipase Production by Yarrowia lipolytica in Solid State Fermentation Using Different Agro Industrial Residues. Ital. Assoc. Chem. Eng. 2014, 38, 301–306. [Google Scholar] [CrossRef]
- Pereira-Meirelles, F.V.; Rocha-Leão, M.H.M.; Anna, G.L.S. A Stable Lipase from Candida lipolytica: Cultivation Conditions and Crude Enzyme Characteristics. Appl. Biochem. Biotechnol. 1997, 63–65, 73–85. [Google Scholar] [CrossRef] [PubMed]
- Charney, J.; Tomarelli, R.M. A Colorimetric Method for the Determination of the Proteolytic Activity of Duodenal Juice. J. Biol. Chem. 1947, 171, 501–505. [Google Scholar] [CrossRef]




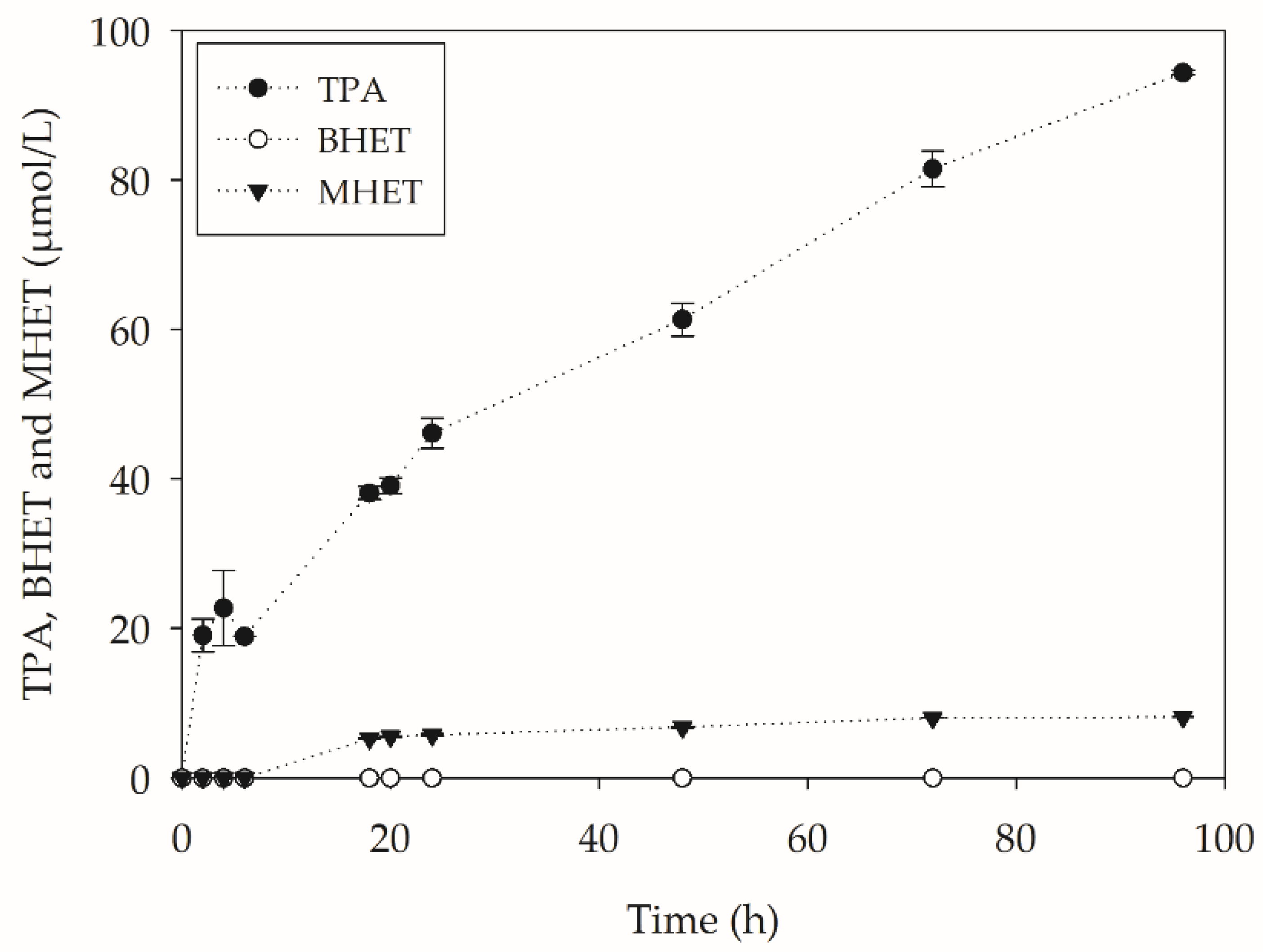
| Assay | χtpa | χbhet | χmhet |
|---|---|---|---|
| CALB + BHET | 1 | 0 | 0 |
| SSF-YL + BHET | 0.084 | 0.284 | 0.632 |
| Assay | χtpa | χbhet | χmhet | Conversion (%) |
|---|---|---|---|---|
| CALB + PC-PET | 1 | 0 | 0 | 0.65 |
| SSF-YL + PC-PET | 0.618 | 0.182 | 0.201 | 1.64 |
| Assay | µ (h−1) | tg (h) | Xmax (g L−1) | Qx (g L−1 h−1) |
|---|---|---|---|---|
| YP | 0.271 | 2.56 | 10.45 | 0.580 |
| YT | 0.262 | 2.65 | 10.08 | 0.560 |
| YPD | 0.302 | 2.29 | 20.54 | 0.716 |
| YTD | 0.304 | 2.28 | 17.03 | 0.618 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sales, J.C.S.; de Castro, A.M.; Ribeiro, B.D.; Coelho, M.A.Z. Post-Consumer Poly(ethylene terephthalate) (PET) Depolymerization by Yarrowia lipolytica: A Comparison between Hydrolysis Using Cell-Free Enzymatic Extracts and Microbial Submerged Cultivation. Molecules 2022, 27, 7502. https://doi.org/10.3390/molecules27217502
Sales JCS, de Castro AM, Ribeiro BD, Coelho MAZ. Post-Consumer Poly(ethylene terephthalate) (PET) Depolymerization by Yarrowia lipolytica: A Comparison between Hydrolysis Using Cell-Free Enzymatic Extracts and Microbial Submerged Cultivation. Molecules. 2022; 27(21):7502. https://doi.org/10.3390/molecules27217502
Chicago/Turabian StyleSales, Julio Cesar Soares, Aline Machado de Castro, Bernardo Dias Ribeiro, and Maria Alice Zarur Coelho. 2022. "Post-Consumer Poly(ethylene terephthalate) (PET) Depolymerization by Yarrowia lipolytica: A Comparison between Hydrolysis Using Cell-Free Enzymatic Extracts and Microbial Submerged Cultivation" Molecules 27, no. 21: 7502. https://doi.org/10.3390/molecules27217502
APA StyleSales, J. C. S., de Castro, A. M., Ribeiro, B. D., & Coelho, M. A. Z. (2022). Post-Consumer Poly(ethylene terephthalate) (PET) Depolymerization by Yarrowia lipolytica: A Comparison between Hydrolysis Using Cell-Free Enzymatic Extracts and Microbial Submerged Cultivation. Molecules, 27(21), 7502. https://doi.org/10.3390/molecules27217502







