Abstract
Metabolomics is an essential method to study the dynamic changes of metabolic networks and products using modern analytical techniques, as well as reveal the life phenomena and their inherent laws. Currently, more and more attention has been paid to the development of metabolic histochemistry in the fungus field. This paper reviews the application of metabolomics in fungal research from five aspects: identification, response to stress, metabolite discovery, metabolism engineering, and fungal interactions with plants.
1. Introduction
Metabolomics is an emerging omics technology following genomics, proteomics, and transcriptomics. The concept of metabolomics first originated from metabolomic profiling proposed in 1971 by Devaux et al. [1]. Metabolomics was put forward as a group of metabolites in organisms by Oliver et al. in 1998 [2]. Nicholson raised the concept of metabolomics on the basis of a statistical analysis of NMR spectroscopic data from mouse urine. These data are defined as “a quantitative measurement of the dynamic multiparametric metabolic response of living systems to pathophysiological stimuli or genetic modifications” [3]. Traditional metabolomics is divided into targeted metabolomics and untargeted metabolomics. Targeted metabolomics is the measurement of a defined set of chemically characterized and biochemically annotated metabolites, usually focusing on one or more relevant metabolic pathways [4]. Recently, it has been subdivided and further developed into widely targeted metabolomics, pseudotargeted metabolomics, quasi-targeted metabolomics, LM precision targeted metabolomics, etc. Although the above methods all use the MRM mode for mass spectrometric data acquisition, widely targeted metabolomics and pseudotargeted metabolomics are performed by qualitatively and relatively quantifying the target through substances in the local library (established on the basis of partial standards, untargeted data, literature data, etc.), LM precision targeted metabolomics can absolutely characterize the substances corresponding to all standards. Even in combination with external standard methods, absolute quantification of metabolites in a sample can be achieved. However, untargeted metabolomics analyzes all measurable metabolites in a sample. The aim was to measure metabolites in the samples whenever possible [5]. According to different research objects and purposes, metabonomics can be divided into four levels: metabolic fingerprinting analysis, metabolic target analysis, metabolic profiling, and metabolomics [6].
Metabolomics, through modern instrumental analytical methods with high throughput, sensitivity, and resolution, combined with chemometric methods, analyzes the change law of metabolites after stimulation or interference in biological systems. Its focus is more on small-molecule metabolites with relative molecular weights of less than 1000 in biological tissues or cells and is often used to study plant and microbial systems [7,8,9]. The existence time of fungi on the Earth is unknown, and no definite conclusions can be drawn about their origin. Fungal cells do not contain chloroplasts and plastids; they are typical heterotrophs with parasitic or saprophytic patterns. The latest research speculates that there are as many as six million species of fungi worldwide, of which more than 600 are closely related to humans. They can participate in the formation of the human micro-ecosystem as resident fungi or cause diseases as pathogens. Fungi profoundly affect human health, agriculture, biodiversity, natural ecology, industry, biomedicine, etc. [10,11,12]. The application of metabolomics to different research fields of fungi, such as the classification and identification of fungi, metabolic pathways of fungi, the discovery of fungal natural products, and plant–fungal interactions [13,14,15,16,17,18], can help researchers entirely mine the potential of fungi. This article mainly reviews the research methods of metabolomics and the latest progress of metabolomics in various research fields of fungi (Table 1), aiming to promote further research on fungal metabolomics.

Table 1.
Application of metabolomics in fungal research in recent years.
2. Fungal Metabolomic Approaches
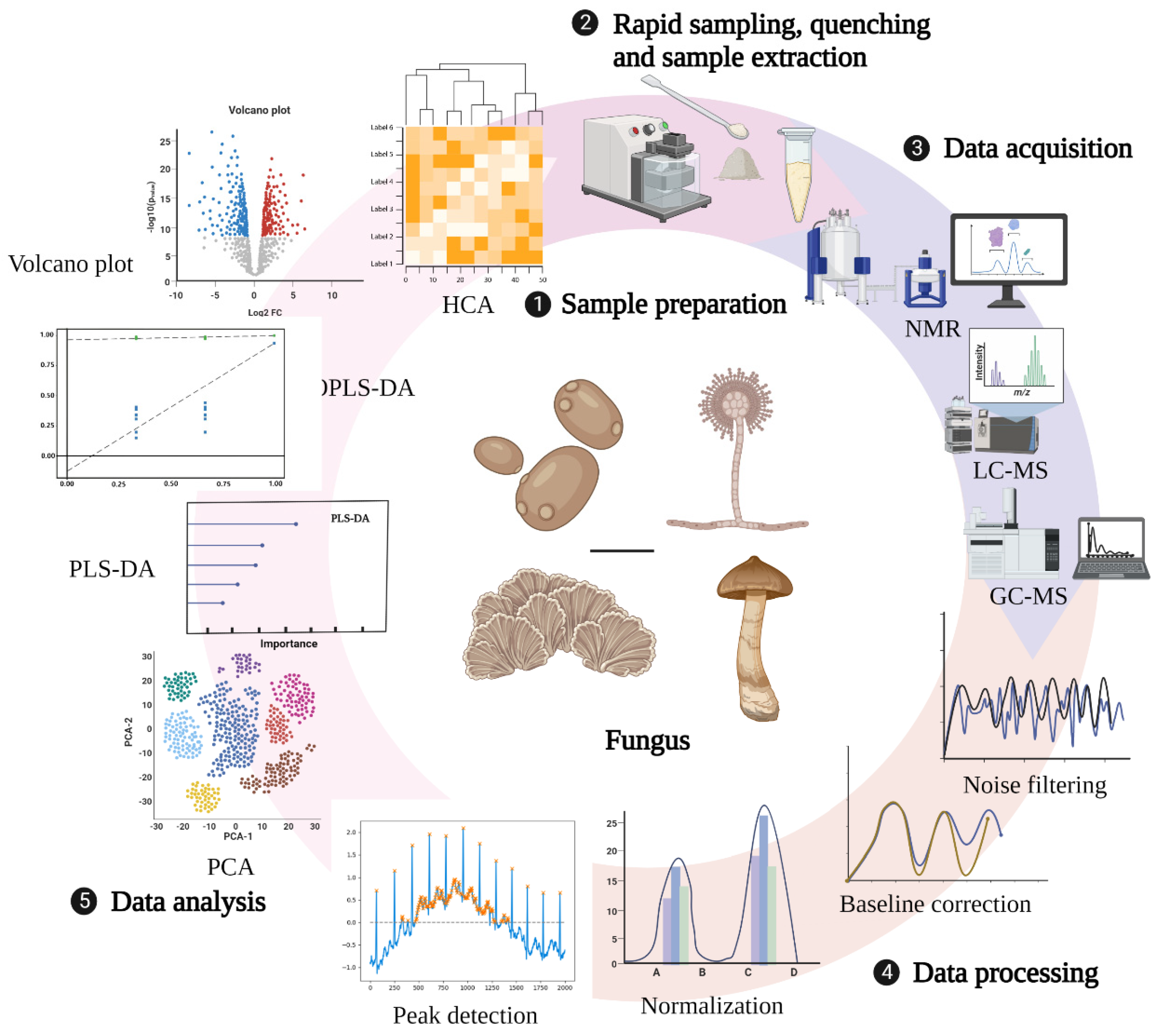
The technical route for metabolomic research in fungi mainly involves three main processes: sample preparation, data collection and processing, and analysis (Figure 1). Sample preparation can affect not only the observed metabolite content but also the biological interpretation of the data. Therefore, appropriate sample collection and preparation steps are required to avoid interfering with the efficient metabolomics analysis. Currently, studies on sample preparation in biological fluids, tissues, mammalian cells, and plants have been relatively comprehensive, but there are few reviews related to fungal sample preparation strategies [61,62,63,64]. An ideal sample preparation method for metabolomics should be as simple, rapid, highly selective, and reproducible as possible and capable of quenching to determine the actual metabolic composition at the sampling time. General sample preparation steps include rapid sampling, quenching, and sample extraction [65].
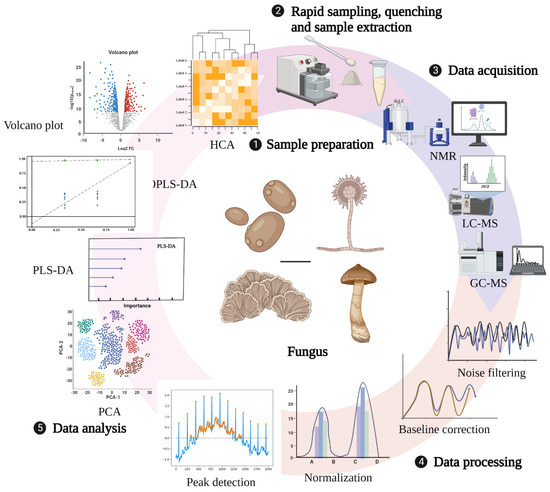
Figure 1.
General technical route of metabolomics. Created with BioRender.com (accessed on 28 October 2022).
2.1. Rapid Sampling
Rapid sampling from fermentation tanks or shake flasks is the first step in performing fungal sample preparation, since 1969 when Harrison et al. first attempted rapid sampling from small-scale laboratory bioreactors [66], to Iversen and Theobald et al., who developed iterations of sampling systems with minimal dead volume for manual feeding [67,68]. Fast sampling systems are currently characterized by motorized sampling, high frequency, negligible dead areas, and efficient inactivation [69,70,71]. The technology developed by van Gulik used adenine nucleotide as an indicator to analyze its dynamic response to changing glucose concentration and quickly sample yeast metabolites. It has a very high sampling frequency, which can also ensure long-term sterility [72]. Although the sampling method developed by Hannes Link et al., which directly injects fungi such as yeast into high-resolution mass spectrometers for real-time metabolomic analysis, achieves automated detection of target compounds [73], this approach is expensive and impractical for the majority of researchers.
2.2. Quenching
The purpose of quenching is to rapidly stop various metabolic activities within the cells, inactivate enzymes, keep the metabolite content stable, and reduce the degradation of metabolites. The cold methanol quenching method is a standard method used in the past, and the research on cell quenching mainly focused on different methanol content and quenching temperature. Specifically, 60% (v/w) methanol quenching at −40 °C is one of the standard methods for microbial metabolomics research. However, this method causes the leakage of cellular metabolites [74,75]. So far, cold methanol use for quenching has remained a controversial topic. Due to the advantage of a high extraction efficiency of representative intracellular metabolites by cold methanol quenching, some researchers have continuously improved the original method of cold methanol quenching proposed by de Koning et al. According to the degree of metabolite leakage depending on the exposure time, temperature, and nature of the methanol solution, under fast sampling equipment, setting the sample/quencher liquid (v/v) ratio to 1:5 or less for quenching in pure methanol at ≤−40 °C effectively prevents metabolite leakage [76]. Of course, other quenching methods are also under continuous development. Rapid filtration of liquid nitrogen is more suitable for cell quenching than cold methanol quenching, with minimal damage to cell integrity and improved recovery of intracellular metabolites [77]. However, ice crystals produced by the freezing of liquid nitrogen can potentially damage the cell membrane, which also leads to the leakage of metabolites inside the cell. Quenching using strong acids such as perchloric acid can lead to the degradation of some compounds in a strong acid environment with severe metabolite reduction [78]. It seems that the perfect quenching method is challenging to achieve. Although Meinert et al. believe that the quenching method of methanol quenching solution (60%, −40 °C) has no metabolite leakage, we can still find that the metabolite investigated using this method is insufficient and far from reaching the level of no leakage for all detection indicators [79]. Moreover, most of the quenching studies focused on Gram-positive bacteria [80], Gram-negative bacteria [81], yeast cells [82,83], etc., whereas research on filamentous fungi is rare.
2.3. Sample Extraction
How to extract the quenching sample is a critical step in the sample preparation stage. Furthermore, the ideal extraction should not change the metabolites’ physical properties and chemical structure and should maximize the sequestration of the metabolite content. Current extraction methods can presumably be grouped into physical and chemical categories. Physical methods often use homogenizers, ultrasonic, microwave, and other tools. Chemical methods mostly use acid, base, water, methanol, ethanol, and different ratios of water and organic solvents, mixed solvents, etc. Researchers can choose different solvents depending on different samples. As mentioned above, strong acids and bases can cause severe metabolite leakage resulting in a low recovery rate of final metabolites. Although this is a classical extraction method, using strong acids and bases such as perchloric acid is still not recommended. Solvents with the highest extraction efficiency can be selected experimentally. Three sample preparation methods and five solvent mixtures of Mortierella alpina were evaluated using gas chromatography/mass spectrometry (GC–MS) [84]. The results showed better reproducibility and recovery of lyophilized. Methanol/water (1:1) was more effective in extracting metabolites of Mortierella alpina. Compared with biphasic extraction at different pH with methanol extraction, which is easy and fast and suitable for the extraction of metabolites from Phanerochaete chrysosporium, biphasic extraction at different pH is more suitable for target analysis [85]. Of note, despite the high efficiency and recovery of metabolites extracted by supercritical fluids, partially unstable metabolites may undergo decomposition due to the pressure ranging between 200 and 500 bar [86].
2.4. Instrumental Analysis Methods
Commonly used metabolomics analysis methods require collecting raw data after sample quenching and extraction. Currently, several analytical methods exist for qualitative and quantitative analysis of metabonomic extracts in metabonomic research. The commonly used analytical methods for fungal metabolomics include gas chromatography/mass spectrometry (GC–MS), liquid chromatography/mass spectrometry (LC–MS), nuclear magnetic resonance spectroscopy/mass spectrometry (NMR–MS), capillary electrophoresis/mass spectrometry (CE–MS), and matrix-assisted laser desorption ionization mass spectrometry (MALDI-MS). GC–MS is often used to analyze substances with excellent thermal stability and volatility, allowing simultaneous analysis of sugars, amino acids, phosphorylated metabolites, organic acids, lipids, amines, and other compounds. It exhibits extraordinary robustness, excellent separation ability, high selectivity, effective sensitivity, and reproducibility [87]. However, there are also some disadvantages, as nonvolatile compounds require derivatization. The main derivatization methods are silanization, acylation, alkylation, and esterification. Koek et al. established a GC–MS spectrometry metabonomic analysis technology suitable for microorganisms and verified the method with various microorganisms [88]. The results showed that the method has good repeatability, effective reproducibility, and fast linear regression characteristics. It can be used for the metabonomic analysis of various components of microorganisms, such as alcohols, aldehydes, amino acids, fatty acids, organic acids, sugars, purines, pyrimidines, and aromatic compounds. HPLC reduces the complexity of samples and offers several advantages, such as simple preparation, high sensitivity, signal reproducibility, minimal noise, and high qualitative and quantitative ability. It is helpful for thermally labile compounds, nonvolatile compounds, polar compounds, and compounds that are macromolecules. With the development of high-performance liquid chromatography (HPLC) and ultra-performance liquid chromatography (UHPLC), the resolution of peaks was improved, and the speed of analysis was accelerated [89]. NMR techniques have the advantages of high reproducibility, accurate quantification, simple sample preparation, measurable analytes in various solvents, clear identification of unknown metabolites, and complete metabolite detection. The disadvantage is low sensitivity, which severely limits the use of NMR in metabolomics [90]. Capillary electrophoresis techniques are relatively new and less applied analytical methods, mainly for studying molecules, but they are preferred when dealing with highly polar, charged metabolites [91]. They allow rapid and high-resolution analysis of charged metabolites such as nucleic acids, amino acids, carboxylic acids, and sugar phosphates. Each analytical tool has advantages and disadvantages. A single analytical platform tool cannot directly and precisely characterize or quantify thousands of small-molecule metabolites involved in fungal metabolic processes. The right combination of tools is often needed depending on the experimental situation to better analyze the target fungi.
2.5. Data Processing and Analysis Methods
The original data obtained by the analytical instrument cannot provide a clean and comparable metabolite spectrum. Therefore, the original data must be preprocessed and generally completed in the experimental system. This mainly includes noise reduction and baseline correction, peak detection and deconvolution, normalization, and data standardization [92,93]. The classical analysis method is to use a single variable, i.e., parameter by parameter, or to use multivariable techniques to evaluate group differences. Although the univariate analysis method is simple and convenient, it cannot accurately distinguish the groups when the difference is small. Multivariate analysis can be used to analyze the changes in single metabolites between different groups and the dependent structure of individual molecules [94]. Multivariate analysis can be divided into two categories; one is the unsupervised learning method, which classifies the original data directly, including principal component analysis (PCA), hierarchical clustering analysis (HCA), and self-organizing maps (SOMs). The other is the supervised learning method, i.e., learning the training samples with a given sample label, such as partial least squares discriminant analysis (PLS-DA), partial least squares discriminant analysis based on orthogonal signal correction (OPLS-DA), artificial neural network (ANN), and support vector machine (SVM). Among them, PCA, PLS-DA, and OPLS-DA are the most frequently used multivariate statistical analysis methods in the field of metabolomics [95,96,97,98]. The generally used analysis software includes MetAlign [99], MZmine [100], XCMS [101], Metabolomic Analysis and Visualization Engine (MAVEN), Metabolite Biological Role (MBRole), MetaCoreTM, MetaboAnalyst, InCroMA, and 3Omics [102,103,104].
3. Application of Metabolomics in the Field of Fungal Research
3.1. Application of Metabolomics in Classification and Identification of Fungal Research
Morphological methods, as the traditional fungal classification method [105], have some limitations, i.e., the classification from appearance characteristics is affected by the fungal growth environment, similar morphology, and other factors, thus affecting the accuracy of classification. Genomic DNA/DNA hybridization [106,107], ribosomal typing [108,109], multilocus sequence typing [110,111], ITS rDNA sequences [112], and lipid profile analysis [113] are popular methods in fungal taxonomic identification. Chemical taxonomy was initially considered complementary to morphological methods based on primary and, more often, secondary metabolites. However, with advances in HPLC and mass spectrometry, the application of metabolomic chemotaxonomic in fungal taxonomic identification has progressed considerably (Table 2). Kang et al. analyzed the secondary metabolites of seven species of Trichoderma (33 strains) using its sequence and metabolome-based chemotaxonomic comparison. They found that the chemical taxonomy based on secondary metabolites was more accurate than its sequence and identified an unknown group of Trichoderma [114]. Chen used HPLC fingerprinting combined with stoichiometric analysis of Ganoderma lucidum fruiting bodies and screened four marker components as discriminative variables to distinguish Ganoderma lucidum [115]. However, Wen et al. believed that the identification of distinguishing Ganoderma lucidum with the NMR metabolomics method was less time-consuming and faster, which was in line with the quality control of large-scale production. Through NMR metabolomics, labeling choline and propionic amino acids as discriminating variables not only successfully differentiated between Chinese and Korean Ganoderma lucidum but also differentiated the cultivated origin of Chinese Ganoderma lucidum [116]. This method of taxonomic identification based on the specific metabolites of fungal species effectively avoids the limitation of low accuracy of traditional methods and identifying fungi from different regions or even different growth stages.

Table 2.
Metabolomics for taxonomic identification of fungi.
Nevertheless, metabolome-based chemotaxonomy also has drawbacks. First, how to overcome the problem of finding specific markers from a plethora of metabolites for optimal biological interpretation is still unanswered. Second, changes in various environmentally relevant regulatory genes may not affect the expression of taxonomic-related genes. Lastly, while it is desirable to analyze metabolites of specific organelles, how organelles can be isolated while maintaining a structural, metabolic state remains unattainable.
3.2. Application of Metabolomics in the Study of Fungal Response to Stress
Fungi can survive only under various stress reactions such as ionizing radiation, hydration activity, acid–base environment, hypoxic stress, solar ultraviolet radiation, agricultural and industrial pollutants, biological stress, nutrient stress, oxidative stress, heat stress, and cold stress to survive. Ecological metabolomics studies changes in endogenous metabolites produced by biological systems that are affected by environmental factors [123]. Using metabolomics to study fungi, we can understand how fungi respond to stress when they grow or infect their hosts. It is helpful to optimize the application of fungi in biotechnology, improve the environment, and even prevent fungal diseases.
Environmental stress, i.e., abiotic stress, is usually the most severe situation faced by fungi. Oliveira et al. found that the nutrient content of low-molecular-weight metabolites of wild mushrooms is higher than that of indoor cultivated ones [19], perhaps because the functions of the low-molecular-weight metabolite gene clusters of these mushrooms and their relative expression have differences in the two environments. Metabolomic analysis of the white rot basidomycete Phanerochaete chrysosporium under air and 100% oxygen revealed that three metabolites associated with the oxygen stress response were veratryl alcohol (VA), threonate, and erythronate. High concentrations of ROS can directly activate the VA synthesis pathway, and the intracellular oxygen concentration is significantly elevated. In contrast, threonate and erythronate resistance to hyperoxia is a process of progressive gradient accumulation [20]. When the fungal Alternaria sp. MG1 grown on grape interiors was subjected to starvation treatment, the shikimate pathway and the phenylpropanoid (PPPN) pathway were strongly activated, and relevant metabolites such as resveratrol were significantly upregulated [21]. The metabolic profiles of Cryptococcus neoformans changed under Cu stress, and the differential metabolites were mainly related to the metabolism of amino acids and carbohydrates. Replacing the carbon source with glycerol and ethanol can counteract the toxic effect of copper on Cryptococcus neoformans and improve urea clearance [22]. Yan et al. analyzed the metabolites of Pleurotus ostreatus under different heat stress times (6, 12, 24, and 48 h) for dynamic metabolite changes. They found that the contents of metabolites such as amino acids, nucleotides, and lipids showed an increasing trend with increased heat stress time [23]. Zhao et al. found that Volvariella volvacea showed little resistance to low temperatures. Nevertheless, under chilling stress, the relative levels of compounds such as amino acids and organic acids inside Volvariella volvacea increase significantly, and soluble sugars such as sorbitol are induced to be produced, improving its osmoregulatory capacity [24]. Interestingly, Aspergillus aculeatus, under drought and heat stress, increased the accumulation of amino acids and sugars and enhanced the total photosynthesis of tall fescue, resulting in a vastly improved ability of tall fescue inoculated with Aspergillus aculeatus to resist cold and heat stress [25].
Aspergillus flavus showed significant changes in carbohydrates, sulfur-containing amino acids and their derivatives, fatty acids, etc. under drought stress [124]. 1-Nonanol can destroy the integrity of the cell membrane in Aspergillus flavus and affect mitochondrial function, which induces apoptosis in Aspergillus flavus [26]. Aspergillus niger resisted copper stress by converting sorbitol from glucose to produce a large amount of mannitol [27]. When Aspergillus niger was exposed to 5% ethanol stress, its growth amount was about 70% less than that under normal growth conditions [28]. Using untargeted metabolomics to study its reaction mechanism, it was found that TAG, DAG, and hTAGs significantly accumulated. These neutral glycerolipids were previously believed to be associated with the fungi’s exposure to abiotic stress factors [125,126]. Whether glycerides in the response of Aspergillus niger strain Es4 to ethanol stress can be used as a new response of the fungus to ethanol stress still needs further confirmation. Hammerl et al. further established a differential offline LC–NMR (DOLC–NMR) method to qualitatively and quantitatively analyze metabolic changes in Penicillium roqueforti when L-tyrosine levels are perturbed. Twenty-three metabolites were affected by the amino-acid perturbation method, among which the amino-acid degradation products 2-(4-hydroxyphenyl) acetic acid and 2-(3,4-dihydroxyphenyl) acetic acid were significantly upregulated [127]. Jiang et al. found that treatment of Ganoderma lucidum with methyl jasmonate (MeJA) for 24 h was the optimal condition to induce the biosynthesis of Ganoderma lucidum. MeJA induction can lead to metabolic rearrangements in Ganoderma lucidum, inhibit its normal glucose metabolism, energy supply, and protein synthesis, and promote cellular secondary metabolic production [29]. After treatment with tributyltin (TBT), the mycelial morphology of Cunninghamella echinulata changed, the metabolic activity was inhibited, and glycolysis and the TCA cycle were dysregulated. This fungus can eliminate the hazard of tributyltin compounds to the organisms by accumulating amino acids with antioxidant functions, such as betaine, proline, and GABA, to recover from the toxic TBT environment [30].
In contrast, benzoic acid derivatives such as sodium benzoate, p-aminobenzoic acid, and p-methylbenzoic acid all promote lipid synthesis in Penicillium sr21. Furthermore, 200 mg/L p-aminobenzoic acid even promoted glucose catabolism during glycolysis, increased the mevalonate pathway, weakened the tricarboxylic acid cycle, and promoted the production of tetrahydrofolate and NADPH [31]. The antibacterial polycationic peptide ε-poly-l-lysine (ε-PL) enhances the freeze–thaw tolerance of industrial yeast by promoting cell membrane-associated fatty-acid synthesis before freeze–thaw and promoting alglucan biosynthesis and glycerophospholipid metabolism after freeze–thaw [32]. From the above studies, it is easy to see that both biotic and abiotic stresses involve a variety of metabolic pathways in fungi, the most important of which are the metabolism of amino acids and their derivatives, glycolytic pathways, etc. Significant marginal changes in the levels of metabolites such as sugars, nucleotides, and lipids are the main mechanisms of their dynamic regulation.
3.3. Application of Metabolomics in the Discovery of Fungal Metabolites
A large number of metabolites such as primary and secondary metabolites exist in fungi. Primary metabolites are monomers synthesized by primary metabolisms, such as monosaccharides or monosaccharide derivatives, nucleotides, vitamins, amino acids, fatty acids, and various macromolecular polymers composed of them, including proteins, nucleic acids, polysaccharides, lipids, and other essential substances. Secondary metabolites refer to substances synthesized by fungi in which primary metabolites serve as precursors with no clear function in their life activities, such as gibberellins, penicillins, aflatoxins, and cordycepin [128]. Fungi can provide diverse and unique secondary metabolites, making them potential drug sources. Traditional assays can easily lead to the rediscovery of known compounds. With the advancement of analytical technology platforms, MS-based metabolomics workflows are mainly suitable for screening hundreds of natural products simultaneously for dereplication studies and extractions of bioactive compounds, which is of great benefit for the comprehensive exploration of potentially useful secondary metabolites. In addition to drug discovery, screening of bioactive compounds or discovery of unknown fungal metabolites that play critical roles in host fungal interactions are also required to identify fungal secondary metabolites. A metabolomics approach can aid in discovering and detecting novel metabolites in fungi (Table 3).

Table 3.
Novel metabolites discovered and detected using metabolomics.
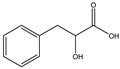
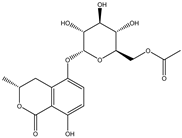
Cordycepin is considered to be an important marker for the identification of Cordyceps sinensis. While nucleosides such as thymine, uracil, adenine, and guanosine are also the main substances used by researchers to identify and analyze Cordyceps sinensis. Mishra reidentified the nucleobases in samples of Ganoderma lucidum and Cordyceps sinensis by HPLC–MS and found that both had abundant nucleosides. Furthermore, the water extract of Ganoderma lucidum and the ethanolic extract of Cordyceps sinensis had the highest nucleobase content [33]. Joshi et al., for the first time, identified the presence of cordycepin using ion mobility mass spectrometry (IMMS), which provided a new method for identifying oridonin [34]. The growth of Cordyceps pupae can be divided into the first to the third stage of growth and the fourth stage of senescence. Principal component analysis found an obvious separation between the first and fourth stages of cordycepin, indicating that cordycepin was significantly enriched in the senescence stage of fruiting bodies [35]. Furthermore, cordycepin, the contents of amino acids and carbohydrates such as glucose, xylitol, and mannose were also obviously increased. The biosynthesis of cordycepin may be regulated by the glutamine and glutamate metabolic pathways. Although Cordyceps militaris and Cordyceps sinensis belong to the same family as Clavicipitaceae, and Cordyceps militaris is even called “northern Cordyceps sinensis” in China, they have drastically different nutritional contents, which Chen et al. confirmed from the metabolite level. The results of utilizing LC–MS technology to analyze natural Cordyceps sinensis and artificial cultured Cordyceps militaris showed significant metabolomic differences between them [36]. Similarly, the chemical composition of Cordyceps sinensis and Cordyceps militaris cultured with tussah pupae was compared, and 25 differential metabolites were found, involving 16 metabolic pathways such as histidine metabolism. Cordyceps sinensis has many healthy nutrients, especially amino acids, unsaturated fatty acids, peptides, and mannitol. Moreover, the superior hemostatic activity and the antioxidant capacity of Cordyceps sinensis cultured with tussah pupae suggest its extreme clinical value as an affordable alternative to oridonin [37]. Cordyceps militaris strains were inoculated on germinated soybean (GSC), and the yield and biological activity of GSCs reached the highest after 1 week. Compounds 1–4, which were highly abundant in GSCs, were identified as four novel isoflavone methyl glycosides (daidzein 7-o-β-D-glucoside 400-o-methide, glycitein 7-o-β-D-glucoside 400-o-methide, genistein 7-o-β-D-glucoside 400-o-methide, and genistein 40-o-β-D-glucoside 400-o-methide) [38]. Apparently, mixed coculture is a good way to improve the nutrients of Cordyceps militaris. Coculture of fungi is often beneficial to induce purposeful fungal differentiation, affect the content of metabolites, and produce multiple metabolic pathways. Coculture of Coriolus versicolor and Ganoderma lucidum with 62 newly synthesized or high-yielding features compared to monoculture. Two new xylosides (compounds 2 and 3) were included. Compound 2 was further identified as N-(4-methoxyphenyl) carboxamide 2-O-β-D-xyloside, which increases the viability of the BEAS-2B human immortalized bronchial epithelial cell line. 3-Phenyllactic acid and orsellinic acid were first detected in malate bacilli [39]. However, fungal interactions also produce antagonistic inhibition. The coculture of Aspergillus oryzae and Zygosaccharomyces rouxii reduced the amounts of imidazoleacetic acid, phenylpyruvic acid, and n-formyl-l-aspartic acid, taurine, and glycolic acid [40]. Obviously, coculture inhibited the growth of Zygosaccharomyces rouxii.
Agaricus bisporus is a worldwide edible mushroom. The surface browning of mushrooms is one of the major factors affecting consumers’ purchase. The nutritional value of Agaricus bisporus changed after UV irradiation, with 47 compounds increasing in concentration and 72 compounds decreasing in concentration [131]. Looking at the difference between browning tolerant cultivars and common Agaricus bisporus cultivars at the metabolic level, genes such as AbPPo were found to be involved in the regulation of mushroom browning, and higher levels of organic acids, such as butyric acid, were found in brown tolerant Agaricus bisporus cultivars [41]. In addition, the pH value and alginate content concentration also affected the activity of AbPPo. Lower pH levels inhibited the expression of AbPPo, and high alginate concentrations may be beneficial for maintaining the activity of AbPPo. The browning of filamentous mushrooms appears to be different. Yu et al. suggested that phenylpropanoid biosynthesis and tyrosine metabolism may promote the browning of filamentous fungi. In addition, dopa melanin accumulation may also be one of the causes for the browning of Flammulina velutipes [42].
Endophytic fungi are ubiquitous in the plants body and should be actively exploited to utilize these resources, whether harmful to the plant or not. Hawary et al. isolated a butenolide derivative from the endophytic soybean fungus Thra terreus, Aspergillide B1, as well as 3a-hydroxy-3,5-dihydromonacolin L [43]. Using computer-aided technology such as CADD, they suggested that Aspergillide B1 and 3a-hydroxy-3,5-dihydromonacolin L are promising candidates for the treatment of COVID-19. Nevertheless, the speculation is limited to computer-assisted approaches, it lacks pharmacological experimental validation, and whether it is effective remains to be proven. Tawfike et al. isolated the endophytic fungus Aspergillus flocculus from Markhamia Platycalyx, and the secondary metabolites cis-4-hydroxymellein, 5-hydroxymellein, diorcinol, bo-tryoisocoumarin A, and mellein had anticancer activity and inhibited the growth of the chronic leukemia cell line K562 3-hydroxymellein. Moreover, diorcinol can suppress sleeping sickness caused by the parasite Trypanosoma brucei [129]. Kamal et al. successfully predicted two compounds, clodospirone B and demethyl laciodilodine, with good anti-trypanosome effects from the endophytic fungus Lasiodiplodia theobromae [132].
The metabolites of fungi would change under different fermentation times. When the fermentation time is too short, the content of the target metabolites might not yet have peaked, whereas, when the fermentation time is too long, the target metabolites might have undergone decomposition. For the first time, Bu et al. showed that anthocyanins could be produced from fungi as a metabolite often thought to exist only in natural plants. They performed a comparative analysis of the metabolome of Aspergillus sydowii H-1 on the second and eighth days of fermentation and found significant differences in the production of five anthocyanins, the chalcone synthase gene, and cinnamic acid-4-hydroxylase gene, which may be associated with the synthesis of anthocyanins [130].
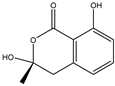
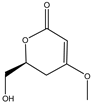
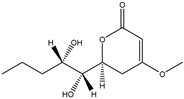
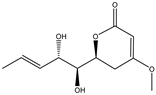
Fan et al. applied HRMS/MS feature-based molecular networking technology (FBMN) to determine Pyr enochaetopsis sp. They identified proteins A, B, and C in FVE-001 and protein D in FVE-087, four novel decaprenylspirotetraenoic acid derivatives with anti-melanoma activity [133]. FBMN has several functions in the identification and directional separation of stereoisomers. They combined UPLC–Q-TOF-MS with FBMN to discover three novel similar desferriferriferrichrome compounds [44] from wild Morchella sp. Le et al. used this approach to investigate the metabolomics of a strain of Penicillium mms417 isolated from blue mussel Mytilus edulis and obtained five new derivatives of natural fungus pyran-2-one derivatives: 5,6-dihydro-6S-hydroxymethyl-4-methoxy-2H-pyran-2-one, (6S, 1′R, 2′S)-LL-P880β, 5,6-dihydro-4-methoxy-6S-(1′S, 2′S-dihydroxy pent-3′(E)-enyl)-2H-pyran-2-one, 4-methoxy-6-(1′R, 2′S-dihydroxy pent-3′(E)-enyl)-2H-pyran-2-one, and 4-methoxy-2H-pyran-2-one [45]. Combining metabolomics and FBMN, compound features can be highlighted and clustered together to achieve efficient dereplication of compounds, greatly reducing the difficulty of discovering new metabolites.
3.4. Application of Metabolomics in Fungal Metabolic Engineering
Bailey defined metabolic engineering as “improving cellular activity by manipulating the enzymatic, transport, and regulatory functions of cells through the use of recombinant DNA technology” [134]. Fungi exhibited a variety of capabilities in industrial applications, including organic acid fermentation, protein production, and secondary metabolism. Advances in genome engineering have expanded the range of potential applications for fungal bioproduction. The development of genetic engineering tools is essential for efficiently utilizing genomic data. Currently, sequencing analyses of many filamentous fungi have revealed an underestimated potential, i.e., the presence of a large number of silent secondary metabolite genes. Metabolomics methods can be used to analyze changes in various metabolites of fungi after DNA recombination.
Huang et al. measured six major metabolites in the isoprene biosynthetic pathway using GC–SIM-MS and detected the changes after gene modification [46]. The roles of the erg9 and CoQ1 genes could be used as targets to aid in redirecting sterol precursors to the phosphorylated isoprenoid pathway. This approach could enhance the understanding of this pathway in many biological systems. To investigate the effect of histone deacetylase activity (HDACi) on the model fungus Aspergillus nidulans, Albright et al. analyzed the changes in more than 1000 small molecules secreted by Aspergillus nidulans. They found that almost the same number of compounds were upregulated and downregulated more than 100-fold after genetic or chemical reduction of HDACi [47]. Fellutamides, the natural product of Aspergillus nidulans, were first detected as a proteasome inhibitor that can be expressed about 100-fold or more upon HDACi induction. When using ionic liquids to stimulate Aspergillus nidulans, choline upregulated the primary metabolism of Aspergillus nidulans, while 1-ethyl-3-methylimidazolium chloride downregulated the primary metabolism, both of which stimulated the production of acetyl CoA and nonproteinogenic amino acids. Twenty-one of 66 known skeleton genes were upregulated [48].
Interactions between fungi and bacteria cause metabolic modifications in fungi. After undergoing in vitro confrontation culture, the metabolome changes of Fusarium verticillioides were much greater than those of Streptomyces sp. Compared with monoculture, many metabolites of Fusarium verticillioides were overproduced under resistant conditions compared to Streptomyces sp., especially 16 proteinogenic amino acids, inosine, and uridine, which means that the corresponding rate of protein synthesis would be slowed down, resulting in slower growth and less toxigenesis of Fusarium verticillioides [49]. Both the environment and the pH affect the biosynthesis of mycotoxins from Fusarium verticillioides. After targeted disruption of Fusarium verticillioides by the pH-responsive transcription factor PAC1, pH and PAC1 interference were found to affect the biosynthesis of arabitol, mannitol, and trehalose. Trehalose biosynthesis is reduced in PAC1-impaired plants. All three genes are downregulated when PAC1 is perturbed [50]. Using a Fusarium graminearum strain deficient for the H3K27 methyltransferase kmt6 to assign metabolites to genes, Ampressa et al. isolated large amounts of fusaristatin A, gibepyrone A, and fusarpyrones A and B from kmt6 mutants by activating silent metabolic pathways through mutations in repressive chromatin modifying enzymes. Triterpenones and trioctanoic acid were found in kmt6fus1 double mutants [51]. GC–EI-MS-based metabolomics has proven to be effective in unraveling the effects of genetic engineering and fungicide toxicity on fungal metabolism. Liu et al. studied the metabolism of Fusarium graminearum strains producing low toxins using a metabolome approach based on NMR and GC–MS and found new possible bactericidal targets [52]. The phenotypic observation and significance of nucleobase transporters in Aspergillus nidulans tolerance to Boscalid were validated by kalampokis et al. through metabolomic analysis of various biosynthetic pathways and metabolites [53].
Relative to other microorganisms, fungi are more classified. Furthermore, filamentous fungi are quite different from fungi such as yeasts in terms of growth mode and genetic characteristics. Multinucleated filamentous fungi are prone to heterokaryotic transformant phenomena. Thus, gene editing on filamentous fungi requires rapid and efficient manipulation techniques. Metabolomics and the construction of metabolic networks benefit the optimization and improvement of fungal metabolic pathways. Compared with transcriptomics and proteomics, metabolomics is able to more keenly analyze the effects of environmental perturbations or stresses on cells. Because there are cases where environmental alterations do not affect changes at the cellular transcriptional or protein level but can be manifested by metabolites.
3.5. Application of Metabolomics in the Field of Plant–Fungal Interaction
As mentioned above, endophytic fungi are ubiquitous in plants in nature. Some endophytic fungi have developed a mutually beneficial symbiosis with the hosts during a long period of evolution. They can regulate the hormone levels of plants, produce secondary metabolites similar to their hosts, assist the host plants in resisting environmental stresses, etc. Others are harmful fungi, and plant diseases caused by harmful fungi pose a significant threat to global food security. Understanding the interactions between fungi and plants is essential for preventing and controlling plant diseases. In the last decade, metabolomics technologies have been widely applied in various research fields on fungal plant interactions, such as identifying fungi, determining the mechanism of infection, and detecting the interaction between fungi and the host. The applications of metabolomics can help us to understand the pathogenesis and plant defense mechanisms of pathogenic fungi and develop effective prevention and treatment strategies for fungal diseases.
The rate of plant primary and secondary metabolite production is limited by its growth cycle, but endophytic fungi can promote the formation of metabolites from parasitic plants, as seen in Diaporthe phaseolorum (DP) and Trichoderma spirale (TS) during their symbiosis with Combretum lanceolatum. DP promotes the biosynthesis of primary metabolites such as threonine, malate, and N-acetylmannosamine of Combretum lanceolatum, which are metabolite precursors that have been shown to be bioprotective [54]. In the case of mutually beneficial symbiotic plants with fungi, plants can provide essential nutrients for fungal survival, and fungi mediate host plant defense responses to stresses such as environmental stress. When Pisolithus tinctorius was parasitized on cork oak roots, the contents of root exudates such as carbohydrates, organic acids, tannins, long-chain fatty acids, and monoacylglycerols were significantly decreased. In contrast, root defense substances such as γ-aminobutyric acid (GABA), a terpenoid, guarantee that the cork oak roots can control the proliferative range of Pisolithus tinctorius while symbiosing with Pisolithus tinctorius [55]. Phytohormones such as salicylic acid (SA) and jasmonic acid (JA) are endogenous regulators used by higher plants to defend against foreign pathogens [135,136]. Metabolic pathways are significantly different in soybean inoculated with Fusarium Verticillium compared to normal soybean. Flavonoid contents are significantly higher in soybean inoculated with molds, and Ja induces the synthesis of biomacromolecules such as glycine to enhance soybean resistance [56]. Moreover, mannitol, threitol and trehalose were significantly enriched in Armillaria luteobuablina-treated roots [57]. Exogenous threitol could promote the colonization of Armillaria luteobuablina in E. grandis roots and trigger hormonal responses in root cells, a phenomenon that was not detected in previous studies.
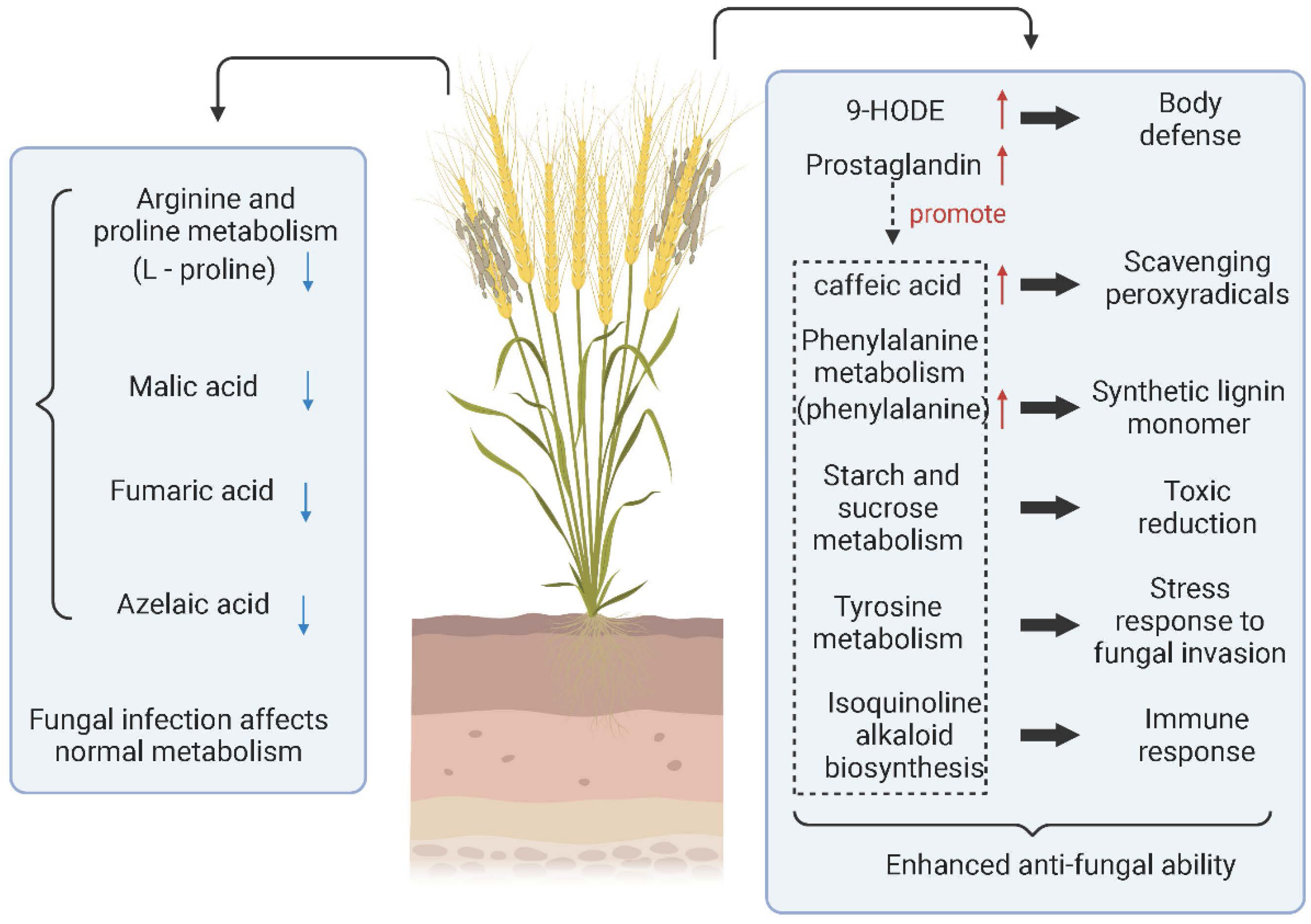
Fungal diseases are not only able to invade host plants initially, but they can also still invade again after rehabilitation. Mainly through spore germination or infecting the orifice through the mycelium, a few fungi can invade directly through the cuticle of plant tissue. Ren et al. performed LC–MS metabolomic analysis of grains infected with Tilletia controverta and normal grains (Figure 2). They found that the expression of 9-HODE, prostaglandin D3, caffeic acid, L-phenylalanine, and tetradecanoic acid was significantly upregulated. In infected samples, prostaglandin D3 was a coordinating factor to promote the increase of other metabolites involved in body defense. L-Phenylalanine promotes the synthesis of lignin monomers, caffeic acid, tetraacetic acid, etc., which are antifungal substances produced by cereals. In addition, the content of grain metabolites such as malate, L-proline, and fumarate decreased, indicating that the level of self-metabolism in Tilletia controverta-invaded wheat was inhibited [58]. In contrast, when Tilletia caries infested wheat, it did not directly change the metabolites of wheat. However, it prompted wheat to change the key metabolites and reduce the defense resistance function of wheat through pathways such as reducing the immune response to the sweet taste of wheat [137].
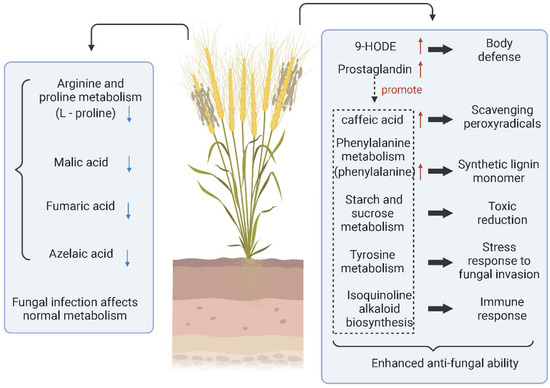
Figure 2.
Metabolite changes and metabolic pathways involved in wheat infected with Tilletia controverta. (↑) represents upregulation and (↓) represents downregulation. Created with BioRender.com (accessed on 17 May 2022).
Metabolomics is more conducive to developing rapid and effective drugs against fungal diseases than traditional chemical methods. In the past, most antifungal disease drugs were synthetic fungicides, based on the international new trend of environmental protection and green health. Developing natural antifungal components is a more reasonable choice. Chen et al. found that pinocembroside (PICB) isolated from Ficus hirta Vahl could significantly inhibit mycelia growth of Penicillium digitatum, a pathogenic bacterium of citrus green mold disease [59]. Metabolomics studies have shown that PICB alters the morphology of mycelium and Penicillium digitatum cells and promotes membrane peroxidation, which may be associated with the disruption of amino-acid metabolism, lipid metabolism, fatty-acid metabolism, TCA cycle, and purine metabolism. In addition, tomatoes were treated with the secondary metabolites 6-pentyl-2H-pyran-2-one and hartstic acid isolated from Trichoderma fungi, and metabolites were studied by HRMAS-NMR. Tomato samples treated with Trichoderma fungi secondary metabolites had significantly increased levels of acetylcholine, GABA, and amino acids [60]. These are well-known metabolites advantageous for plant growth, illustrating that developing antifungal disease drugs from a certain endophytic fungus is also a feasible approach.
4. Conclusions
As can be seen from the above analysis, metabolomics is a potent and effective tool. Through instrument analysis and data processing, we can gain insight into the changes of small-molecule metabolic components in test samples caused by biotic or abiotic factors. Furthermore, they can be associated with related metabolic pathways, metabolic networks, and metabolically related enzyme gene sets, transcriptomes, and proteomes. Metabolomics is widely used in fungal research and can provide a comprehensive and systematic analytical approach for fungal research. With the continuous development and improvement of sample preparation methods and analytical techniques, fungal metabolomics has made great progress in recent years. However, there are still some urgent issues to be solved in fungal metabolomics. For example, there is no standard method to quench and extract fungal metabolites, the data processing is complicated, and the automatic data processing platform technology is imperfect. The fungal metabolomics database is rare and incomplete, which is a crucial factor constraining the further development of fungal metabolomics.
On the other hand, although more than a few thousand metabolites have been identified, this is still only the tip of the iceberg for fungal metabolites. More importantly, many researchers still have an insufficient understanding of the metabolic pathways for fungal metabolomics and are only limited to primary and secondary metabolite studies in fungi. Compared with the application of metabolomics in disease diagnosis and drug research development, fungal metabolomics is still at an early stage of development. It is believed that metabolomics technology will be continuously improved with the continuous development of science and technology. Greater progress will also be made in fungal metabolomics studies. In conclusion, metabolomics has provided new insights into fungal research from different perspectives, which can be tightly integrated with other studies so that metabolic pathways, regulatory responses, and homeostatic mechanisms can be deeply investigated. This contributes to a better and deeper understanding of fungi’s complex interactions and their responses to environmental and genetic changes.
Author Contributions
Conceptualization, formal analysis, and writing—original draft, G.L.; visualization, T.J.; writing—review and editing, X.L.; investigation, Q.L.; funding acquisition, resources, supervision, and writing—review and editing, G.Z. and J.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (No. 21877075, 21807066). The authors also gratefully acknowledge the financial support of the Key Research and Development Program of Shandong Province (No. 2019GSF107003).
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
NMR, nuclear magnetic resonance; GC–MS, gas chromatography/mass spectrometry; UHPLC, ultrahigh-performance liquid chromatography; LC–MS, liquid chromatography/mass spectroscopy; PCA, principal component analysis; HCA, hierarchical clustering analysis; SOMS, self-organizing maps; PLS-DA, partial least squares discriminant analysis; OPLS-DA, partial least squares discriminant analysis; ANN, artificial neural network; SVM, support vector machine; MAVEN, Metabolomic Analysis and Visualization Engine; MBRole, Metabolite Biological Role; GAs, ganoderic acid; MeJA, methyl jasmonate; TCA, tricarboxylic acid; VA, veratryl alcohol; TBT, tributyltin; IMMS, ion migration mass spectrometry; HPTLC, high-performance thin-layer chromatography; GSC, germinated soybean cultivated; HDACi, histone deacetylase activity; VIE, Venturia inaequalis elicitor; PiCB, pinocembroside; FBMN, feature-based molecular networking technology; NADPH, nicotinamide adenine dinucleotide phosphate; ROS, reactive oxygen species; DAG, diacylglycerol; TAG, triacylglycerol; hTAGs, hydroxy triacylglycerol; DOLC-NMR, differential offline liquid chromatography/nuclear magnetic resonance; PPPN, phenylpropanoid; GABA, γ-aminobutyric acid; ε-PL, ε-poly-l-lysine; ABPPO, agaricus bisporus polyphenol oxidase; CADD, computer-aided drug design; GC–SIM-MS, gas chromatography/selective ion monitoring mass spectrometry.
References
- Devaux, P.G.; Horning, M.G.; Horning, E.C. Benzyloxime Derivatives of Steroids. A New Metabolic Profile Procedure for Human Urinary Steroids Human Urinary Steroids. Anal. Lett. 1971, 4, 151–160. [Google Scholar] [CrossRef]
- Oliver, S.G.; Winson, M.K.; Kell, D.B.; Baganz, F. Systematic functional analysis of the yeast genome. Trends Biotechnol. 1998, 16, 373–378. [Google Scholar] [CrossRef]
- Nicholson, J.K.; Lindon, J.C.; Holmes, E. ‘Metabonomics’: Understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica 1999, 29, 1181–1189. [Google Scholar] [CrossRef] [PubMed]
- Roberts, L.D.; Souza, A.L.; Gerszten, R.E.; Clish, C.B. Targeted Metabolomics. Curr. Protoc. Mol. Biol. 2012, 98, 30.2.1–30.2.24. [Google Scholar] [CrossRef] [PubMed]
- Patti, G.J.; Yanes, O.; Siuzdak, G. Innovation: Metabolomics: The apogee of the omics trilogy. Nat. Rev. Mol. Cell Biol. 2012, 13, 263–269. [Google Scholar] [CrossRef]
- Fiehn, O.; Kopka, J.; Trethewey, R.N.; Willmitzer, L. Identification of uncommon plant metabolites based on calculation of elemental compositions using gas chromatography and quadrupole mass spectrometry. Anal. Chem. 2000, 72, 3573–3580. [Google Scholar] [CrossRef]
- Nicholson, J.K.; Wilson, I.D. Opinion: Understanding ‘global’ systems biology: Metabonomics and the continuum of metabolism. Nat. Rev. Drug Discov. 2003, 2, 668–676. [Google Scholar] [CrossRef]
- Fernie, A.R.; Trethewey, R.N.; Krotzky, A.J.; Willmitzer, L. Metabolite profiling: From diagnostics to systems biology. Nat. Rev. Mol. Cell Biol. 2004, 5, 763–769. [Google Scholar] [CrossRef] [PubMed]
- Sumner, L.; Mendes, P.; Dixon, R. Plant Metabolomics: Large-Scale Phytochemistry in the Functional Genomics Era. Phytochemistry 2003, 62, 817–836. [Google Scholar] [CrossRef]
- Taylor, D.L.; Hollingsworth, T.N.; McFarland, J.W.; Lennon, N.J.; Nusbaum, C.; Ruess, R.W. A first comprehensive census of fungi in soil reveals both hyperdiversity and fine-scale niche partitioning. Ecol. Monogr. 2014, 84, 3–20. [Google Scholar] [CrossRef]
- Fisher, M.C.; Gow, N.A.R.; Gurr, S.J. Tackling emerging fungal threats to animal health, food security and ecosystem resilience. Philos. Trans. R. Soc. B Biol. Sci. 2016, 371, 20160332. [Google Scholar] [CrossRef] [PubMed]
- Fisher, M.C.; Henk, D.A.; Briggs, C.J.; Brownstein, J.S.; Madoff, L.C.; McCraw, S.L.; Gurr, S.J. Emerging fungal threats to animal, plant and ecosystem health. Nature 2012, 484, 186–194. [Google Scholar] [CrossRef] [PubMed]
- Tang, J. Microbial Metabolomics. Curr. Genom. 2011, 12, 391–403. [Google Scholar] [CrossRef] [PubMed]
- Mashego, M.R.; Rumbold, K.; De Mey, M.; Vandamme, E.; Soetaert, W.; Heijnen, J.J. Microbial metabolomics: Past, present and future methodologies. Biotechnol. Lett. 2007, 29, 1–16. [Google Scholar] [CrossRef]
- Kluger, B.; Lehner, S.; Schuhmacher, R. Metabolomics and Secondary Metabolite Profiling of Filamentous Fungi. In Biosynthesis and Molecular Genetics of Fungal Secondary Metabolites; Zeilinger, S., Martín, J.-F., García-Estrada, C., Eds.; Springer: New York, NY, USA, 2015; Volume 2, pp. 81–101. [Google Scholar]
- Keller, N.P. Fungal secondary metabolism: Regulation, function and drug discovery. Nat. Rev. Microbiol. 2019, 17, 167–180. [Google Scholar] [CrossRef]
- Chen, F.; Ma, R.; Chen, X.-L. Advances of Metabolomics in Fungal Pathogen–Plant Interactions. Metabolites 2019, 9, 169. [Google Scholar] [CrossRef]
- Jewett, M.C.; Hofmann, G.; Nielsen, J. Fungal metabolite analysis in genomics and phenomics. Curr. Opin. Biotechnol. 2006, 17, 191–197. [Google Scholar] [CrossRef]
- Silva, C.; Raisa Barbosa Cunha, J.; Almeida Conceição, A.; Gonzaga Almeida, E.; Cunha Zied, D.; Gonçalves Vieira Junior, W.; Souza Dias, E.; Isikhuemhen, O.S.; Verardi Abdelnur, P.; Gonçalves de Siqueira, F. Outdoor versus indoor cultivation: Effects on the metabolite profile of Agaricus subrufescens strains analyzed by untargeted metabolomics. Food Chem. 2022, 374, 131740. [Google Scholar] [CrossRef]
- Miura, D.; Tanaka, H.; Wariishi, H. Metabolomic differential display analysis of the white-rot basidiomycete Phanerochaete chrysosporium grown under air and 100% oxygen. FEMS Microbiol. Lett. 2004, 234, 111–116. [Google Scholar] [CrossRef][Green Version]
- Lu, Y.; Che, J.; Xu, X.; Pang, B.; Zhao, X.; Liu, Y.; Shi, J. Metabolomics Reveals the Response of the Phenylpropanoid Biosynthesis Pathway to Starvation Treatment in the Grape endophyte sp. MG1. J. Agric. Food Chem. 2020, 68, 1126–1135. [Google Scholar] [CrossRef]
- Sun, T.; Li, X.; Song, W.; Yu, S.; Wang, L.; Ding, C.; Xu, Y. Metabolomic alterations associated with copper stress in Cryptococcus neoformans. Future Microbiol. 2021, 16, 305–316. [Google Scholar] [CrossRef] [PubMed]
- Yan, Z.; Zhao, M.; Wu, X.; Zhang, J. Metabolic Response of to Continuous Heat Stress. Front. Microbiol. 2019, 10, 3148. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Chen, M.; Li, Z.; Zhao, Y.; Yang, H.; Zha, L.; Yu, C.; Wu, Y.; Song, X. The Response of to Low-Temperature Stress Based on Metabonomics. Front. Microbiol. 2020, 11, 1787. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Sun, X.; Feng, Q.; Luo, H.; Wassie, M.; Amee, M.; Amombo, E.; Chen, L. Comparative physiological and metabolomic analyses reveal mechanisms of Aspergillus aculeatus-mediated abiotic stress tolerance in tall fescue. Plant Physiol. Biochem. PPB 2019, 142, 342–350. [Google Scholar] [CrossRef]
- Zhang, S.-B.; Qin, Y.-L.; Li, S.-F.; Lv, Y.-Y.; Zhai, H.-C.; Hu, Y.-S.; Cai, J.-P. Antifungal mechanism of 1-nonanol against Aspergillus flavus growth revealed by metabolomic analyses. Appl. Microbiol. Biotechnol. 2021, 105, 7871–7888. [Google Scholar] [CrossRef]
- Perdigão Cota de Almeida, S.; Rozas, E.E.; Oller do Nascimento, C.A.; Dias, M.; Mendes, M.A. Metabolomic and secretomic approach to the resistance features of the fungus Aspergillus niger IOC 4687 to copper stress. Met. Integr. Biomet. Sci. 2021, 13, mfaa010. [Google Scholar] [CrossRef]
- Vinayavekhin, N.; Kongchai, W.; Piapukiew, J.; Chavasiri, W. Aspergillus niger upregulated glycerolipid metabolism and ethanol utilization pathway under ethanol stress. Microbiologyopen 2020, 9, e00948. [Google Scholar] [CrossRef]
- Jiang, A.-L.; Liu, Y.-N.; Liu, R.; Ren, A.; Ma, H.-Y.; Shu, L.-B.; Shi, L.; Zhu, J.; Zhao, M.-W. Integrated Proteomics and Metabolomics Analysis Provides Insights into Ganoderic Acid Biosynthesis in Response to Methyl Jasmonate in. Int. J. Mol. Sci. 2019, 20, 6116. [Google Scholar] [CrossRef]
- Soboń, A.; Szewczyk, R.; Rozalska, S.; Dlugonski, J. Metabolomics of the recovery of the filamentous fungus Cunninghamella echinulata exposed to tributyltin. Int. Biodeterior. Biodegrad. 2018, 127, 130–138. [Google Scholar] [CrossRef]
- Li, Z.; Ling, X.; Zhou, H.; Meng, T.; Zeng, J.; Hang, W.; Shi, Y.; He, N. Screening chemical modulators of benzoic acid derivatives to improve lipid accumulation in SR21 with metabolomics analysis. Biotechnol. Biofuels 2019, 12, 209. [Google Scholar] [CrossRef]
- Lu, L.; Zhu, K.-X.; Yang, Z.; Guo, X.-N.; Xing, J.-J. Metabolomics analysis of freeze-thaw tolerance enhancement mechanism of ε-poly-l-lysine on industrial yeast. Food Chem. 2022, 382, 132315. [Google Scholar] [CrossRef] [PubMed]
- Mishra, J.; Bhardwaj, A.; Pal, M.; Rajput, R.; Misra, K. High performance thin layer chromatography hyphenated with electrospray mass spectrometry for evaluation of nucleobases in two traditional Chinese medicinal mushrooms: A metabolomic approach. J. Liq. Chromatogr. Relat. Technol. 2018, 41, 910–918. [Google Scholar] [CrossRef]
- Joshi, R.; Sharma, A.; Thakur, K.; Kumar, D.; Nadda, G. Metabolite analysis and nucleoside determination using reproducible UHPLC-Q-ToF-IMS in Ophiocordyceps sinensis. J. Liq. Chromatogr. Relat. Technol. 2018, 41, 927–936. [Google Scholar] [CrossRef]
- Oh, J.; Yoon, D.-H.; Shrestha, B.; Choi, H.-K.; Sung, G.-H. Metabolomic profiling reveals enrichment of cordycepin in senescence process of Cordyceps militaris fruit bodies. J. Microbiol. 2019, 57, 54–63. [Google Scholar] [CrossRef]
- Chen, L.; Liu, Y.; Guo, Q.; Zheng, Q.; Zhang, W. Metabolomic comparison between wild Ophiocordyceps sinensis and artificial cultured Cordyceps militaris. Biomed. Chromatogr. BMC 2018, 32, e4279. [Google Scholar] [CrossRef]
- Liu, Y.; Xiao, K.; Wang, Z.; Wang, S.; Xu, F. Comparison of metabolism substances in Cordyceps sinensis and Cordyceps militaris cultivated with tussah pupa based on LC-MS. J. Food Biochem. 2021, 45, e13735. [Google Scholar] [CrossRef]
- Choi, J.N.; Kim, J.; Lee, M.Y.; Park, D.K.; Hong, Y.-S.; Lee, C.H. Metabolomics revealed novel isoflavones and optimal cultivation time of Cordyceps militaris fermentation. J. Agric. Food Chem. 2010, 58, 4258–4267. [Google Scholar] [CrossRef]
- Yao, L.; Zhu, L.-P.; Xu, X.-Y.; Tan, L.-L.; Sadilek, M.; Fan, H.; Hu, B.; Shen, X.-T.; Yang, J.; Qiao, B.; et al. Discovery of novel xylosides in co-culture of basidiomycetes Trametes versicolor and Ganoderma applanatum by integrated metabolomics and bioinformatics. Sci. Rep. 2016, 6, 33237. [Google Scholar] [CrossRef]
- Liu, Z.; Kang, B.; Duan, X.; Hu, Y.; Li, W.; Wang, C.; Li, D.; Xu, N. Metabolomic profiles of the liquid state fermentation in co-culture of A. oryzae and Z. rouxii. Food Microbiol. 2022, 103, 103966. [Google Scholar] [CrossRef]
- Cai, Z.-X.; Chen, M.-Y.; Lu, Y.-P.; Guo, Z.-J.; Zeng, Z.-H.; Liao, J.-H.; Zeng, H. Metabolomics and transcriptomics unravel the mechanism of browning resistance in Agaricus bisporus. PLoS ONE 2022, 17, e0255765. [Google Scholar] [CrossRef]
- Fu, Y.; Yu, Y.; Tan, H.; Wang, B.; Peng, W.; Sun, Q. Metabolomics reveals dopa melanin involved in the enzymatic browning of the yellow cultivars of East Asian golden needle mushroom (Flammulina filiformis). Food Chem. 2022, 370, 131295. [Google Scholar] [CrossRef] [PubMed]
- El-Hawary, S.S.; Mohammed, R.; Bahr, H.S.; Attia, E.Z.; El-Katatny, M.H.; Abelyan, N.; Al-Sanea, M.M.; Moawad, A.S.; Abdelmohsen, U.R. Soybean-associated endophytic fungi as potential source for anti-COVID-19 metabolites supported by docking analysis. J. Appl. Microbiol. 2021, 131, 1193–1211. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Hengchao, E.; Dong, H.; Zhang, Y.; Qiu, J.; Qian, Y.; Zhou, C. Combination of untargeted metabolomics approach and molecular networking analysis to identify unique natural components in wild Morchella sp. by UPLC-Q-TOF-MS. Food Chem. 2022, 366, 130642. [Google Scholar] [CrossRef] [PubMed]
- Le, V.-T.; Bertrand, S.; Robiou du Pont, T.; Fleury, F.; Caroff, N.; Bourgeade-Delmas, S.; Gentil, E.; Logé, C.; Genta-Jouve, G.; Grovel, O. Untargeted Metabolomics Approach for the Discovery of Environment-Related Pyran-2-ones Chemodiversity in a Marine-Sourced. Mar. Drugs 2021, 19, 378. [Google Scholar] [CrossRef]
- Huang, B.; Zeng, H.; Dong, L.; Li, Y.; Sun, L.; Zhu, Z.; Chai, Y.; Chen, W. Metabolite target analysis of isoprenoid pathway in Saccharomyces cerevisiae in response to genetic modification by GC-SIM-MS coupled with chemometrics. Metabolomics 2011, 7, 134–146. [Google Scholar] [CrossRef]
- Albright, J.C.; Henke, M.T.; Soukup, A.A.; McClure, R.A.; Thomson, R.J.; Keller, N.P.; Kelleher, N.L. Large-scale metabolomics reveals a complex response of Aspergillus nidulans to epigenetic perturbation. ACS Chem. Biol. 2015, 10, 1535–1541. [Google Scholar] [CrossRef]
- Alves, P.C.; Hartmann, D.O.; Núñez, O.; Martins, I.; Gomes, T.L.; Garcia, H.; Galceran, M.T.; Hampson, R.; Becker, J.D.; Silva Pereira, C. Transcriptomic and metabolomic profiling of ionic liquid stimuli unveils enhanced secondary metabolism in Aspergillus nidulans. BMC Genom. 2016, 17, 284. [Google Scholar] [CrossRef]
- Nguyen, P.-A.; Strub, C.; Lagrée, M.; Bertrand-Michel, J.; Schorr-Galindo, S.; Fontana, A. Study of in vitro interaction between Fusarium verticillioides and Streptomyces sp. using metabolomics. Folia Microbiol. 2020, 65, 303–314. [Google Scholar] [CrossRef]
- Smith, J.E.; Lay, J.O.; Bluhm, B.H. Metabolic fingerprinting reveals a new genetic linkage between ambient pH and metabolites associated with desiccation tolerance in Fusarium verticillioides. Metabolomics 2012, 8, 376–385. [Google Scholar] [CrossRef]
- Adpressa, D.A.; Connolly, L.R.; Konkel, Z.M.; Neuhaus, G.F.; Chang, X.L.; Pierce, B.R.; Smith, K.M.; Freitag, M.; Loesgen, S. A metabolomics-guided approach to discover Fusarium graminearum metabolites after removal of a repressive histone modification. Fungal Genet. Biol. FG B 2019, 132, 103256. [Google Scholar] [CrossRef]
- Liu, C.; Chen, F.; Zhang, J.; Liu, L.; Lei, H.; Li, H.; Wang, Y.; Liao, Y.-C.; Tang, H. Metabolic Changes of Fusarium graminearum Induced by Gene Deletion. J. Proteome Res. 2019, 18, 3317–3327. [Google Scholar] [CrossRef] [PubMed]
- Kalampokis, I.F.; Kapetanakis, G.C.; Aliferis, K.A.; Diallinas, G. Multiple nucleobase transporters contribute to boscalid sensitivity in Aspergillus nidulans. Fungal Genet. Biol. FG B 2018, 115, 52–63. [Google Scholar] [CrossRef] [PubMed]
- Lacerda, J.W.F.; Siqueira, K.A.; Vasconcelos, L.G.; Bellete, B.S.; Dall’Oglio, E.L.; Sousa Junior, P.T.; Faraggi, T.M.; Vieira, L.C.C.; Soares, M.A.; Sampaio, O.M. Metabolomic Analysis of Combretum lanceolatum Plants Interaction with Diaporthe phaseolorum and Trichoderma spirale Endophytic Fungi through H-NMR. Chem. Biodiversity 2021, 18, e2100350. [Google Scholar] [CrossRef]
- Sebastiana, M.; Gargallo-Garriga, A.; Sardans, J.; Pérez-Trujillo, M.; Monteiro, F.; Figueiredo, A.; Maia, M.; Nascimento, R.; Silva, M.S.; Ferreira, A.N.; et al. Metabolomics and transcriptomics to decipher molecular mechanisms underlying ectomycorrhizal root colonization of an oak tree. Sci. Rep. 2021, 11, 8576. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yang, C.; Chen, J.; He, Y.; Deng, J.; Xie, C.; Xiao, X.; Long, X.; Wu, X.; Liu, W.; et al. Changing light promotes isoflavone biosynthesis in soybean pods and enhances their resistance to mildew infection. Plant Cell Environ. 2021, 44, 2536–2550. [Google Scholar] [CrossRef]
- Wong, J.W.H.; Plett, K.L.; Natera, S.H.A.; Roessner, U.; Anderson, I.C.; Plett, J.M. Comparative metabolomics implicates threitol as a fungal signal supporting colonization of Armillaria luteobubalina on eucalypt roots. Plant Cell Environ. 2020, 43, 374–386. [Google Scholar] [CrossRef]
- Ren, Z.; Fang, M.; Muhae-Ud-Din, G.; Gao, H.; Yang, Y.; Liu, T.; Chen, W.; Gao, L. Metabolomics analysis of grains of wheat infected and noninfected with Tilletia controversa Kühn. Sci. Rep. 2021, 11, 18876. [Google Scholar] [CrossRef]
- Chen, C.; Cai, N.; Chen, J.; Wan, C. UHPLC-Q-TOF/MS-Based Metabolomics Approach Reveals the Antifungal Potential of Pinocembroside against Green Mold Phytopathogen. Plants 2019, 9, 17. [Google Scholar] [CrossRef]
- Mazzei, P.; Vinale, F.; Woo, S.L.; Pascale, A.; Lorito, M.; Piccolo, A. Metabolomics by Proton High-Resolution Magic-Angle-Spinning Nuclear Magnetic Resonance of Tomato Plants Treated with Two Secondary Metabolites Isolated from Trichoderma. J. Agric. Food Chem. 2016, 64, 3538–3545. [Google Scholar] [CrossRef]
- Ceglarek, U.; Leichtle, A.; Brügel, M.; Kortz, L.; Brauer, R.; Bresler, K.; Thiery, J.; Fiedler, G.M. Challenges and developments in tandem mass spectrometry based clinical metabolomics. Mol. Cell. Endocrinol. 2009, 301, 266–271. [Google Scholar] [CrossRef]
- Griffiths, W.J.; Wang, Y. Mass spectrometry: From proteomics to metabolomics and lipidomics. Chem. Soc. Rev. 2009, 38, 1882–1896. [Google Scholar] [CrossRef] [PubMed]
- Theodoridis, G.; Gika, H.G.; Wilson, I.D. LC-MS-based methodology for global metabolite profiling in metabonomics/metabolomics. TrAC Trends Anal. Chem. 2008, 27, 251–260. [Google Scholar] [CrossRef]
- Li, N.; Song, Y.P.; Tang, H.; Wang, Y. Recent developments in sample preparation and data pre-treatment in metabonomics research. Arch. Biochem. Biophys. 2016, 589, 4–9. [Google Scholar] [CrossRef] [PubMed]
- Vuckovic, D. Current trends and challenges in sample preparation for global metabolomics using liquid chromatography–mass spectrometry. Anal. Bioanal. Chem. 2012, 403, 1523–1548. [Google Scholar] [CrossRef] [PubMed]
- Harrison, D.E.; Maitra, P.K. Control of respiration and metabolism in growing Klebsiella aerogenes. Role Adenine Nucleotides Biochem. J. 1969, 112, 647–656. [Google Scholar] [CrossRef]
- Iversen, J.J.L. A rapid sampling valve with minimal dead space for laboratory scale fermenters. Biotechnol. Bioeng. 1981, 23, 437–440. [Google Scholar] [CrossRef]
- Theobald, U.; Mailinger, W.; Reuss, M.; Rizzi, M. In vivo analysis of glucose-induced fast changes in yeast adenine nucleotide pool applying a rapid sampling technique. Anal. Biochem. 1993, 214, 31–37. [Google Scholar] [CrossRef]
- Buziol, S.; Bashir, I.; Baumeister, A.; Claassen, W.; Noisommit-Rizzi, N.; Mailinger, W.; Reuss, M. New bioreactor-coupled rapid stopped-flow sampling technique for measurements of metabolite dynamics on a subsecond time scale. Biotechnol. Bioeng. 2002, 80, 632–636. [Google Scholar] [CrossRef]
- Schädel, F.; Franco-Lara, E. Rapid sampling devices for metabolic engineering applications. Appl. Microbiol. Biotechnol. 2009, 83, 199–208. [Google Scholar] [CrossRef]
- Lameiras, F.; Heijnen, J.J.; van Gulik, W.M. Development of tools for quantitative intracellular metabolomics of chemostat cultures. Metab. Off. J. Metab. Soc. 2015, 11, 1253–1264. [Google Scholar] [CrossRef][Green Version]
- Van Gulik, W.M. Fast sampling for quantitative microbial metabolomics. Curr. Opin. Biotechnol. 2010, 21, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Link, H.; Fuhrer, T.; Gerosa, L.; Zamboni, N.; Sauer, U. Real-time metabolome profiling of the metabolic switch between starvation and growth. Nat. Meth. 2015, 12, 1091–1097. [Google Scholar] [CrossRef] [PubMed]
- De Koning, W.; van Dam, K. A method for the determination of changes of glycolytic metabolites in yeast on a subsecond time scale using extraction at neutral pH. Anal. Biochem. 1992, 204, 118–123. [Google Scholar] [CrossRef]
- Link, H.; Anselment, B.; Weuster-Botz, D. Leakage of adenylates during cold methanol/glycerol quenching of Escherichia coli. Metabolomics 2008, 4, 240–247. [Google Scholar] [CrossRef]
- Canelas, A.B.; Ras, C.; ten Pierick, A.; van Dam, J.C.; Heijnen, J.J.; van Gulik, W.M. Leakage-free rapid quenching technique for yeast metabolomics. Metabolomics 2008, 4, 226–239. [Google Scholar] [CrossRef]
- Zheng, X.; Yu, J.; Cairns, T.C.; Zhang, L.; Zhang, Z.; Zhang, Q.; Zheng, P.; Sun, J.; Ma, Y. Comprehensive Improvement of Sample Preparation Methodologies Facilitates Dynamic Metabolomics of Aspergillus niger. Biotechnol. J. 2019, 14, 1800315. [Google Scholar] [CrossRef]
- Chassagnole, C.; Noisommit-Rizzi, N.; Schmid, J.W.; Mauch, K.; Reuss, M. Dynamic modeling of the central carbon metabolism of Escherichia coli. Biotechnol. Bioeng. 2002, 79, 53–73. [Google Scholar] [CrossRef]
- Meinert, S.; Rapp, S.; Schmitz, K.; Noack, S.; Kornfeld, G.; Hardiman, T. Quantitative quenching evaluation and direct intracellular metabolite analysis in Penicillium chrysogenum. Anal. Biochem. 2013, 438, 47–52. [Google Scholar] [CrossRef]
- Moritz, B.; Striegel, K.; De Graaf, A.A.; Sahm, H. Kinetic properties of the glucose-6-phosphate and 6-phosphogluconate dehydrogenases from Corynebacterium glutamicum and their application for predicting pentose phosphate pathway flux in vivo. Eur. J. Biochem. 2000, 267, 3442–3452. [Google Scholar] [CrossRef]
- Spura, J.; Reimer, L.C.; Wieloch, P.; Schreiber, K.; Buchinger, S.; Schomburg, D. A method for enzyme quenching in microbial metabolome analysis successfully applied to gram-positive and gram-negative bacteria and yeast. Anal. Biochem. 2009, 394, 192–201. [Google Scholar] [CrossRef]
- Loret, M.O.; Pedersen, L.; François, J. Revised procedures for yeast metabolites extraction: Application to a glucose pulse to carbon-limited yeast cultures, which reveals a transient activation of the purine salvage pathway. Yeast 2007, 24, 47–60. [Google Scholar] [CrossRef] [PubMed]
- Hans, M.A.; Heinzle, E.; Wittmann, C. Quantification of intracellular amino acids in batch cultures of Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2001, 56, 776–779. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Chen, H.; Tang, X.; Yang, Q.; Zhang, H.; Chen, Y.Q.; Chen, W. Evaluation of metabolome sample preparation and extraction methodologies for oleaginous filamentous fungi Mortierella alpina. Metab. Off. J. Metab. Soc. 2019, 15, 50. [Google Scholar] [CrossRef]
- Madla, S.; Miura, D.; Wariishi, H. Optimization of Extraction Method for GC-MS based Metabolomics for Filamentous Fungi. J. Microb. Biochem. Technol. 2012, 4, 005–009. [Google Scholar] [CrossRef]
- Lim, G.B.; Lee, S.Y.; Lee, E.K.; Haam, S.J.; Kim, W.S. Separation of astaxanthin from red yeast Phaffia rhodozyma by supercritical carbon dioxide extraction. Biochem. Eng. J. 2002, 11, 181–187. [Google Scholar] [CrossRef]
- Beale, D.J.; Pinu, F.R.; Kouremenos, K.A.; Poojary, M.M.; Narayana, V.K.; Boughton, B.A.; Kanojia, K.; Dayalan, S.; Jones, O.A.H.; Dias, D.A. Review of recent developments in GC–MS approaches to metabolomics-based research. Metabolomics 2018, 14, 152. [Google Scholar] [CrossRef]
- Koek, M.M.; Muilwijk, B.; van der Werf, M.J.; Hankemeier, T. Microbial metabolomics with gas chromatography/mass spectrometry. Anal. Chem. 2006, 78, 1272–1281. [Google Scholar] [CrossRef]
- Xiao, J.F.; Zhou, B.; Ressom, H.W. Metabolite identification and quantitation in LC-MS/MS-based metabolomics. TrAC Trends Anal. Chem. 2012, 32, 1–14. [Google Scholar] [CrossRef]
- Markley, J.L.; Brüschweiler, R.; Edison, A.S.; Eghbalnia, H.R.; Powers, R.; Raftery, D.; Wishart, D.S. The future of NMR-based metabolomics. Curr. Opin. Biotechnol. 2017, 43, 34–40. [Google Scholar] [CrossRef]
- Xiao, C.; Chi, Q.; Wang, X. Recent Progress in Mass Spectrometry-based Metabolomics for Colorectal Cancer. Chem. Res. Chin. Univ. 2022, 38, 886–893. [Google Scholar] [CrossRef]
- Goodacre, R.; Broadhurst, D.; Smilde, A.K.; Kristal, B.S.; Baker, J.D.; Beger, R.; Bessant, C.; Connor, S.; Capuani, G.; Craig, A.; et al. Proposed minimum reporting standards for data analysis in metabolomics. Metabolomics 2007, 3, 231–241. [Google Scholar] [CrossRef]
- Di Guida, R.; Engel, J.; Allwood, J.W.; Weber, R.J.M.; Jones, M.R.; Sommer, U.; Viant, M.R.; Dunn, W.B. Non-targeted UHPLC-MS metabolomic data processing methods: A comparative investigation of normalisation, missing value imputation, transformation and scaling. Metabolomics 2016, 12, 93. [Google Scholar] [CrossRef] [PubMed]
- Bartel, J.; Krumsiek, J.; Theis, F.J. Statistical methods for the analysis of high-throughput metabolomics data. Comput. Struct. Biotechnol. J. 2013, 4, e201301009. [Google Scholar] [CrossRef] [PubMed]
- Trygg, J.; Holmes, E.; Lundstedt, T. Chemometrics in Metabonomics. J. Proteome Res. 2007, 6, 469–479. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S. Computational Approaches to Metabolomics. Methods Mol. Biol. 2010, 593, 283. [Google Scholar] [CrossRef] [PubMed]
- Westerhuis, J.A.; van Velzen, E.J.J.; Hoefsloot, H.C.J.; Smilde, A.K. Multivariate paired data analysis: Multilevel PLSDA versus OPLSDA. Metab. Off. J. Metab. Soc. 2010, 6, 119–128. [Google Scholar] [CrossRef]
- Gorrochategui, E.; Jaumot, J.; Lacorte, S.; Tauler, R. Data analysis strategies for targeted and untargeted LC-MS metabolomic studies: Overview and workflow. TrAC Trends Anal. Chem. 2016, 82, 425–442. [Google Scholar] [CrossRef]
- Lommen, A. MetAlign: Interface-driven, versatile metabolomics tool for hyphenated full-scan mass spectrometry data preprocessing. Anal. Chem. 2009, 81, 3079–3086. [Google Scholar] [CrossRef]
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Oresic, M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform. 2010, 11, 395. [Google Scholar] [CrossRef]
- Tautenhahn, R.; Patti, G.J.; Rinehart, D.; Siuzdak, G. XCMS Online: A web-based platform to process untargeted metabolomic data. Anal. Chem. 2012, 84, 5035–5039. [Google Scholar] [CrossRef]
- Melamud, E.; Vastag, L.; Rabinowitz, J.D. Metabolomic analysis and visualization engine for LC-MS data. Anal. Chem. 2010, 82, 9818–9826. [Google Scholar] [CrossRef] [PubMed]
- Chagoyen, M.; Pazos, F. MBRole: Enrichment analysis of metabolomic data. Bioinformatics 2011, 27, 730–731. [Google Scholar] [CrossRef] [PubMed]
- Cambiaghi, A.; Ferrario, M.; Masseroli, M. Analysis of metabolomic data: Tools, current strategies and future challenges for omics data integration. Brief. Bioinform. 2017, 18, 498–510. [Google Scholar] [CrossRef] [PubMed]
- Hibbett, D.; Abarenkov, K.; Kõljalg, U.; Öpik, M.; Chai, B.; Cole, J.; Wang, Q.; Crous, P.; Robert, V.; Helgason, T.; et al. Sequence-based classification and identification of Fungi. Mycologia 2016, 108, 1049–1068. [Google Scholar] [CrossRef]
- Watanabe, M.; Goto, K.; Sugita-Konishi, Y.; Kamata, Y.; Hara-Kudo, Y. Sensitive detection of whole-genome differentiation among closely-related species of the genus Fusarium using DNA-DNA hybridization and a microplate technique. J. Vet. Med. Sci. 2012, 74, 1333–1336. [Google Scholar] [CrossRef]
- Bleykasten-Grosshans, C.; Fabrizio, R.; Friedrich, A.; Schacherer, J. Species-Wide Transposable Element Repertoires Retrace the Evolutionary History of the Saccharomyces cerevisiae Host. Mol. Biol. Evol. 2021, 38, 4334–4345. [Google Scholar] [CrossRef]
- Ferrer, C.; Colom, F.; Frasés, S.; Mulet, E.; Abad, J.L.; Alió, J.L. Detection and identification of fungal pathogens by PCR and by ITS2 and 5.8S ribosomal DNA typing in ocular infections. J. Clin. Microbiol. 2001, 39, 2873–2879. [Google Scholar] [CrossRef]
- Nakamura, S.; Sato, H.; Tanaka, R.; Kusuya, Y.; Takahashi, H.; Yaguchi, T. Ribosomal subunit protein typing using matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) for the identification and discrimination of Aspergillus species. BMC Microbiol. 2017, 17, 100. [Google Scholar] [CrossRef]
- Wu, S.-Y.; Kang, M.; Liu, Y.; Chen, Z.-X.; Xiao, Y.-L.; He, C.; Ma, Y. Molecular epidemiology and antifungal susceptibilities of Cryptococcus species isolates from HIV and non-HIV patients in Southwest China. Eur. J. Clin. Microbiol. Infect. Dis. 2021, 40, 287–295. [Google Scholar] [CrossRef]
- Chowdhary, A.; Prakash, A.; Randhawa, H.S.; Kathuria, S.; Hagen, F.; Klaassen, C.H.; Meis, J.F. First environmental isolation of Cryptococcus gattii, genotype AFLP5, from India and a global review. Mycoses 2013, 56, 222–228. [Google Scholar] [CrossRef]
- Bonito, G.M.; Gryganskyi, A.P.; Trappe, J.M.; Vilgalys, R. A global meta-analysis of Tuber ITS rDNA sequences: Species diversity, host associations and long-distance dispersal. Mol. Ecol. 2010, 19, 4994–5008. [Google Scholar] [CrossRef] [PubMed]
- Keymer, A.; Pimprikar, P.; Wewer, V.; Huber, C.; Brands, M.; Bucerius, S.L.; Delaux, P.-M.; Klingl, V.; Röpenack-Lahaye, E.v.; Wang, T.L.; et al. Lipid transfer from plants to arbuscular mycorrhiza fungi. eLife 2017, 6, e29107. [Google Scholar] [CrossRef] [PubMed]
- Kang, D.; Kim, J.; Choi, J.N.; Liu, K.-H.; Lee, C.H. Chemotaxonomy of Trichoderma spp. using mass spectrometry-based metabolite profiling. J. Microbiol. Biotechnol. 2011, 21, 5–13. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhu, S.-B.; Xie, M.-Y.; Nie, S.-P.; Liu, W.; Li, C.; Gong, X.-F.; Wang, Y.-X. Quality control and original discrimination of Ganoderma lucidum based on high-performance liquid chromatographic fingerprints and combined chemometrics methods. Anal. Chim. Acta 2008, 623, 146–156. [Google Scholar] [CrossRef] [PubMed]
- Wen, H.; Kang, S.; Song, Y.; Song, Y.; Sung, S.H.; Park, S. Differentiation of cultivation sources of Ganoderma lucidum by NMR-based metabolomics approach. Phytochem. Anal. PCA 2010, 21, 73–79. [Google Scholar] [CrossRef]
- Aliferis, K.A.; Cubeta, M.A.; Jabaji, S. Chemotaxonomy of fungi in the Rhizoctonia solani species complex performing GC/MS metabolite profiling. Metabolomics 2013, 9, 159–169. [Google Scholar] [CrossRef]
- Hettick, J.M.; Green, B.J.; Buskirk, A.D.; Kashon, M.L.; Slaven, J.E.; Janotka, E.; Blachere, F.M.; Schmechel, D.; Beezhold, D.H. Discrimination of Aspergillus isolates at the species and strain level by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry fingerprinting. Anal. Biochem. 2008, 380, 276–281. [Google Scholar] [CrossRef]
- Qian, J.; Cutler, J.E.; Cole, R.B.; Cai, Y. MALDI-TOF mass signatures for differentiation of yeast species, strain grouping and monitoring of morphogenesis markers. Anal. Bioanal. Chem. 2008, 392, 439–449. [Google Scholar] [CrossRef]
- Green, K.A.; Berry, D.; Feussner, K.; Eaton, C.J.; Ram, A.; Mesarich, C.H.; Solomon, P.; Feussner, I.; Scott, B. Lolium perenne apoplast metabolomics for identification of novel metabolites produced by the symbiotic fungus Epichloë festucae. New Phytol. 2020, 227, 559–571. [Google Scholar] [CrossRef]
- Yang, F.; Zhao, M.; Zhou, L.; Zhang, M.; Liu, J.; Marchioni, E. Identification and Differentiation of Wide Edible Mushrooms Based on Lipidomics Profiling Combined with Principal Component Analysis. J. Agric. Food Chem. 2021, 69, 9991–10001. [Google Scholar] [CrossRef]
- Kaewnarin, K.; Limjiasahapong, S.; Jariyasopit, N.; Anekthanakul, K.; Kurilung, A.; Wong, S.C.C.; Sirivatanauksorn, Y.; Visessanguan, W.; Khoomrung, S. High-Resolution QTOF-MRM for Highly Accurate Identification and Quantification of Trace Levels of Triterpenoids in Mycelium. J. Am. Soc. Mass Spectrom. 2021, 32, 2451–2462. [Google Scholar] [CrossRef] [PubMed]
- Sardans, J.; Peñuelas, J.; Rivas-Ubach, A. Ecological metabolomics: Overview of current developments and future challenges. Chemoecology 2011, 21, 191–225. [Google Scholar] [CrossRef]
- Fountain, J.C.; Yang, L.; Pandey, M.K.; Bajaj, P.; Alexander, D.; Chen, S.; Kemerait, R.C.; Varshney, R.K.; Guo, B. Carbohydrate, glutathione, and polyamine metabolism are central to Aspergillus flavus oxidative stress responses over time. BMC Microbiol. 2019, 19, 209. [Google Scholar] [CrossRef] [PubMed]
- Tan, W.-J.; Yang, Y.-C.; Zhou, Y.; Huang, L.-P.; Xu, L.; Chen, Q.-F.; Yu, L.-J.; Xiao, S. Diacylglycerol Acyltransferase and Diacylglycerol Kinase Modulate Triacylglycerol and Phosphatidic Acid Production in the Plant Response to Freezing Stress. Plant Physiol. 2018, 177, 1303–1318. [Google Scholar] [CrossRef]
- Fan, J.; Yu, L.; Xu, C. A Central Role for Triacylglycerol in Membrane Lipid Breakdown, Fatty Acid-Oxidation, and Plant Survival under Extended Darkness. Plant Physiol. 2017, 174, 1517–1530. [Google Scholar] [CrossRef]
- Hammerl, R.; Frank, O.; Dietz, M.; Hirschmann, J.; Hofmann, T. Tyrosine Induced Metabolome Alterations of and Quantitation of Secondary Key Metabolites in Blue-Mold Cheese. J. Agric. Food Chem. 2019, 67, 8500–8509. [Google Scholar] [CrossRef]
- Keller, N.P.; Turner, G.; Bennett, J.W. Fungal secondary metabolism—From biochemistry to genomics. Nat. Rev. Microbiol. 2005, 3, 937–947. [Google Scholar] [CrossRef]
- Tawfike, A.F.; Romli, M.; Clements, C.; Abbott, G.; Young, L.; Schumacher, M.; Diederich, M.; Farag, M.; Edrada-Ebel, R. Isolation of anticancer and anti-trypanosome secondary metabolites from the endophytic fungus Aspergillus flocculus via bioactivity guided isolation and MS based metabolomics. J. Chromatogr. B Analyt. Technol. Biomed. Life. Sci. 2019, 1106–1107, 71–83. [Google Scholar] [CrossRef]
- Bu, C.; Zhang, Q.; Zeng, J.; Cao, X.; Hao, Z.; Qiao, D.; Cao, Y.; Xu, H. Identification of a novel anthocyanin synthesis pathway in the fungus Aspergillus sydowii H-1. BMC Genom. 2020, 21, 29. [Google Scholar] [CrossRef]
- Pandohee, J.; Stevenson, P.G.; Conlan, X.A.; Zhou, X.-R.; Jones, O.A.H. Off-line two-dimensional liquid chromatography for metabolomics: An example using Agaricus bisporus mushrooms exposed to UV irradiation. Metabolomics 2015, 11, 939–951. [Google Scholar] [CrossRef]
- Kamal, N.; Viegelmann, C.V.; Clements, C.J.; Edrada-Ebel, R. Metabolomics-Guided Isolation of Anti-trypanosomal Metabolites from the Endophytic Fungus Lasiodiplodia theobromae. Planta Med. 2017, 83, 565–573. [Google Scholar] [CrossRef] [PubMed]
- Fan, B.; Grauso, L.; Li, F.; Scarpato, S.; Mangoni, A.; Tasdemir, D. Application of Feature-Based Molecular Networking for Comparative Metabolomics and Targeted Isolation of Stereoisomers from Algicolous fungi. Mar. Drugs 2022, 20, 210. [Google Scholar] [CrossRef] [PubMed]
- Bailey, J.E. Toward a science of metabolic engineering. Science 1991, 252, 1668–1675. [Google Scholar] [CrossRef] [PubMed]
- Mishra, A.; Singh, S.P.; Mahfooz, S.; Singh, S.P.; Bhattacharya, A.; Mishra, N.; Nautiyal, C.S. Endophyte-Mediated Modulation of Defense-Related Genes and Systemic Resistance in Withania somnifera (L.) Dunal under Alternaria alternata Stress. Appl. Environ. Microbiol. 2018, 84, e02845-17. [Google Scholar] [CrossRef] [PubMed]
- Pappas, M.L.; Liapoura, M.; Papantoniou, D.; Avramidou, M.; Kavroulakis, N.; Weinhold, A.; Broufas, G.D.; Papadopoulou, K.K. The Beneficial Endophytic Fungus Strain K Alters Tomato Responses against Spider Mites to the Benefit of the Plant. Front. Plant Sci. 2018, 9, 1603. [Google Scholar] [CrossRef]
- Weed, R.A.; Savchenko, K.G.; Lessin, L.M.; Carris, L.M.; Gang, D.R. Untargeted Metabolomic Investigation of Wheat Infected with Stinking Smut. Phytopathology 2021, 111, 2343–2354. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).