Streptomyces rochei MS-37 as a Novel Marine Actinobacterium for Green Biosynthesis of Silver Nanoparticles and Their Biomedical Applications
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Nanoparticle Biosynthesis
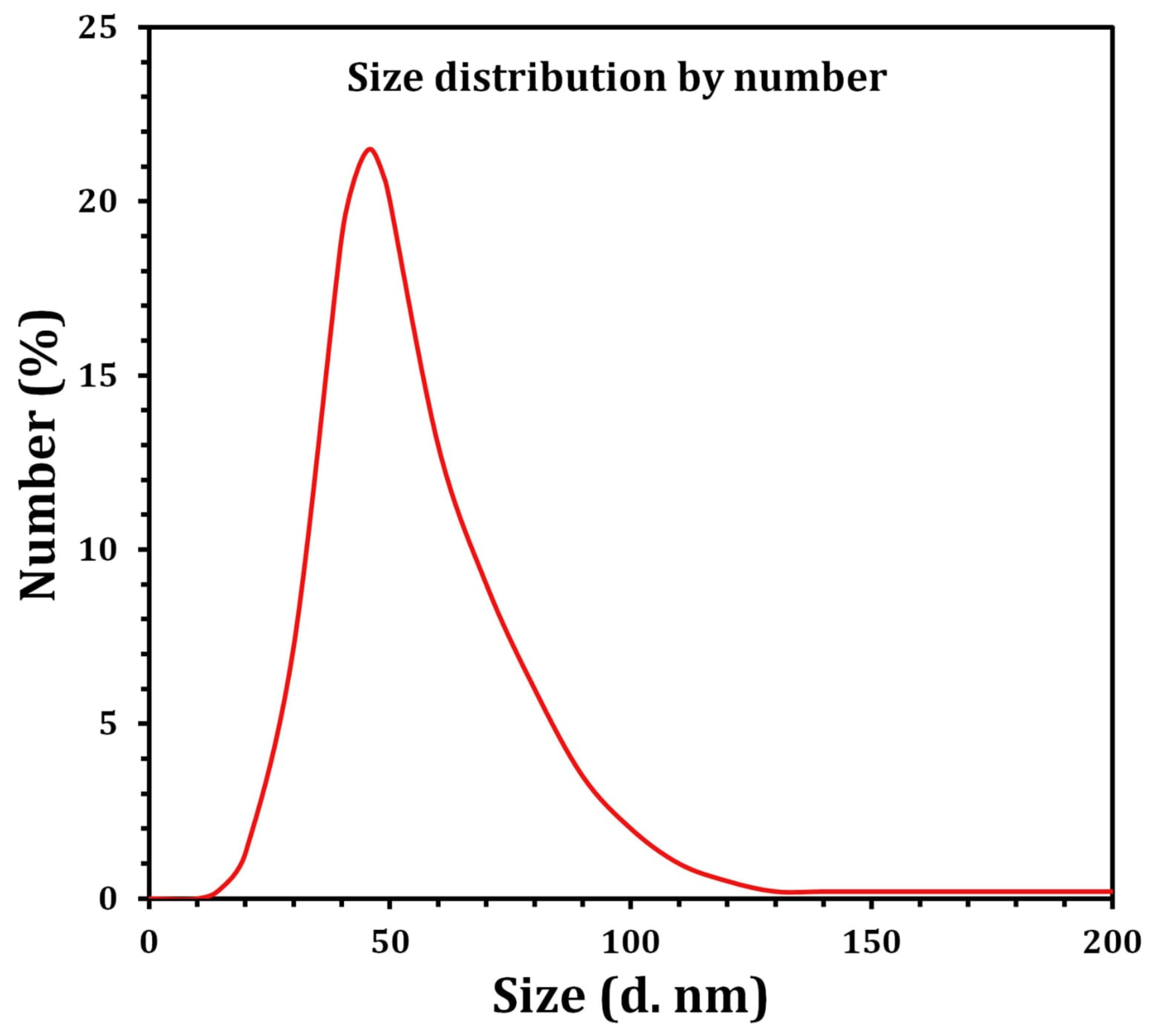
2.3. Characterization of SNPs
2.4. Antibacterial Assays
2.4.1. Isolation and Identification of Oral Pathogenic Bacterial Strains
2.4.2. Susceptibility Testing
2.5. Antibiofilm Activity
2.6. Nitric Oxide Radical Scavenging Activity
2.7. Inhibition of Protein Denaturation
2.8. Cytotoxicity Assessment
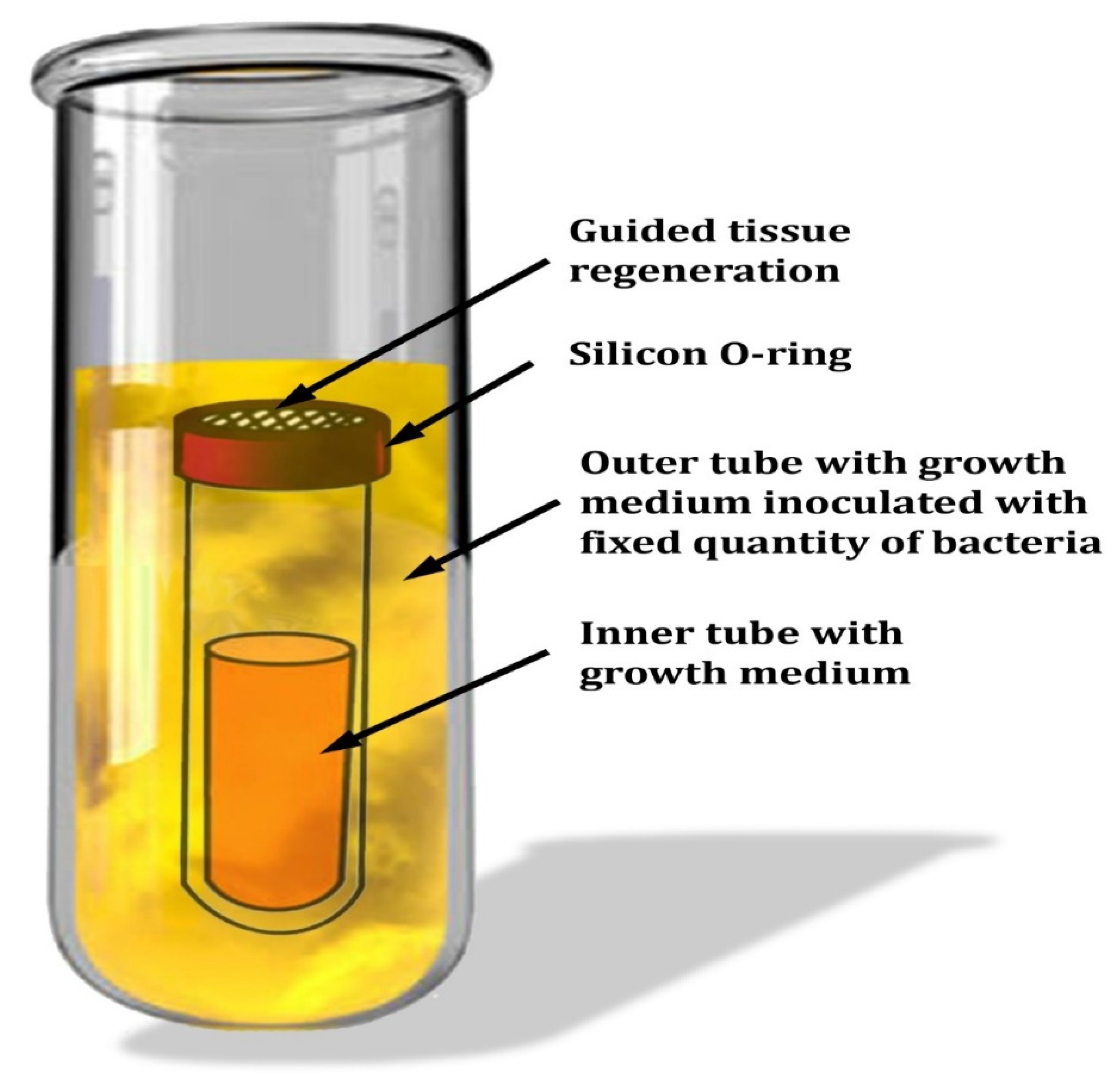
2.9. Evaluation of SNPs on the Membrane of Guided Tissue Regeneration
2.10. Statistical Analysis
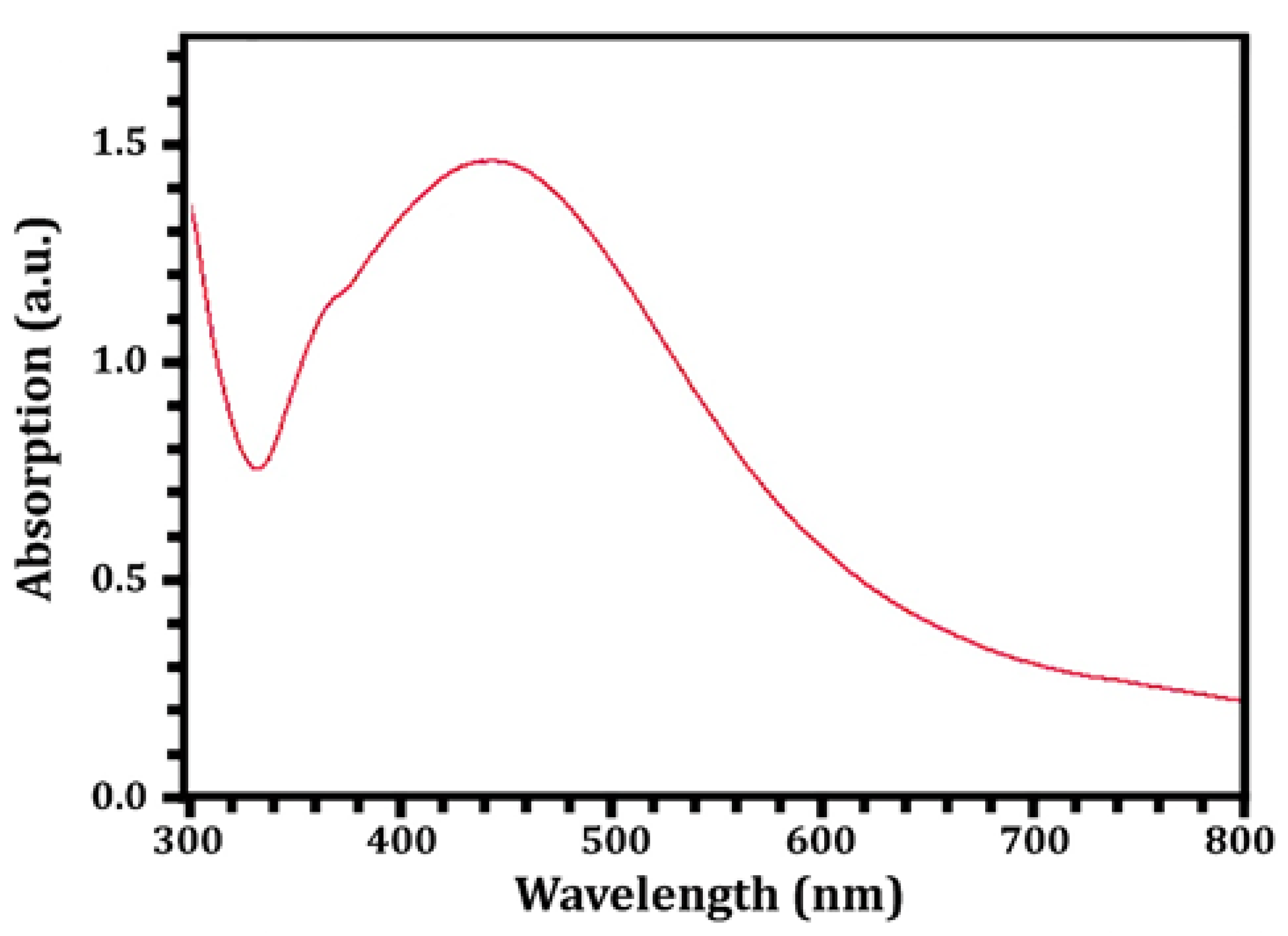
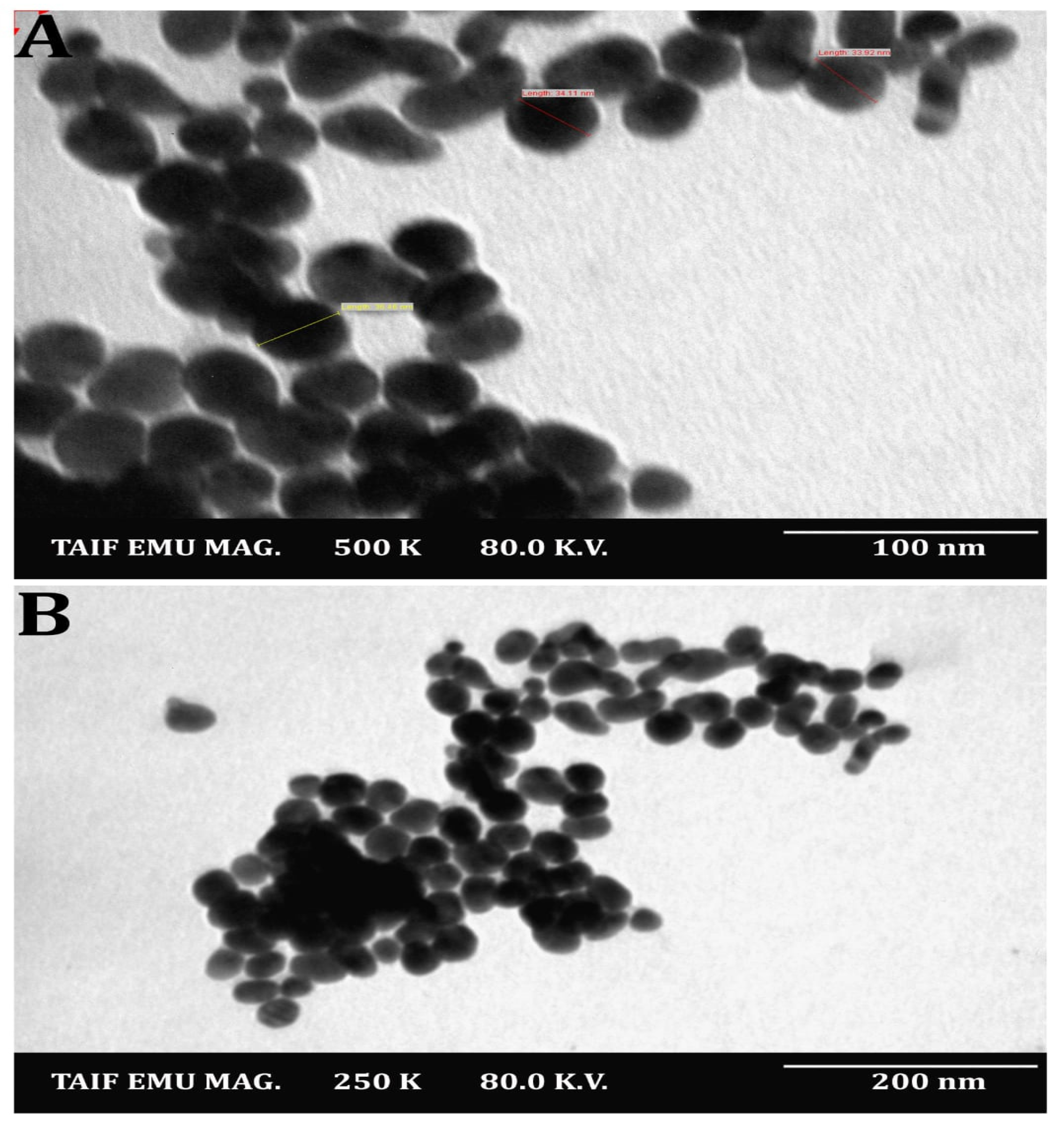
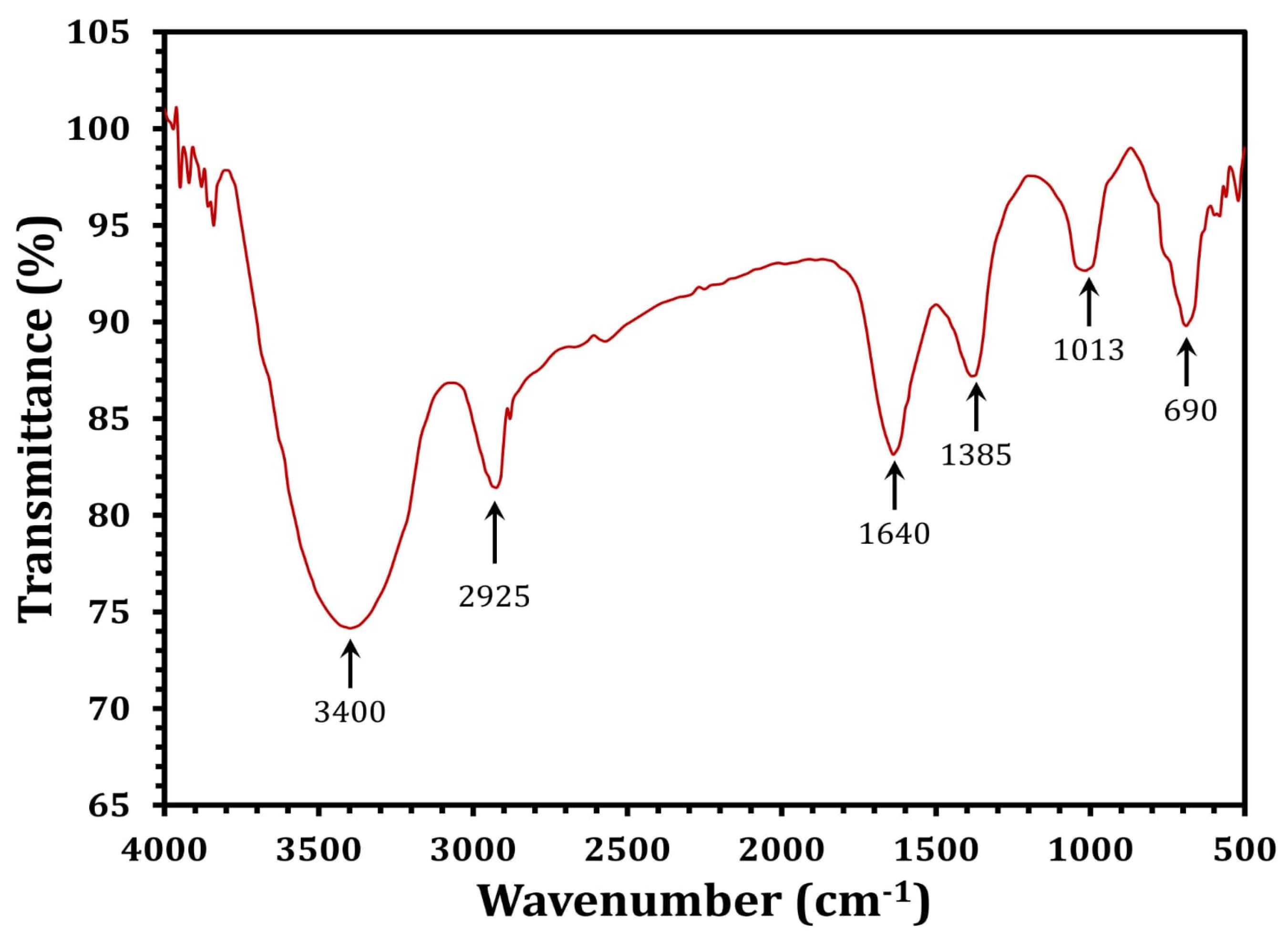
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- AlJasser, R.; AlAqeely, R.; AlKenani, M.; AlQahtani, S.; AlZahrani, A.; Lambarte, R. The effect of systemic Isotretinoin on salivary tissue inhibitors of metalloproteinases 1 and 2 and salivary flow rate in periodontal disease. Saudi J. Biol. Sci. 2022, 29, 148–153. [Google Scholar] [CrossRef]
- Yadalam, P.K.; Rengaraj, S.; Mugri, M.H.; Sayed, M.; Porwal, A.; Alahmari, N.M.; Alzahrani, K.M.; Robaian, A.; Baeshen, H.A.; Patil, S. Designing an immunoinformatic vaccine for peri-implantitis using a structural biology approach. Saudi J. Biol. Sci. 2022, 29, 622–629. [Google Scholar] [CrossRef] [PubMed]
- Johansson, I.; Witkowska, E.; Kaveh, B.; Lif Holgerson, P.; Tanner, A.C.R. The microbiome in populations with a low and high prevalence of caries. J. Dent. Res. 2016, 95, 80–86. [Google Scholar] [CrossRef] [PubMed]
- Khalil, M.; El-Sabbagh, M.; El Naggar, E.; El-Erian, R. Antibacterial activity of Salvadora persica against oral pathogenic bacterial isolates. Niger. J. Clin. Pract. 2019, 22, 1378–1387. [Google Scholar] [CrossRef]
- Bhatia, A.; Bains, S.K.; Singh, M.P. To assess knowledge and awareness of North Indian population towards periodontal therapy and oral-systemic disease link: A cross-sectional survey. J. Interdiscip. Dent. 2013, 3, 79. [Google Scholar] [CrossRef]
- Moghadam, E.T.; Yazdanian, M.; Tahmasebi, E.; Tebyanian, H.; Ranjbar, R.; Yazdanian, A.; Seifalian, A.; Tafazoli, A. Current herbal medicine as an alternative treatment in dentistry: In vitro, in vivo and clinical studies. Eur. J. Pharmacol. 2020, 889, 173665. [Google Scholar] [CrossRef] [PubMed]
- Rani, S.; Chandra, R.V.; Reddy, A.; Reddy, B.; Nagarajan, S.; Naveen, A. Evaluation of the antibacterial effect of silver nanoparticles on guided tissue regeneration membrane colonization—An in vitro study. J. Int. Acad. Periodontol. 2015, 17, 66–76. [Google Scholar] [PubMed]
- Rossa, M.L.; Lima, L.A.; Pustiglioni, F.E.; Hespanhol, A.M.; Kon, S.; Grigolli Filho, J. SEM analyses of bacterial contamination of e-PTFE membranes and GTR clinical results. J. Inter. Acad. Periodontol. 2006, 8, 115–124. [Google Scholar]
- Cheng, C.F.; Lee, Y.Y.; Chi, L.Y.; Chen, Y.T.; Hung, S.L.; Ling, L.J. Bacterial penetration through antibiotic-loaded guided tissue regeneration membranes. J. Periodontol. 2009, 80, 1471–1478. [Google Scholar] [CrossRef]
- Wang, Z.; Liu, X.; Duan, Y.; Huang, Y. Infection microenvironment-related antibacterial nanotherapeutic strategies. Biomaterials 2021, 280, 121249. [Google Scholar] [CrossRef]
- Wei, Y.; Yang, L.; Pandeya, A.; Cui, J.; Zhang, Y.; Li, Z. Pyroptosis-induced inflammation and tissue damage. J. Mol. Biol. 2021, 434, 167301. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Tsao, R. Dietary polyphenols, oxidative stress and antioxidant and anti-inflammatory effects. Curr. Opin. Food Sci. 2016, 8, 33–42. [Google Scholar] [CrossRef]
- Ahamad, N.; Kar, A.; Mehta, S.; Dewani, M.; Ravichandran, V.; Bhardwaj, P.; Sharma, S.; Banerjee, R. Immunomodulatory nanosystems for treating inflammatory diseases. Biomaterials 2021, 274, 120875. [Google Scholar] [CrossRef]
- Elisha, I.L.; Dzoyem, J.P.; McGaw, L.J.; Botha, F.S.; Eloff, J.N. The anti-arthritic, anti-inflammatory, antioxidant activity and relationships with total phenolics and total flavonoids of nine South African plants used traditionally to treat arthritis. BMC Complement. Altern. Med. 2016, 16, 307. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.S.; Morsy, R.; El-Zawawy, N.A.; Fareed, M.F.; Bedaiwy, M.Y. Synthesized zinc peroxide nanoparticles (ZnO2-NPs): A novel antimicrobial, anti-elastase, anti-keratinase, and anti-inflammatory approach toward polymicrobial burn wounds. Int. J. Nanomed. 2017, 12, 6059. [Google Scholar] [CrossRef]
- Ali, S.S.; Sonbol, F.I.; Sun, J.; Hussein, M.A.; Hafez, A.E.E.; Abdelkarim, E.A.; Kornaros, M.; Ali, A.; Azab, M. Molecular characterization of virulence and drug resistance genes-producing Escherichia coli isolated from chicken meat: Metal oxide nanoparticles as novel antibacterial agents. Microb. Pathog. 2020, 143, 104164. [Google Scholar] [CrossRef]
- Ali, S.S.; Al-Tohamy, R.; Koutra, E.; Moawad, M.S.; Kornaros, M.; Mustafa, A.M.; Mahmoud, Y.A.G.; Badr, A.; Osman, M.E.; Elsamahy, T.; et al. Nanobiotechnological advancements in agriculture and food industry: Applications, nanotoxicity, and future perspectives. Sci. Total Environ. 2021, 792, 148359. [Google Scholar] [CrossRef]
- Ali, S.S.; Moawad, M.S.; Hussein, M.A.; Azab, M.; Abdelkarim, E.A.; Badr, A.; Sun, J.; Khalil, M. Efficacy of metal oxide nanoparticles as novel antimicrobial agents against multi-drug and multi-virulent Staphylococcus aureus isolates from retail raw chicken meat and giblets. Int. J. Food Microbiol. 2021, 344, 109116. [Google Scholar] [CrossRef]
- Khalil, M.A.; El-Shanshoury, A.E.; Alghamdi, M.A.; Mohamed, S.F.; Sun, J.; Ali, S.S. Biosynthesis of silver nanoparticles by marine actinobacterium Nocardiopsis dassonvillei and exploring their therapeutic potentials. Front. Microbiol. 2022, 12, 705673. [Google Scholar] [CrossRef]
- Halkai, K.R.; Halkai, R.; Mudda, J.A.; Shivanna, V.; Rathod, V. Antibiofilm efficacy of biosynthesized silver nanoparticles against endodontic-periodontal pathogens: An in vitro study. J. Conserv. Dent. 2018, 21, 662. [Google Scholar] [CrossRef]
- Khalil, M.A.; El Maghraby, G.M.; Sonbol, F.I.; Allam, N.G.; Ateya, P.S.; Ali, S.S. Enhanced efficacy of some antibiotics in presence of silver nanoparticles against Multidrug resistant Pseudomonas aeruginosa recovered from burn wound infections. Front. Microbiol. 2021, 12, 648560. [Google Scholar] [CrossRef] [PubMed]
- Khalil, M.A.; Sonbol, F.I.; Al-Madboly, L.A.; Aboshady, T.A.; Alqurashi, A.S.; Ali, S.S. Exploring the therapeutic potentials of exopolysaccharides derived from lactic acid bacteria and bifidobacteria: Antioxidant, antitumor, and periodontal regeneration. Front. Microbiol. 2022, 13, 803688. [Google Scholar] [CrossRef] [PubMed]
- Ong, W.T.J.; Nyam, K.L. Evaluation of silver nanoparticles in cosmeceutical and potential biosafety complications. Saudi J. Biol. Sci. 2022, 29, 2085–2094. [Google Scholar] [CrossRef] [PubMed]
- Panpaliya, N.P.; Dahake, P.T.; Kale, Y.J.; Dadpe, M.V.; Kendre, S.B.; Siddiqi, A.G.; Maggavi, U.R. In vitro evaluation of antimicrobial property of silver nanoparticles and chlorhexidine against five different oral pathogenic bacteria. Saudi Dent. J. 2019, 31, 76–83. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-López, E.; Gomes, D.; Esteruelas, G.; Bonilla, L.; Lopez-Machado, A.L.; Galindo, R.; Cano, A.; Espina, M.; Ettcheto, M.; Camins, A.; et al. Metal-based nanoparticles as antimicrobial agents: An overview. Nanomaterials 2020, 10, 292. [Google Scholar] [CrossRef] [PubMed]
- Bertoglio, F.; De Vita, L.; D’Agostino, A.; Fernandez, Y.D.; Falqui, A.; Casu, A.; Merli, D.; Milanese, C.; Rossi, S.; Taglietti, A.; et al. Increased antibacterial and antibiofilm properties of silver nanoparticles using silver fluoride as precursor. Molecules 2020, 25, 3494. [Google Scholar] [CrossRef]
- Di Federico, A.; Rizzo, A.; Carloni, R.; De Giglio, A.; Bruno, R.; Ricci, D.; Brandi, G. Atezolizumab-bevacizumab plus Y-90 TARE for the treatment of hepatocellular carcinoma: Preclinical rationale and ongoing clinical trials. Expert Opin. Investig. Drugs 2022, 31, 361–369. [Google Scholar] [CrossRef]
- Bahlol, H.S.; Foda, M.F.; Ma, J.; Han, H. Robust synthesis of size-dispersal triangular silver nanoprisms via chemical reduction route and their cytotoxicity. Nanomaterials 2019, 9, 674. [Google Scholar] [CrossRef] [PubMed]
- Veeraraghavan, V.P.; Periadurai, N.D.; Karunakaran, T.; Hussain, S.; Surapaneni, K.M.; Jiao, X. Green synthesis of silver nanoparticles from aqueous extract of Scutellaria barbata and coating on the cotton fabric for antimicrobial applications and wound healing activity in fibroblast cells (L929). Saudi J. Biol. Sci. 2021, 28, 3633–3640. [Google Scholar] [CrossRef]
- Suriyakala, G.; Sathiyaraj, S.; Devanesan, S.; AlSalhi, M.S.; Rajasekar, A.; Maruthamuthu, M.K.; Babujanarthanam, R. Phytosynthesis of silver nanoparticles from Jatropha integerrima Jacq. flower extract and their possible applications as antibacterial and antioxidant agent. Saudi J. Biol. Sci. 2022, 29, 680–688. [Google Scholar] [CrossRef]
- Monteiro, D.R.; Gorup, L.F.; Takamiya, A.S.; Ruvollo-Filho, A.C.; de Camargo, E.R.; Barbosa, D.B. The growing importance of materials that prevent microbial adhesion: Antimicrobial effect of medical devices containing silver. Int. J. Antimicrob. Agents 2009, 34, 103–110. [Google Scholar] [CrossRef] [PubMed]
- Sharma, D.; Kanchi, S.; Bisetty, K. Biogenic synthesis of nanoparticles: A review. Arab. J. Chem. 2019, 12, 3576–3600. [Google Scholar] [CrossRef]
- Makvandi, P.; Ghomi, M.; Padil, V.V.; Shalchy, F.; Ashrafizadeh, M.; Askarinejad, S. Biofabricated nanostructures and their composites in regenerative medicine. ACS Appl. Nano Mater. 2020, 3, 6210–6238. [Google Scholar] [CrossRef]
- Darwesh, O.M.; Ali, S.S.; Matter, I.A.; Elsamahy, T. Nanotextiles waste management: Controlling of release and remediation of wastes. In Nanosensors and Nanodevices for Smart Multifunctional Textiles; Elsevier: Amsterdam, The Netherlands, 2021; pp. 267–286. [Google Scholar]
- Chernousova, S.; Epple, M. Silver as antibacterial agent: Ion, nanoparticle, and metal. Angew. Chem. Int. Ed. 2013, 52, 1636–1653. [Google Scholar] [CrossRef] [PubMed]
- Pallavicini, P.; Dacarro, G.; Taglietti, A. Self-assembled monolayers of silver nanoparticles: From intrinsic to switchable inorganic antibacterial surfaces. Eur. J. Inorg. Chem. 2018, 2018, 4846–4855. [Google Scholar] [CrossRef]
- Pandit, C.; Roy, A.; Ghotekar, S.; Khusro, A.; Islam, M.N.; Emran, T.B.; Lam, S.E.; Khandaker, M.U.; Bradley, D.A. Biological agents for synthesis of nanoparticles and their applications. J. King Saud Univ. Sci. 2022, 34, 101869. [Google Scholar] [CrossRef]
- Mabrouk, M.; Elkhooly, T.A.; Amer, S.K. Actinomycete strain type determines the monodispersity and antibacterial properties of biogenically synthesized silver nanoparticles. J. Genet. Eng. Biotechnol. 2021, 19, 57. [Google Scholar] [CrossRef]
- Mohseni, M.; Norouzi, H.; Hamedi, J.; Roohi, A. Screening of antibacterial producing actinomycetes from sediments of the Caspian Sea. Int. J. Mol. Cell Med. 2013, 2, 64. [Google Scholar]
- Veerasamy, R.; Xin, T.Z.; Gunasagaran, S.; Xiang, T.F.W.; Yang, E.F.C.; Jeyakumar, N.; Dhanaraj, S.A. Biosynthesis of silver nanoparticles using mangosteen leaf extract and evaluation of their antimicrobial activities. J. Saudi Chem. Soc. 2011, 15, 113–120. [Google Scholar] [CrossRef]
- Khalil, M.; Abdelhafez, Y.G.; Atta, H.; Kandeel, A.A. SPECT/CT is equivalent to diffusion-weighted MRI in characterizing equivocal osseous lesions detected by planar bone scintigraphy. Eur. J. Nuclear Med. Mol. Imaging 2017, 44, S218–S219. [Google Scholar]
- Liu, X.; Atwater, M.; Wang, J.; Huo, Q. Extinction coefficient of gold nanoparticles with different sizes and different capping ligands. Colloids Surf. B 2007, 58, 3–7. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.S.; El-Sheekh, M.; Manni, A.; Ruiz, H.A.; Elsamahy, T.; Sun, J.; Schagerl, M. Microalgae-mediated wastewater treatment for biofuels production: A comprehensive review. Microbiol. Res. 2022, 265, 127187. [Google Scholar] [CrossRef] [PubMed]
- Goel, N.; Ahmad, R.; Singh, R.; Sood, S.; Khare, S.K. Biologically synthesized silver nanoparticles by Streptomyces sp. EMB24 extracts used against the drug-resistant bacteria. Bioresour. Technol. Rep. 2021, 15, 100753. [Google Scholar] [CrossRef]
- CLSI. Performance Standards for Antimicrobial Susceptibility Tests, 13th ed.; CLSI Standard M02; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018. [Google Scholar]
- Christensen, G.D.; Simpson, W.A.; Younger, J.J.; Baddour, L.M.; Barrett, F.F.; Melton, D.M.; Beachey, E.H. Adherence of coagulase-negative staphylococci to plastic tissue culture plates: A quantitative model for the adherence of staphylococci to medical devices. J. Clin. Microbiol. 1985, 22, 996–1006. [Google Scholar] [CrossRef] [PubMed]
- Stepanovic, S.; Vukovic, D.; DakicSavic, I.B.; Svabic-Vlahovic, M. A modified microtiter-plate for quantification of staphylococcal biofilm formation. J. Microbiol. Methods 2000, 40, 175–179. [Google Scholar] [CrossRef]
- Boora, F.; Chirisa, E.; Mukanganyama, S. Evaluation of nitrite radical scavenging properties of selected Zimbabwean plant extracts and their phytoconstituents. J. Food Process. 2014, 2014, 918018. [Google Scholar] [CrossRef]
- Tonisi, S.; Okaiyeto, K.; Mabinya, L.V.; Okoh, A.I. Evaluation of bioactive compounds, free radical scavenging and anticancer activities of bulb extracts of Boophone disticha from Eastern Cape Province, South Africa. Saudi J. Biol. Sci. 2020, 27, 3559–3569. [Google Scholar] [CrossRef]
- Hmidani, A.; Bouhlali, E.D.T.; Khouya, T.; Ramchoun, M.; Filali-Zegzouti, Y.; Alem, C.; Benlyas, M. Antioxidant, anti-inflammatory and anticoagulant activities of three Thymus species grown in southeastern Morocco. Future J. Pharm. Sci. 2019, 5, 4. [Google Scholar] [CrossRef]
- Vijayakumar, N.; Bhuvaneshwari, V.K.; Ayyadurai, G.K.; Jayaprakash, R.; Gopinath, K.; Nicoletti, M.; Alarifi, S.; Govindarajan, M. Green synthesis of zinc oxide nanoparticles using Anoectochilus elatus, and their biomedical applications. Saudi J. Biol. Sci. 2021, 29, 2270–2279. [Google Scholar] [CrossRef]
- Ahmed, S.A.; Gogal Jr, R.M.; Walsh, J.E. A new rapid and simple non-radioactive assay to monitor and determine the proliferation of lymphocytes: An alternative to [3H] thymidine incorporation assay. J. Immunol. Methods 1994, 170, 211–224. [Google Scholar] [CrossRef]
- Chang, C.Y.; Yamada, S. Evaluation of the regenerative effect of a 25% doxycycline-loaded biodegradable membrane for guided tissue regeneration. J. Periodontol. 2000, 71, 1086–1093. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, M.; Duan, X.; Mujumdar, A. Effect of nano-silver coating on microbial control of microwave-freeze combined dried sea cucumber. Int. Agrophys. 2011, 25, 181–186. [Google Scholar]
- Yazdimamaghani, M.; Vashaee, D.; Assefa, S.; Walker, K.J.; Madihally, S.V.; Köhler, G.A. Hybrid macroporous gelatin/bioactive-glass/nanosilver scaffolds with controlled degradation behavior and antimicrobial activity for bone tissue engineering. J. Biomed. Nanotechnol. 2014, 10, 911–931. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Hu, C.; Shao, L. The antimicrobial activity of nanoparticles: Present situation and prospects for the future. Int. J. Nanomed. 2017, 12, 1227. [Google Scholar] [CrossRef]
- Espinosa-Cristóbal, L.F.; Holguín-Meráz, C.; Zaragoza-Contreras, E.A.; Martínez-Martínez, R.E.; Donohue-Cornejo, A.; Loyola-Rodríguez, J.P.; Cuevas-González, J.C.; Reyes-López, S.Y. Antimicrobial and substantivity properties of silver nanoparticles against oral microbiomes clinically isolated from young and young-adult patients. J. Nanomater. 2019, 2019, 3205971. [Google Scholar] [CrossRef]
- Hall-Stoodley, L.; Costerton, J.W.; Stoodley, P. Bacterial biofilms: From the natural environment to infectious diseases. Nat. Rev. Microbiol. 2004, 2, 95–108. [Google Scholar] [CrossRef]
- Al-Dhabi, N.A.; Ghilan AK, M.; Esmail, G.A.; Arasu, M.V.; Duraipandiyan, V.; Ponmurugan, K. Environmental friendly synthesis of silver nanomaterials from the promising Streptomyces parvus strain Al-Dhabi-91 recovered from the Saudi Arabian marine regions for antimicrobial and antioxidant properties. J. Photochem. Photobiol. B Biol. 2019, 197, 111529. [Google Scholar] [CrossRef]
- Salem, S.S.; Fouda, A. Green synthesis of metallic nanoparticles and their prospective biotechnological applications: An overview. Biol. Trace Elem. Res. 2021, 199, 344–370. [Google Scholar] [CrossRef]
- Al-Otibi, F.; Alfuzan, S.A.; Alharbi, R.I.; Al-Askar, A.A.; AL-Otaibi, R.M.; Al Subaie, H.F.; Moubayed, N.M. Comparative Study of Antifungal Activity of Two Preparations of Green Silver Nanoparticles from Portulaca oleracea Extract. Saudi J. Biol. Sci. 2022, 29, 2772–2781. [Google Scholar] [CrossRef]
- Manivasagan, P.; Venkatesan, J.; Senthilkumar, K.; Sivakumar, K.; Kim, S.-K. Biosynthesis, antimicrobial and cytotoxic effect of silver nanoparticles using a novel Nocardiopsis sp. MBRC-1. Biomed. Res. Int. 2013, 2013, 287638. [Google Scholar] [CrossRef]
- Singh, H.; Du, J.; Singh, P.; Yi, T.H. Ecofriendly synthesis of silver and gold nanoparticles by Euphrasia officinalis leaf extract and its biomedical applications. Artif. Cells Nanomed. Biotechnol. 2018, 46, 1163–1170. [Google Scholar] [CrossRef]
- Navya, P.N.; Daima, H.K. Rational engineering of physicochemical properties of nanomaterials for biomedical applications with nanotoxicological perspectives. Nano Converg. 2016, 3, 1. [Google Scholar] [CrossRef] [PubMed]
- Chithrani, B.D.; Chan, W.C. Elucidating the mechanism of cellular uptake and removal of protein-coated gold nanoparticles of different sizes and shapes. Nano Lett. 2007, 7, 1542–1550. [Google Scholar] [CrossRef] [PubMed]
- Chithrani, B.D.; Ghazani, A.A.; Chan, W.C. Determining the size and shape dependence of gold nanoparticle uptake into mammalian cells. Nano Lett. 2006, 6, 662–668. [Google Scholar] [CrossRef]
- Dacarro, G.; Pallavicini, P.; Bertani, S.M.; Chirico, G.; D’Alfonso, L.; Falqui, A.; Marchesi, N.; Pascale, A.; Sironi, L.; Taglietti, A.; et al. Synthesis of reduced-size gold nanostars and internalization in SH-SY5Y cells. J. Colloid Interface Sci. 2017, 505, 1055–1064. [Google Scholar] [CrossRef]
- Thirunavoukkarasu, M.; Balaji, U.; Behera, S.; Panda, P.K.; Mishra, B.K. Biosynthesis of silver nanoparticle from leaf extract of Desmodium gangeticum (L.) DC. and its biomedical potential. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2013, 116, 424–427. [Google Scholar] [CrossRef]
- Seyedeh, M.G.; Sepideh, H.; Shojaosadati, S.A. Green synthesis of silver nanoparticles by a novel method: Comparative study of their properties. Carbohydr. Polym. 2012, 89, 467–472. [Google Scholar]
- Kora, A.J.; Sashidhar, R.B.; Arunachalam, J. Aqueous extract of gumolibanum (Boswellia serrata): A reductant and stabilizer for the biosynthesis of antibacterial silver nanoparticles. Process. Biochem. 2012, 47, 1516–1520. [Google Scholar] [CrossRef]
- Saranyaadevi, K.; Subha, V.; Ravindran, R.S.E.; Renganathan, S. Green synthesis and characterization of silver nanoparticle using leaf extract of Capparis zeylanica. Asian J. Pharm. Clin. Res. 2014, 7, 44–48. [Google Scholar]
- Rai, M.K.; Deshmukh, S.D.; Ingle, A.P.; Gade, A.K. Silver nanoparticles: The powerful nanoweapon against multidrug-resistant bacteria. J. Appl. Microbiol. 2012, 112, 841–852. [Google Scholar] [CrossRef]
- Verma, A.; Sanghi, S.; Grover, D.; Aggarwal, S.; Gupta, R.; Pandit, N. Effect of insertion of xanthan-based chlorhexidine gel in the maintenance phase following the treatment of chronic periodontitis. J. Indian Soc. Periodontol. 2012, 16, 381–385. [Google Scholar] [CrossRef] [PubMed]
- Knight, E.T.; Liu, J.; Seymour, G.J.; Faggion Jr, C.M.; Cullinan, M.P. Risk factors that may modify the innate and adaptive immune responses in periodontal diseases. Periodontology 2016, 71, 22–51. [Google Scholar] [CrossRef]
- Ali, S.S.; Shaaban, M.T.; Abomohra, A.E.F.; El-Safity, K. Macroalgal activity against multiple drug resistant Aeromonas hydrophila: A novel treatment study towards enhancement of fish growth performance. Microb. Pathog. 2016, 101, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Freitas, R.D.; Dias, R.B.; Vidal, M.T.A.; Valverde, L.D.F.; Costa, R.G.A.; Damasceno, A.K.A.; Sales, C.B.S.; de Oliveira Siquara da Rocha, L.; Dos Reis, M.G.; Soares, M.B.P.; et al. Inhibition of CAL27 oral squamous carcinoma cell by targeting hedgehog pathway with vismodegib or itraconazole. Front. Oncol. 2020, 2469, 563838. [Google Scholar] [CrossRef] [PubMed]
- Franci, G.; Falanga, A.; Galdiero, S.; Palomba, L.; Rai, M.; Morelli, G.; Galdiero, M. Silver nanoparticles as potential antibacterial agents. Molecules 2015, 20, 8856–8874. [Google Scholar] [CrossRef] [PubMed]
- Khalil, M.A.E.F.; Sonbol, F.I.; Badr, A.; Ali, S.S. Comparative study of virulence factors among ESβL-producing and nonproducing Pseudomonas aeruginosa clinical isolates. Turk. J. Med. Sci. 2015, 45, 60–69. [Google Scholar] [CrossRef]
- Khalil, M.A.; El-Sheekh, M.M.; El-Adawi, H.I.; El-Deeb, N.M.; Hussein, M.Z. Efficacy of microencapsulated lactic acid bacteria in Helicobater pylori eradication therapy. J. Res. Med. Sci. 2015, 20, 950. [Google Scholar] [CrossRef]
- Khalil, M.; Ismail, M.M.; El Shafay, S.M. Evaluation of antibacterial activity of macroalgae extracts as adjunctive therapy in neonates sepsis induced by Klebsiella pneumoniae. Arab. J. Sci. Eng. 2020, 45, 4599–4607. [Google Scholar] [CrossRef]
- Qing, Y.; Cheng, L.; Li, R.; Liu, G.; Zhang, Y.; Tang, X.; Wang, J.; Liu, H.; Qin, Y. Potential antibacterial mechanism of silver nanoparticles and the optimization of orthopedic implants by advanced modification technologies. Int. J. Nanomed. 2018, 13, 3311–3327. [Google Scholar] [CrossRef]
- Abbaszadegan, A.; Ghahramani, Y.; Gholami, A.; Hemmateenejad, B.; Dorostkar, S.; Nabavizadeh, M.; Sharghi, H. The effect of charge at the surface of silver nanoparticles on antimicrobial activity against gram-positive and gram-negative bacteria: A preliminary study. J. Nanomater. 2015, 16, 53. [Google Scholar] [CrossRef]
- Pandey, J.K.; Swarnkar, R.K.; Soumya, K.K.; Dwivedi, P.; Singh, M.K.; Sundaram, S.; Gopal, R. Silver nanoparticles synthesized by pulsed laser ablation: As a potent antibacterial agent for human enteropathogenic gram-positive and gram-negative bacterial strains. Appl. Biochem. Biotechnol. 2014, 174, 1021–1031. [Google Scholar] [CrossRef] [PubMed]
- Deshmukh, S.P.; Patil, S.M.; Mullani, S.B.; Delekar, S.D. Silver nanoparticles as an effective disinfectant: A review. Mater. Sci. Eng. C 2019, 97, 954–965. [Google Scholar] [CrossRef] [PubMed]
- Panáček, A.; Kvítek, L.; Prucek, R.; Kolář, M.; Večeřová, R.; Pizúrová, N.; Sharma, V.K.; Nevěčná, T.; Zbořil, R. Silver colloid nanoparticles: Synthesis, characterization, and their antibacterial activity. J. Phys. Chem. B 2006, 110, 16248–16253. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Patil, S.; Ahire, M.; Kitture, R.; Kale, S.; Pardesi, K.; Cameotra, S.S.; Bellare, J.; Dhavale, D.D.; Jabgunde, A.; et al. Synthesis of silver nanoparticles using Dioscorea bulbifera tuber extract and evaluation of its synergistic potential in combination with antimicrobial agents. Int. J. Nanomed. 2012, 7, 483. [Google Scholar]
- Su, H.L.; Chou, C.C.; Hung, D.J.; Lin, S.H.; Pao, I.C.; Lin, J.H.; Huang, F.-L.; Dong, R.-X.; Lin, J.-J. The disruption of bacterial membrane integrity through ROS generation induced by nanohybrids of silver and clay. Biomaterials 2009, 30, 5979–5987. [Google Scholar] [CrossRef]
- Quinteros, M.A.; Aristizábal, V.C.; Dalmasso, P.R.; Paraje, M.G.; Páez, P.L. Oxidative stress generation of silver nanoparticles in three bacterial genera and its relationship with the antimicrobial activity. Toxicol. Vitr. 2016, 36, 216–223. [Google Scholar] [CrossRef]
- Morones, J.R.; Elechiguerra, J.L.; Camacho, A.; Holt, K.; Kouri, J.B.; Ramírez, J.T.; Yacaman, M.J. The bactericidal effect of silver nanoparticles. Nanotechnology 2005, 16, 2346. [Google Scholar] [CrossRef]
- Manivasagan, P.; Venkatesan, J.; Sivakumar, K.; Kim, S.K. Actinobacteria mediated synthesis of nanoparticles and their biological properties: A review. Crit. Rev. Microbiol. 2016, 42, 209–221. [Google Scholar] [CrossRef]
- Liao, C.; Li, Y.; Tjong, S.C. Bactericidal and cytotoxic properties of silver nanoparticles. Int. J. Mol. Sci. 2019, 20, 449. [Google Scholar] [CrossRef]
- Chauhan, R.; Kumar, A.; Abraham, J. A biological approach to the synthesis of silver nanoparticles with Streptomyces sp. JAR1 and its antimicrobial activity. Sci. Pharm. 2013, 81, 607–624. [Google Scholar] [CrossRef]
- Li, J.; Rong, K.; Zhao, H.; Li, F.; Lu, Z.; Chen, R. Highly selective antibacterial activities of silver nanoparticles against Bacillus subtilis. J. Nanosci. Nanotechnol. 2013, 13, 6806–6813. [Google Scholar] [CrossRef] [PubMed]
- Jung, W.K.; Koo, H.C.; Kim, K.W.; Shin, S.; Kim, S.H.; Park, Y.H. Antibacterial activity and mechanism of action of the silver ion in Staphylococcus aureus and Escherichia coli. Appl. Environ. Microbiol. 2008, 74, 2171–2178. [Google Scholar] [CrossRef] [PubMed]
- Mhaske, A.R.; Shetty, P.C.; Bhat, N.S.; Ramachandra, C.S.; Laxmikanth, S.M.; Nagarahalli, K.; Tekale, P.D. Antiadherent and antibacterial properties of stainless steel and NiTi orthodontic wires coated with silver against Lactobacillus acidophilus—An in vitro study. Prog. Orthod. 2015, 16, 40. [Google Scholar] [CrossRef] [PubMed]
- Zaki, A.; Aziz, M.N.; Ahmad, R.; Ahamad, I.; Ali, M.S.; Yasin, D.; Afzal, B.; Ali, S.M.; Chopra, A.; Hadda, V.; et al. Synthesis, purification and characterization of Plectonema derived AgNPs with elucidation of the role of protein in nanoparticle stabilization. RSC Adv. 2022, 12, 2497–2510. [Google Scholar] [CrossRef] [PubMed]
- Jiménez-Ramírez, A.J.; Martínez-Martínez, R.E.; Ayala-Herrera, J.L.; Zaragoza-Contreras, E.A.; Domínguez-Pérez, R.A.; Reyes-López, S.Y.; Donohue-Cornejo, A.; Cuevas-González, J.C.; Silva-Benítez, E.L.; Espinosa-Cristóbal, L.F. Antimicrobial activity of silver nanoparticles against clinical biofilms from patients with and without dental Caries. J. Nanomater. 2021, 2021, 5587455. [Google Scholar] [CrossRef]
- Charulatha, V.; Rajaram, A. Influence of different crosslinking treatments on the physical properties of collagen membranes. Biomaterials 2003, 24, 759–767. [Google Scholar] [CrossRef]


| Strain No. | Nomenclature | Accession No. | Closest Relative Yeast | Sequence Identity (%) |
|---|---|---|---|---|
| M0601 | Staphylococcus aureus | LN899797 | Staphylococcus aureus strain WO53 (LC107797) | 100.0 |
| M0901 | Staphylococcus aureus | LN899798 | Staphylococcus aureus strain GR41 (LC107809) | 99.57 |
| M1102 | Staphylococcus aureus | LN899800 | Staphylococcus aureus isolate H3 (LN899816) | 99.90 |
| M0201 | Staphylococcus epidermidis | LN899795 | Staphylococcus epidermidis strain TMPC 9023C (OM265429) | 100.0 |
| M0401 | Staphylococcus hominis | LN899796 | Staphylococcus hominis strain PL562 (MK015863) | 99.68 |
| Periodontal Pathogens | Antibiotic Resistance Pattern | # SNPs (μg/mL) | |
|---|---|---|---|
| MIC | MBC | ||
| S. aureus M1102 | VA, E, PB, OX, CAZ | 8 | 32 |
| S. aureus M0601 | TE, VA, E, PB, AMC, OX, FOX | 16 | 32 |
| S. aureus M0901 | VA, GN, NOR, E, PB, AMC, OX, FOX, CAZ | 64 | 64 |
| S. hominis M0401 | TE, VA, GN, CIP, NOR, E, C, PB, AMC, OX, CAZ | 64 | 128 |
| S. epidermidis M0201 | TE, VA, GN, AK, CIP, NOR, E, C, PB, AMC, OX, FOX, CAZ | 128 | 256 |
| Bacterial Strain | Biofilm Formation before Treatment | Biofilm Formation after Treatment with SNPs | ||
|---|---|---|---|---|
| Producing Category | A630nm | Producing Category | A630nm | |
| S. aureus M1102 | +++ | 0.272 ± 0.00 | - | 0.10 ± 0.002 |
| S. aureus M0601 | + | 0.110 ± 0.01 | - | 0.08 ± 0.003 |
| S. aureus M0901 | ++ | 0.210 ± 0.01 | - | 0.07 ± 0.004 |
| S. hominis M0401 | +++ | 0.255 ± 0.00 | - | 0.11 ± 0.001 |
| S. epidermidis M0201 | +++ | 0.56 ± 0.02 | +++ | 0.45 ± 0.002 |
| Tested Materials | IC50 (μg/mL) | |||
|---|---|---|---|---|
| Nitric Oxide Radical Scavenging Activity | Inhibition of Protein Denaturation | * Cytotoxicity | ||
| CAL27 | PMBC | |||
| SNPs | 110.7 ± 6.15 | 89.44 ± 5.52 | 34.03 | 81.16 |
| Standard | 80.07 ± 4.2 # | 215.5 ± 4.90 ## | - | - |
| Group | Day | Adherence Score | ||||
|---|---|---|---|---|---|---|
| S. aureus M1102 | S. aureus M0601 | S. aureus M0901 | S. hominis M0401 | S. epidermidis M0201 | ||
| GTR-C | 1 | 1.1 ± 0.3 | 1.0 ± 0.0 | 1.1 ± 0.34 | 1.3 ± 0.6 | 1.4 ± 0.8 |
| 3 | 1.99 ± 0.33 * | 1.1 ± 0.3 | 1.2 ± 0.24 | 1.5 ± 0.5 | 2.05 ± 0.2 * | |
| 5 | 2.04 ± 0.24 * | 2.2 ± 0.45 * | 2.14 ± 0.4 * | 3.0 ± 0.24 * | 3.1 ± 0.3 * | |
| 7 | 2.9 ± 0.3 * | 3.2 ± 0.1 * | 2.95 ± 0.23 * | 3.1 ± 0.1 * | 3.3 ± 0.21 * | |
| GTR-NS | 1 | 0.08 ± 0.1 | 0.05 ± 0.01 | 0.08 ± 0.04 | 0.9 ± 0.1 | 1.0 ± 0.02 |
| 3 | 1.0 ± 0.2 * | 1.1 ± 0.4 * | 1.3 ± 0.5 * | 1.3 ± 0.5 * | 1.4 ± 0.3 * | |
| 5 | 1.5 ± 0.3 * | 1.4 ± 0.6 * | 1.6 ± 0.4 * | 1.4 ± 0.2 * | 1.6 ± 0.4 * | |
| 7 | 1.8 ± 0.8 * | 1.5 ± 0.72 * | 1.7 ± 0.48 * | 1.5 ± 0.5 * | 1.8 ± 0.43 * | |
| Group | Day | CFU/ml | ||||
|---|---|---|---|---|---|---|
| S. aureus M1102 | S. aureus M0601 | S. aureus M0901 | S. hominis M0401 | S. epidermidis M0201 | ||
| GTR-C | 1 | 285.2 ± 3.0 | 125.0 ± 0.0 | 95.1 ± 0.0 | 121.17 ± 3.8 | 160.4 ± 0.8 |
| 3 | 255.5 ± 2.9 * | 155.58 ± 0.3 | 198.6 ± 1.2 | 199.8 ± 1.6 | 210.2 ± 0.2 * | |
| 5 | 240.1 ± 0.24 * | 278.2 ± 8.45 * | 198.8 ± 0.4 * | 235.17 ± 6.24 * | 270.0 ± 5.3 * | |
| 7 | 179.58 ± 7.3 * | 269.4 ± 4.51 * | 300.0 ± 3.3 * | 285.2 ± 3.1 * | 330 ± 4.81 * | |
| GTR-NS | 1 | 146.78 ± 8.1 | 99.0 ± 0.1 | 82.7 ± 1.4 | 90.0 ± 8.1 | 100.0 ± 7. 2 |
| 3 | 126.54 ± 9.2 * | 84.9 ± 3.4 * | 71.2 ± 6.5 * | 71.2 ± 4.5 * | 94.2 ± 5.3 * | |
| 5 | 115.0 ± 0.3 * | 70.0 ± 6.6 * | 51.4 ± 4.4 * | 54.0 ± 2.2 * | 46.0 ± 4.4 * | |
| 7 | 74.47 ± 10.8 * | 40.8 ± 7.72 * | 32.17 ± 6.8 * | 0.0 ± 0.0 * | 0.0 ± 0.0 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Elsilk, S.E.; Khalil, M.A.; Aboshady, T.A.; Alsalmi, F.A.; Ali, S.S. Streptomyces rochei MS-37 as a Novel Marine Actinobacterium for Green Biosynthesis of Silver Nanoparticles and Their Biomedical Applications. Molecules 2022, 27, 7296. https://doi.org/10.3390/molecules27217296
Elsilk SE, Khalil MA, Aboshady TA, Alsalmi FA, Ali SS. Streptomyces rochei MS-37 as a Novel Marine Actinobacterium for Green Biosynthesis of Silver Nanoparticles and Their Biomedical Applications. Molecules. 2022; 27(21):7296. https://doi.org/10.3390/molecules27217296
Chicago/Turabian StyleElsilk, Sobhy E., Maha A. Khalil, Tamer A. Aboshady, Fatin A. Alsalmi, and Sameh S. Ali. 2022. "Streptomyces rochei MS-37 as a Novel Marine Actinobacterium for Green Biosynthesis of Silver Nanoparticles and Their Biomedical Applications" Molecules 27, no. 21: 7296. https://doi.org/10.3390/molecules27217296
APA StyleElsilk, S. E., Khalil, M. A., Aboshady, T. A., Alsalmi, F. A., & Ali, S. S. (2022). Streptomyces rochei MS-37 as a Novel Marine Actinobacterium for Green Biosynthesis of Silver Nanoparticles and Their Biomedical Applications. Molecules, 27(21), 7296. https://doi.org/10.3390/molecules27217296







