Exosomes as New Generation Vehicles for Drug Delivery: Biomedical Applications and Future Perspectives
Abstract
1. Introduction
1.1. Background
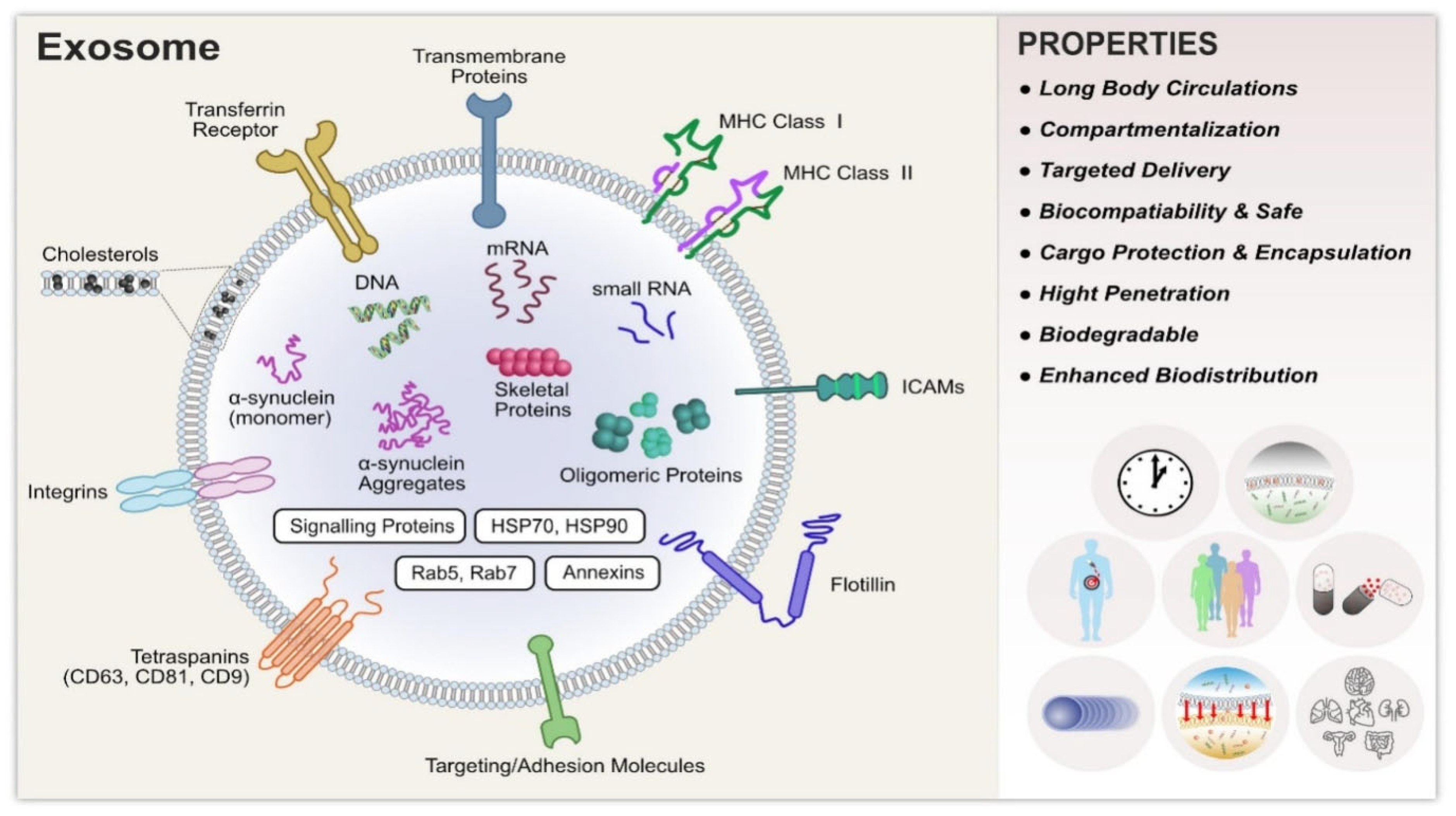
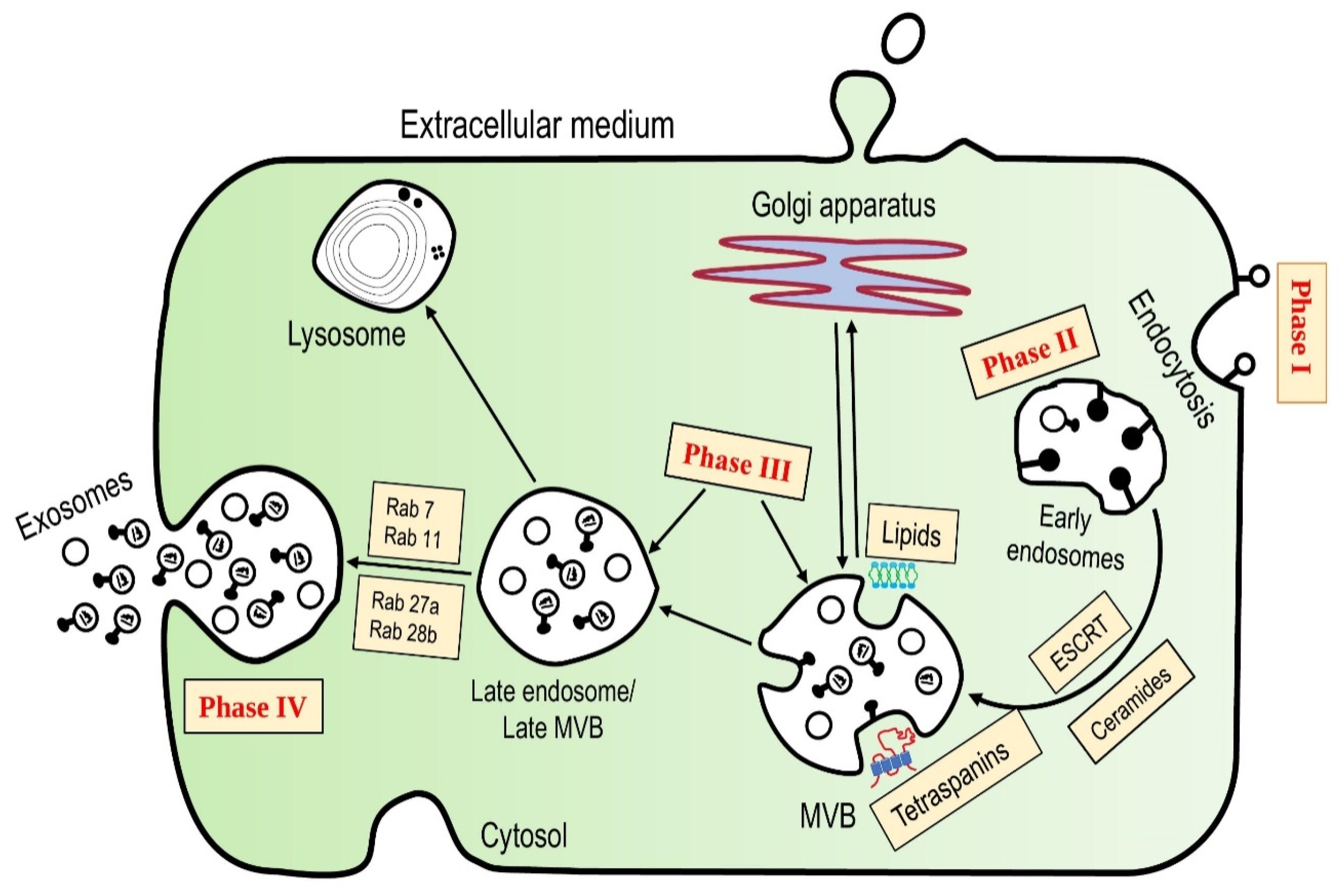
1.2. Structure and Biogenesis
1.2.1. Structure
1.2.2. Biogenesis
| Composition | Function | Examples |
|---|---|---|
| Lipids and Metabolites | Vesicle formation | Glycosphingolipids Monosialotetrahexosyl ganglioside (GM3) Sphingomyelin Cholesterol Phosphatidylserine Prostaglandins Glycerophospholipids organic acids Alcohols, Steroids, Phenols, amino acids conjugates, Sugar and conjugates |
| Biogenesis | ||
| Release & interaction with target cells | ||
| Pathophysiological conditions | ||
| Inflammatory processes | ||
| Rigidity | ||
| Proteins | Transporters | ATP7A, ATP7B, MRP2, SLC1A4, SLC16A1, CLIC1 |
| Receptors | CD46, CD55 | |
| Heat shock proteins | Hsc70, Hsp70, Hsp90 | |
| Tetraspanins | CD9, CD81, CD82 | |
| Metabolic enzymes | GAPDH, Pyruvate | |
| Antigen presentation proteins | HLA Class I & II, Peptide complexes | |
| Lysosomal markers | CD63, Lysosome membrane protein 2 | |
| Membrane adhesion proteins | Integrins | |
| Nucleic Acids | Mediator of horizontal transfer of genetic information | mRNA |
| Gene regulation | Non-Coding RNA | |
| Target cells gene silencing | miRNA | |
| Carcinogenesis and cancer progression | Long Non-Coding RNAs |
1.3. Role of Exosomes
1.4. Types of EVs
- (a)
- Exosomes (30–150 nm);
- (b)
- Oncosomes (100–1000 nm);
- (c)
- Ectoderms/Microvesicles (100–1000 nm);
- (d)
- Apoptotic bodies (200–2000 nm) [51].
2. Isolation and Separation Techniques for Exosomes
2.1. Ultrafiltration
2.2. Immunological Separation
2.3. Ultracentrifugation
2.4. Size Exclusion Chromatography
2.5. Polymer-Based Precipitation Separation
2.6. Magnetic Separation
2.7. Acoustic Fluidic Separation
2.8. Dielectrophoretic Separation
2.9. Deterministic Lateral Displacement (DLD) Separation
2.10. Microfluidic Devices
3. Characterization of Exosomes
3.1. Nanoparticle Tracking Analysis (NTA)
3.2. Dynamic Light Scattering (DLS)
3.3. Atomic Force Microscopy (AFM)
3.4. Microscopy Study
3.4.1. Transmission Electron MICROSCOPY (TEM)
3.4.2. Scanning Electron Microscopy (SEM)
3.5. Enzyme-Linked Immunosorbent Assay (ELISA)
3.6. Fluorescence Correlation Microscopy (FCM)
3.7. Colorimetric Detection
3.8. Surface Plasmon Resonance (SPR) Detection
3.9. Nuclear Magnetic Resonance (NMR) Detection
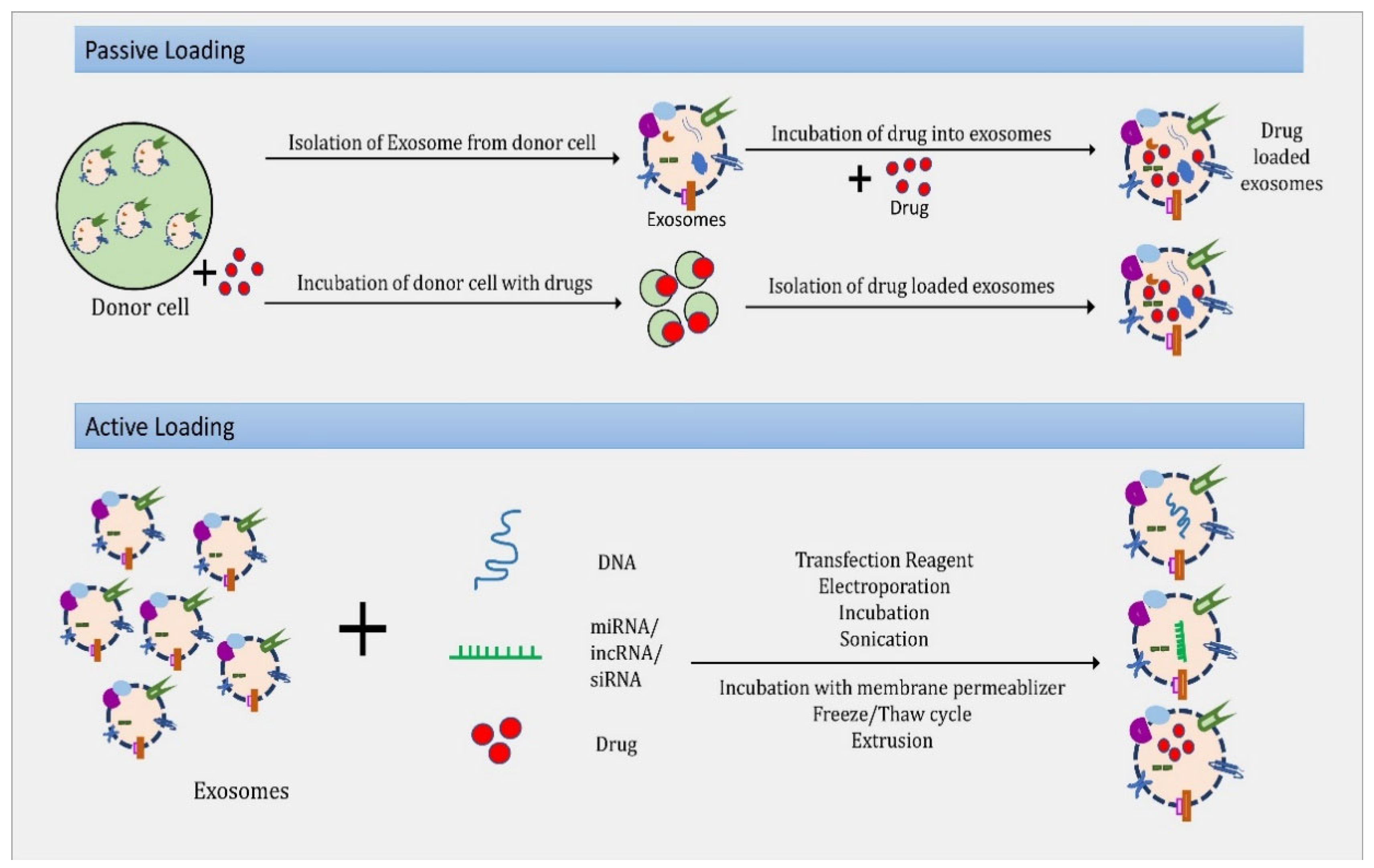
4. Exosomes as Drug Delivery Vehicle
4.1. Small Molecules
4.2. Large Molecule (Protein and Peptide Delivery)
4.3. Nucleic Acids
4.4. Small Interfering RNAs (siRNAs)
4.5. MicroRNA (miRNA)
4.6. Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/CRISPR Associated Protein 9 (Cas9) System
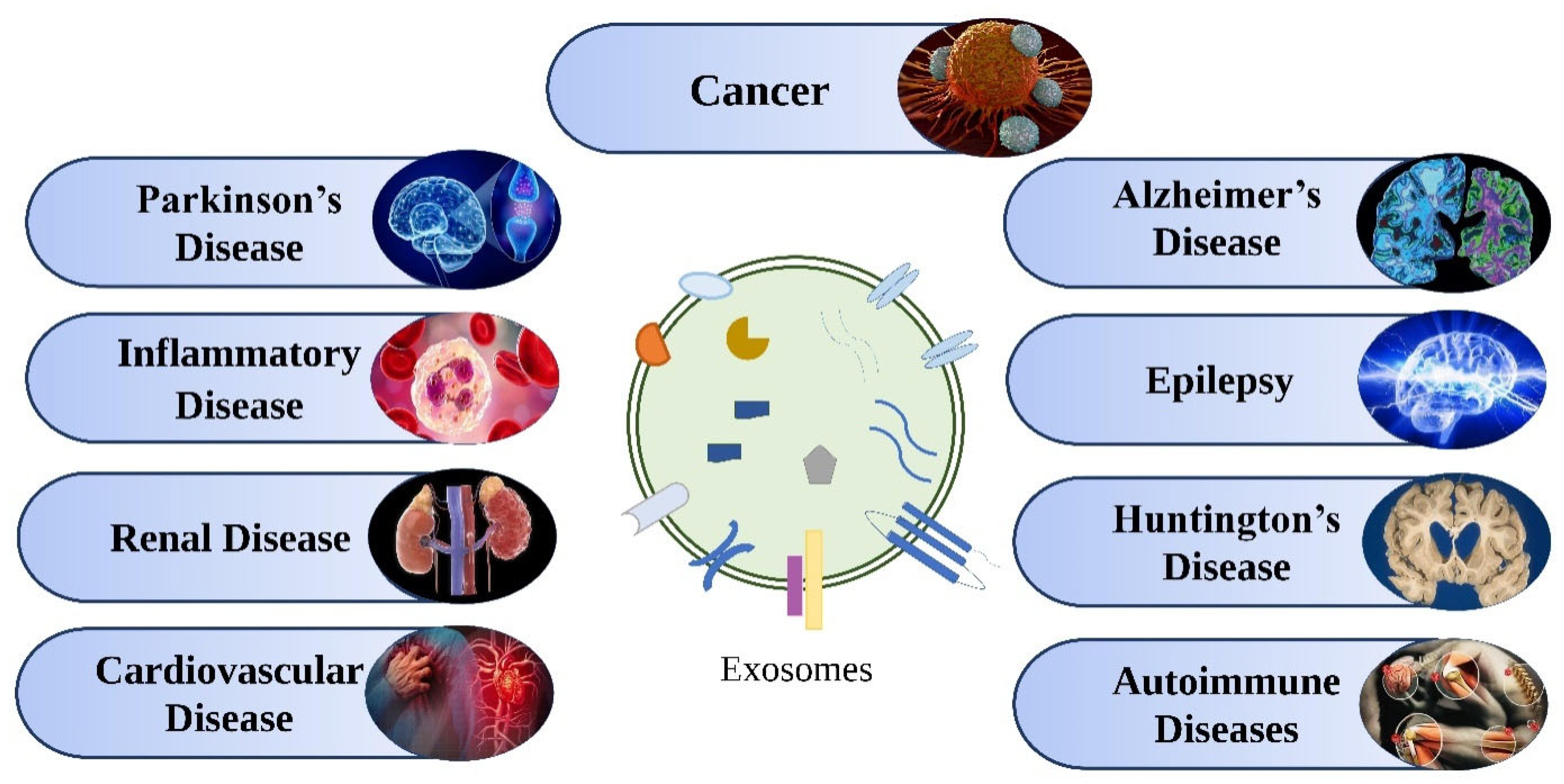
5. Therapeutic Applications of Exosomes
5.1. In Cancer
5.2. In neurological Diseases
5.2.1. Parkinson’s Disease
5.2.2. Alzheimer’s Disease (AD)
5.2.3. Epilepsy
5.2.4. Huntington’s Disease
5.2.5. Stroke
5.2.6. Amyotrophic Lateral Sclerosis
5.3. Inflammatory Disease
5.4. Autoimmune Disease
5.4.1. Exosome’s Role in Rheumatoid Arthritis and Joint Diseases
5.4.2. Exosome’s Role in other Autoimmune and Chronic Inflammatory Diseases
5.5. Renal Diseases
5.6. Cardiovascular Diseases
6. Challenges Associated with Exosome-Based Drug Delivery
7. Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| Term | Full-Form |
| AC | Alternating current |
| AD | Alzheimer’s disease |
| AFM | Atomic Force Microscopy |
| BBB | Blood-brain barrier |
| BSE | Backscattered Electrons |
| CNS | Central nervous system |
| Cryo-TEM | Cryogenic transmission electron microscopy |
| CSF | Cerebrospinal fluid |
| CSP | Cysteine string protein |
| DEP | Dielectrophoretic |
| DLD | Deterministic lateral displacement |
| DLS | Dynamic Light Scattering |
| ELISA | Enzyme-linked Immunosorbent Assay |
| EOAD | Early-onset Alzheimer’s disease |
| ERT | Enzyme-replacement therapy |
| ESCRT | Endosomal sorting complex required for transport |
| EVs | Extracellular vesicles |
| FCM | Fluorescence Correlation Microscopy |
| GMP | Good manufacturing practices |
| GTPase | guanosine triphosphatase |
| HD | Huntington’s disease |
| KI | Knock in |
| LOAD | Late-onset Alzheimer’s disease |
| MHC | Major histocompatibility complex |
| miRNA | MicroRNA |
| MPS | Mononuclear phagocyte system |
| MVB | Multivesicular body |
| MVs | Microvesicles |
| NFTs | Neurofibrillary tangles |
| nPLEX | Nano plasmonic exosome |
| NSCLC | Non-Small-Cell Lung Cancer |
| NTA | Nanoparticle Tracking Analysis |
| PD | Parkinson’s disease |
| PEG | Polyethylene glycol |
| PM | Plasma membrane |
| SEC | Size exclusion chromatography |
| SEM | Scanning Electron Microscopy |
| siRNAs | Small interfering RNAs |
| SPR | Surface Plasmon Resonance |
| TEM | Transmission Electron Microscopy |
| TFF | Tangential flow filtration |
| TSC | Tuberous sclerosis complex |
References
- Anderson, H.C. Vesicles associated with calcification in the matrix of epiphyseal cartilage. J. Cell Biol. 1969, 41, 59–72. [Google Scholar] [CrossRef] [PubMed]
- Bonucci, E. Fine structure of early cartilage calcification. J. Ultrastruct. Res. 1967, 20, 33–50. [Google Scholar] [CrossRef]
- Wolf, P. The nature and significance of platelet products in human plasma. Br. J. Haematol. 1967, 13, 269–288. [Google Scholar] [CrossRef]
- Pan, B.T.; Teng, K.; Wu, C.; Adam, M.; Johnstone, R.M. Electron microscopic evidence for externalization of the transferrin receptor in vesicular form in sheep reticulocytes. J. Cell Biol. 1985, 101, 942–948. [Google Scholar] [CrossRef] [PubMed]
- Anderson, H.C.; Garimella, R.; Tague, S.E. The role of matrix vesicles in growth plate development and biomineralization. Front. Biosci. 2005, 10, 822–837. [Google Scholar] [CrossRef] [PubMed]
- Bakhshian Nik, A.; Hutcheson, J.D.; Aikawa, E. Extracellular vesicles as mediators of cardiovascular calcification. Front. Cardiovasc. Med. 2017, 4, 78. [Google Scholar] [CrossRef] [PubMed]
- Trams, E.G.; Lauter, C.J.; Salem, J.N.; Heine, U. Exfoliation of membrane ecto-enzymes in the form of micro-vesicles. Biochim. Et Biophys. Acta (BBA)-Biomembr. 1981, 645, 63–70. [Google Scholar] [CrossRef]
- Arienti, G.; Carlini, E.; Verdacchi, R.; Cosmi, E.V.; Palmerini, C.A. Prostasome to sperm transfer of CD13/aminopeptidase N (EC 3.4. 11.2). Biochim. Biophys. Acta 1997, 1336, 533–538. [Google Scholar] [CrossRef]
- Stegmayr, B.; Brody, I.; Ronquist, G. A biochemical and ultrastructural study on the endogenous protein kinase activity of secretory granule membranes of prostatic origin in human seminal plasma. J. Ultrastruct. Res. 1982, 78, 206–214. [Google Scholar] [CrossRef]
- Schwechheimer, C.; Kuehn, M.J. Outer-membrane vesicles from Gram-negative bacteria: Biogenesis and functions. Nat. Rev. Microbiol. 2015, 13, 605–619. [Google Scholar] [CrossRef]
- Woith, E.; Fuhrmann, G.; Melzig, M.F. Extracellular vesicles—Connecting kingdoms. Int. J. Mol. Sci. 2019, 20, 5695. [Google Scholar] [CrossRef] [PubMed]
- Buschmann, D.; Mussack, V.; Byrd, J.B. Separation, characterization, and standardization of extracellular vesicles for drug delivery applications. Adv. Drug Deliv. Rev. 2021, 174, 348–368. [Google Scholar] [CrossRef]
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef]
- Tarasov, V.V.; Svistunov, A.A.; Chubarev, V.N.; Dostdar, S.A.; Sokolov, A.V.; Brzecka, A.; Sukocheva, O.; Neganova, M.E.; Klochkov, S.G.; Somasundaram, S.G.; et al. Extracellular vesicles in cancer nanomedicine. Semin. Cancer Biol. 2021, 69, 212–225. [Google Scholar] [CrossRef]
- Chulpanova, D.S.; Kitaeva, K.V.; James, V.; Rizvanov, A.A.; Solovyeva, V.V. Therapeutic prospects of extracellular vesicles in cancer treatment. Front. Immunol. 2018, 9, 1534. [Google Scholar] [CrossRef]
- Sun, Y.-Z.; Ruan, J.-S.; Jiang, Z.-S.; Wang, L.; Wang, S.-M. Extracellular Vesicles: A New Perspective in Tumor Therapy. BioMed Res. Int. 2018, 2018, 2687954. [Google Scholar] [CrossRef]
- Akers, J.C.; Gonda, D.; Kim, R.; Carter, B.S.; Chen, C.C. Biogenesis of extracellular vesicles (EV): Exosomes, microvesicles, retrovirus-like vesicles, and apoptotic bodies. J. Neuro-Oncol. 2013, 113, 1–11. [Google Scholar] [CrossRef]
- Chung, I.-M.; Rajakumar, G.; Venkidasamy, B.; Subramanian, U.; Thiruvengadam, M. Exosomes: Current use and future applications. Clin. Chim. Acta 2020, 500, 226–232. [Google Scholar] [CrossRef] [PubMed]
- Gurunathan, S.; Kang, M.-H.; Jeyaraj, M.; Qasim, M.; Kim, J.-H. Review of the isolation, characterization, biological function, and multifarious therapeutic approaches of exosomes. Cells 2019, 8, 307. [Google Scholar] [CrossRef]
- Araujo-Abad, S.; Saceda, M.; de Juan Romero, C. Biomedical application of small extracellular vesicles in cancer treatment. Adv. Drug Deliv. Rev. 2022, 182, 114117. [Google Scholar] [CrossRef] [PubMed]
- Abels, E.R.; Breakefield, X.O. Introduction to extracellular vesicles: Biogenesis, RNA cargo selection, content, release, and uptake. Cell. Mol. Neurobiol. 2016, 36, 301–312. [Google Scholar] [CrossRef]
- Kalra, H.; Simpson, R.J.; Ji, H.; Aikawa, E.; Altevogt, P.; Askenase, P.; Bond, V.C.; Borràs, F.E.; Breakefield, X.; Budnik, V. Vesiclepedia: A compendium for extracellular vesicles with continuous community annotation. PLoS Biol. 2012, 10, e1001450. [Google Scholar] [CrossRef] [PubMed]
- Kowal, J.; Tkach, M.; Théry, C. Biogenesis and secretion of exosomes. Curr. Opin. Cell Biol. 2014, 29, 116–125. [Google Scholar] [CrossRef] [PubMed]
- Braccioli, L.; Van Velthoven, C.; Heijnen, C.J. Exosomes: A new weapon to treat the central nervous system. Mol. Neurobiol. 2014, 49, 113–119. [Google Scholar] [CrossRef] [PubMed]
- Yuan, D.; Zhao, Y.; Banks, W.A.; Bullock, K.M.; Haney, M.; Batrakova, E.; Kabanov, A.V. Macrophage exosomes as natural nanocarriers for protein delivery to inflamed brain. Biomaterials 2017, 142, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Tian, T.; Zhu, Y.; Jaffar Ali, D.; Hu, F.; Qi, Y.; Sun, B.; Xiao, Z. Exosomes transfer among different species cells and mediating miRNAs delivery. J. Cell. Biochem. 2017, 118, 4267–4274. [Google Scholar] [CrossRef] [PubMed]
- Mathivanan, S.; Ji, H.; Simpson, R.J. Exosomes: Extracellular organelles important in intercellular communication. J. Proteom. 2010, 73, 1907–1920. [Google Scholar] [CrossRef] [PubMed]
- Schorey, J.S.; Harding, C.V. Extracellular vesicles and infectious diseases: New complexity to an old story. J. Clin. Investig. 2016, 126, 1181–1189. [Google Scholar] [CrossRef]
- Exosomes. Exocarta. Available online: http://www.exocarta.org (accessed on 14 August 2022).
- Xie, F.; Zhou, X.; Fang, M.; Li, H.; Su, P.; Tu, Y.; Zhang, L.; Zhou, F. Extracellular vesicles in cancer immune microenvironment and cancer immunotherapy. Adv. Sci. 2019, 6, 1901779. [Google Scholar] [CrossRef]
- Mimeault, M.; Batra, S.K. Molecular biomarkers of cancer stem/progenitor cells associated with progression, metastases, and treatment resistance of aggressive cancers. Cancer Epidemiol. Prev. Biomark. 2014, 23, 234–254. [Google Scholar] [CrossRef] [PubMed]
- Bashyal, S.; Thapa, C.; Lee, S. Recent progresses in exosome-based systems for targeted drug delivery to the brain. J. Control. Release 2022, 348, 723–744. [Google Scholar] [CrossRef] [PubMed]
- Pegtel, D.M.; Gould, S.J. Exosomes. Annu. Rev. Biochem. 2019, 88, 487–514. [Google Scholar] [CrossRef] [PubMed]
- Raposo, G.; Nijman, H.W.; Stoorvogel, W.; Liejendekker, R.; Harding, C.V.; Melief, C.; Geuze, H.J. B lymphocytes secrete antigen-presenting vesicles. J. Exp. Med. 1996, 183, 1161–1172. [Google Scholar] [CrossRef]
- Liao, W.; Du, Y.; Zhang, C.; Pan, F.; Yao, Y.; Zhang, T.; Peng, Q. Exosomes: The next generation of endogenous nanomaterials for advanced drug delivery and therapy. Acta Biomater. 2019, 86, 1–14. [Google Scholar] [CrossRef] [PubMed]
- McGough, I.J.; Vincent, J.P. Exosomes in developmental signalling. Development 2016, 143, 2482–2493. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, Y.; Liu, H.; Tang, W.H. Exosomes: Biogenesis, biologic function and clinical potential. Cell Biosci. 2019, 9, 19. [Google Scholar] [CrossRef]
- Gurung, S.; Perocheau, D.; Touramanidou, L.; Baruteau, J. The exosome journey: From biogenesis to uptake and intracellular signalling. Cell Commun. Signal. 2021, 19, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Simona, F.; Laura, S.; Simona, T.; Riccardo, A. Contribution of proteomics to understanding the role of tumor-derived exosomes in cancer progression: State of the art and new perspectives. Proteomics 2013, 13, 1581–1594. [Google Scholar] [CrossRef] [PubMed]
- Harding, C.; Heuser, J.; Stahl, P. Receptor-mediated endocytosis of transferrin and recycling of the transferrin receptor in rat reticulocytes. J. Cell Biol. 1983, 97, 329–339. [Google Scholar] [CrossRef]
- Pan, B.-T.; Johnstone, R.M. Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: Selective externalization of the receptor. Cell 1983, 33, 967–978. [Google Scholar] [CrossRef]
- Robbins, P.D.; Dorronsoro, A.; Booker, C.N. Regulation of chronic inflammatory and immune processes by extracellular vesicles. J. Clin. Investig. 2016, 126, 1173–1180. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Holmes, C. Tissue regeneration capacity of extracellular vesicles isolated from bone marrow-derived and adipose-derived mesenchymal stromal/stem cells. Front. Cell Dev. Biol. 2021, 9, 648098. [Google Scholar] [CrossRef] [PubMed]
- Qu, P.; Qing, S.; Liu, R.; Qin, H.; Wang, W.; Qiao, F.; Ge, H.; Liu, J.; Zhang, Y.; Cui, W. Effects of embryo-derived exosomes on the development of bovine cloned embryos. PLoS ONE 2017, 12, e0174535. [Google Scholar] [CrossRef]
- Raab-Traub, N.; Dittmer, D.P. Viral effects on the content and function of extracellular vesicles. Nat. Rev. Microbiol. 2017, 15, 559–572. [Google Scholar] [CrossRef] [PubMed]
- Ramakrishnaiah, V.; Thumann, C.; Fofana, I.; Habersetzer, F.; Pan, Q.; de Ruiter, P.E.; Willemsen, R.; Demmers, J.A.; Stalin Raj, V.; Jenster, G. Exosome-mediated transmission of hepatitis C virus between human hepatoma Huh7. 5 cells. Proc. Natl. Acad. Sci. USA 2013, 110, 13109–13113. [Google Scholar] [CrossRef]
- Costa-Silva, B.; Aiello, N.M.; Ocean, A.J.; Singh, S.; Zhang, H.; Thakur, B.K.; Becker, A.; Hoshino, A.; Mark, M.T.; Molina, H. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat. Cell Biol. 2015, 17, 816–826. [Google Scholar] [CrossRef]
- Melo, S.A.; Sugimoto, H.; O’Connell, J.T.; Kato, N.; Villanueva, A.; Vidal, A.; Qiu, L.; Vitkin, E.; Perelman, L.T.; Melo, C.A. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell 2014, 26, 707–721. [Google Scholar] [CrossRef]
- Fiandaca, M.S.; Kapogiannis, D.; Mapstone, M.; Boxer, A.; Eitan, E.; Schwartz, J.B.; Abner, E.L.; Petersen, R.C.; Federoff, H.J.; Miller, B.L. Identification of preclinical Alzheimer’s disease by a profile of pathogenic proteins in neurally derived blood exosomes: A case-control study. Alzheimer’s Dement. 2015, 11, 600–607.e601. [Google Scholar] [CrossRef]
- Ngolab, J.; Trinh, I.; Rockenstein, E.; Mante, M.; Florio, J.; Trejo, M.; Masliah, D.; Adame, A.; Masliah, E.; Rissman, R.A. Brain-derived exosomes from dementia with Lewy bodies propagate α-synuclein pathology. Acta Neuropathol. Commun. 2017, 5, 1–10. [Google Scholar] [CrossRef]
- Lener, T.; Gimona, M.; Aigner, L.; Börger, V.; Buzas, E.; Camussi, G.; Chaput, N.; Chatterjee, D.; Court, F.A.; Portillo, H.A.d. Applying extracellular vesicles based therapeutics in clinical trials—An ISEV position paper. J. Extracell. Vesicles 2015, 4, 30087. [Google Scholar] [CrossRef]
- Cappello, F.; Logozzi, M.; Campanella, C.; Bavisotto, C.C.; Marcilla, A.; Properzi, F.; Fais, S. Exosome levels in human body fluids: A tumor marker by themselves? Eur. J. Pharm. Sci. 2017, 96, 93–98. [Google Scholar] [CrossRef] [PubMed]
- Keller, S.; Ridinger, J.; Rupp, A.-K.; Janssen, J.W.; Altevogt, P. Body fluid derived exosomes as a novel template for clinical diagnostics. J. Transl. Med. 2011, 9, 86. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Kim, E.H.; Kwak, G.; Chi, S.-G.; Kim, S.H.; Yang, Y. Exosomes: Cell-derived nanoplatforms for the delivery of cancer therapeutics. Int. J. Mol. Sci. 2020, 22, 14. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, A.; Jain, S.M.; S Abrar, S.; Kumar, M.M.; Mathew, C.; Pathak, S. Sources, isolation strategies and therapeutic outcome of exosomes at a glance. Regen. Med. 2020, 15, 2361–2378. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Zhang, W.; Zhang, H.; Zhang, F.; Chen, L.; Ma, L.; Larcher, L.M.; Chen, S.; Liu, N.; Zhao, Q.; et al. Progress, opportunity, and perspective on exosome isolation—Efforts for efficient exosome-based theranostics. Theranostics 2020, 10, 3684–3707. [Google Scholar] [CrossRef]
- Blázquez, R.; Sánchez-Margallo, F.M.; Álvarez, V.; Usón, A.; Marinaro, F.; Casado, J.G. Fibrin glue mesh fixation combined with mesenchymal stem cells or exosomes modulates the inflammatory reaction in a murine model of incisional hernia. Acta Biomater. 2018, 71, 318–329. [Google Scholar] [CrossRef]
- Cheruvanky, A.; Zhou, H.; Pisitkun, T.; Kopp, J.B.; Knepper, M.A.; Yuen, P.S.T.; Star, R.A. Rapid isolation of urinary exosomal biomarkers using a nanomembrane ultrafiltration concentrator. Am. J. Physiol. -Ren. Physiol. 2007, 292, F1657–F1661. [Google Scholar] [CrossRef]
- Busatto, S.; Vilanilam, G.; Ticer, T.; Lin, W.-L.; Dickson, D.W.; Shapiro, S.; Bergese, P.; Wolfram, J. Tangential flow filtration for highly efficient concentration of extracellular vesicles from large volumes of fluid. Cells 2018, 7, 273. [Google Scholar] [CrossRef]
- Chen, C.; Skog, J.; Hsu, C.-H.; Lessard, R.T.; Balaj, L.; Wurdinger, T.; Carter, B.S.; Breakefield, X.O.; Toner, M.; Irimia, D. Microfluidic isolation and transcriptome analysis of serum microvesicles. Lab A Chip 2010, 10, 505–511. [Google Scholar] [CrossRef]
- Théry, C.; Amigorena, S.; Raposo, G.; Clayton, A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. 2006, 30, 23.22.21–23.22.29. [Google Scholar] [CrossRef]
- Clayton, A.; Court, J.; Navabi, H.; Adams, M.; Mason, M.D.; Hobot, J.A.; Newman, G.R.; Jasani, B. Analysis of antigen presenting cell derived exosomes, based on immuno-magnetic isolation and flow cytometry. J. Immunol. Methods 2001, 247, 163–174. [Google Scholar] [CrossRef]
- Greening, D.W.; Xu, R.; Ji, H.; Tauro, B.J.; Simpson, R.J. A protocol for exosome isolation and characterization: Evaluation of ultracentrifugation, density-gradient separation, and immunoaffinity capture methods. Methods Mol. Biol. (Clifton N.J.) 2015, 1295, 179–209. [Google Scholar] [CrossRef]
- Lobb, R.; Becker, M.; Wen, S.; Wong, C.; Wiegmans, A.; Leimgruber, A.; Moller, A. Optimized exosome isolation protocol for cell culture supernatant and human plasma. J. Extracell. Vesicles 2015, 4, 27031. [Google Scholar] [CrossRef] [PubMed]
- Baranyai, T.; Herczeg, K.; Onódi, Z.; Voszka, I.; Módos, K.; Marton, N.; Nagy, G.; Mäger, I.; Wood, M.J.; El Andaloussi, S. Isolation of exosomes from blood plasma: Qualitative and quantitative comparison of ultracentrifugation and size exclusion chromatography methods. PLoS ONE 2015, 10, e0145686. [Google Scholar] [CrossRef] [PubMed]
- Boing, A.N.; Van Der Pol, E.; Grootemaat, A.; Coumans, F.; Sturk, A.; Nieuwland, R. Single-step isolation of extracellular vesicles by size-exclusion chromatography. J. Extracell. Vesicles 2014, 3, 23430. [Google Scholar] [CrossRef]
- Gámez-Valero, A.; Monguió-Tortajada, M.; Carreras-Planella, L.; Franquesa, M.; Beyer, K.; Borràs, F.E. Size-Exclusion Chromatography-based isolation minimally alters Extracellular Vesicles’ characteristics compared to precipitating agents. Sci. Rep. 2016, 6, 33641. [Google Scholar] [CrossRef]
- Yang, X.-X.; Sun, C.; Wang, L.; Guo, X.-L. New insight into isolation, identification techniques and medical applications of exosomes. J. Control. Release 2019, 308, 119–129. [Google Scholar] [CrossRef]
- Zeringer, E.; Barta, T.; Li, M.; Vlassov, A.V. Strategies for isolation of exosomes. Cold Spring Harb. Protoc. 2015, 2015, 319–323. [Google Scholar] [CrossRef]
- Friend, J.; Yeo, L.Y. Microscale acoustofluidics: Microfluidics driven via acoustics and ultrasonics. Rev. Mod. Phys. 2011, 83, 647. [Google Scholar] [CrossRef]
- Lee, S.; Tae, S.; Jee, N.; Shin, S. LDA-based model for measuring impact of change orders in apartment projects and its application for prerisk assessment and postevaluation. J. Constr. Eng. Manag. 2015, 141, 04015011. [Google Scholar] [CrossRef]
- Ramos, A.; Morgan, H.; Green, N.G.; Castellanos, A. Ac electrokinetics: A review of forces in microelectrode structures. J. Phys. D: Appl. Phys. 1998, 31, 2338–2353. [Google Scholar] [CrossRef]
- Ibsen, S.D.; Wright, J.; Lewis, J.M.; Kim, S.; Ko, S.-Y.; Ong, J.; Manouchehri, S.; Vyas, A.; Akers, J.; Chen, C.C. Rapid isolation and detection of exosomes and associated biomarkers from plasma. ACS Nano 2017, 11, 6641–6651. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.R.; Cox, E.C.; Austin, R.H.; Sturm, J.C. Continuous particle separation through deterministic lateral displacement. Science 2004, 304, 987–990. [Google Scholar] [CrossRef] [PubMed]
- Zeming, K.K.; Thakor, N.V.; Zhang, Y.; Chen, C.-H. Real-time modulated nanoparticle separation with an ultra-large dynamic range. Lab A Chip 2016, 16, 75–85. [Google Scholar] [CrossRef] [PubMed]
- Gholizadeh, S.; Draz, M.S.; Zarghooni, M.; Sanati-Nezhad, A.; Ghavami, S.; Shafiee, H.; Akbari, M. Microfluidic approaches for isolation, detection, and characterization of extracellular vesicles: Current status and future directions. Biosens. Bioelectron. 2017, 91, 588–605. [Google Scholar] [CrossRef] [PubMed]
- Contreras-Naranjo, J.C.; Wu, H.-J.; Ugaz, V.M. Microfluidics for exosome isolation and analysis: Enabling liquid biopsy for personalized medicine. Lab A Chip 2017, 17, 3558–3577. [Google Scholar] [CrossRef] [PubMed]
- Tayebi, M.; Zhou, Y.; Tripathi, P.; Chandramohanadas, R.; Ai, Y. Exosome purification and analysis using a facile microfluidic hydrodynamic trapping device. Anal. Chem. 2020, 92, 10733–10742. [Google Scholar] [CrossRef] [PubMed]
- Lu, B.; Ku, J.; Flojo, R.; Olson, C.; Bengford, D.; Marriott, G. Exosome- and extracellular vesicle-based approaches for the treatment of lysosomal storage disorders. Adv. Drug Deliv. Rev. 2022, 188, 114465. [Google Scholar] [CrossRef] [PubMed]
- Boriachek, K.; Islam, M.N.; Möller, A.; Salomon, C.; Nguyen, N.T.; Hossain, M.S.A.; Yamauchi, Y.; Shiddiky, M.J. Biological functions and current advances in isolation and detection strategies for exosome nanovesicles. Small 2018, 14, 1702153. [Google Scholar] [CrossRef] [PubMed]
- Patil, S.M.; Sawant, S.S.; Kunda, N.K. Exosomes as drug delivery systems: A brief overview and progress update. Eur. J. Pharm. Biopharm. 2020, 154, 259–269. [Google Scholar] [CrossRef]
- Li, P.; Kaslan, M.; Lee, S.H.; Yao, J.; Gao, Z. Progress in exosome isolation techniques. Theranostics 2017, 7, 789. [Google Scholar] [CrossRef] [PubMed]
- Batrakova, E.V.; Kim, M.S. Using exosomes, naturally-equipped nanocarriers, for drug delivery. J. Control Release 2015, 219, 396–405. [Google Scholar] [CrossRef]
- Momen-Heravi, F.; Balaj, L.; Alian, S.; Mantel, P.-Y.; Halleck, A.E.; Trachtenberg, A.J.; Soria, C.E.; Oquin, S.; Bonebreak, C.M.; Saracoglu, E.; et al. Current methods for the isolation of extracellular vesicles. Biol. Chem. 2013, 394, 1253–1262. [Google Scholar] [CrossRef]
- Li, Y.; Zheng, Q.; Bao, C.; Li, S.; Guo, W.; Zhao, J.; Chen, D.; Gu, J.; He, X.; Huang, S. Circular RNA is enriched and stable in exosomes: A promising biomarker for cancer diagnosis. Cell Res. 2015, 25, 981–984. [Google Scholar] [CrossRef] [PubMed]
- Muller, L.; Hong, C.-S.; Stolz, D.B.; Watkins, S.C.; Whiteside, T.L. Isolation of biologically-active exosomes from human plasma. J. Immunol. Methods 2014, 411, 55–65. [Google Scholar] [CrossRef] [PubMed]
- Yakimchuk, K. Exosomes: Isolation and characterization methods and specific markers. Mater. Methods 2015, 5, 1450. [Google Scholar] [CrossRef]
- Chen, B.-Y.; Sung, C.W.-H.; Chen, C.; Cheng, C.-M.; Lin, D.P.-C.; Huang, C.-T.; Hsu, M.-Y. Advances in exosomes technology. Clin. Chim. Acta 2019, 493, 14–19. [Google Scholar] [CrossRef]
- Martins, T.S.; Catita, J.; Rosa, I.M.; da Cruz e SilvaSilva, O.A.B.; Henriques, A.G. Exosome isolation from distinct biofluids using precipitation and column-based approaches. PLoS ONE 2018, 13, e0198820. [Google Scholar] [CrossRef]
- Yamashita, T.; Takahashi, Y.; Nishikawa, M.; Takakura, Y. Effect of exosome isolation methods on physicochemical properties of exosomes and clearance of exosomes from the blood circulation. Eur. J. Pharm. Biopharm. 2016, 98, 1–8. [Google Scholar] [CrossRef]
- Panagiotara, A.; Markou, A.; Lianidou, E.S.; Patrinos, G.P.; Katsila, T. Exosomes: A Cancer Theranostics Road Map. Public Heal. Genom. 2017, 20, 116–125. [Google Scholar] [CrossRef]
- Dragovic, R.A.; Gardiner, C.; Brooks, A.S.; Tannetta, D.S.; Ferguson, D.J.P.; Hole, P.; Carr, B.; Redman, C.W.G.; Harris, A.L.; Dobson, P.J.; et al. Sizing and phenotyping of cellular vesicles using Nanoparticle Tracking Analysis. Nanomed. Nanotechnol. Biol. Med. 2011, 7, 780–788. [Google Scholar] [CrossRef] [PubMed]
- Walker, J.G. Improved nano-particle tracking analysis. Meas. Sci. Technol. 2012, 23, 065605. [Google Scholar] [CrossRef]
- Dieckmann, Y.; Cölfen, H.; Hofmann, H.; Petri-Fink, A. Particle Size Distribution Measurements of Manganese-Doped ZnS Nanoparticles. Anal. Chem. 2009, 81, 3889–3895. [Google Scholar] [CrossRef]
- Hoo, C.M.; Starostin, N.; West, P.; Mecartney, M.L. A comparison of atomic force microscopy (AFM) and dynamic light scattering (DLS) methods to characterize nanoparticle size distributions. J. Nanoparticle Res. 2008, 10, 89–96. [Google Scholar] [CrossRef]
- Skliar, M.; Chernyshev, V.S. Imaging of Extracellular Vesicles by Atomic Force Microscopy. J. Vis. Exp. 2019. [Google Scholar] [CrossRef]
- Hardij, J.; Cecchet, F.; Berquand, A.; Gheldof, D.; Chatelain, C.; Mullier, F.; Chatelain, B.; Dogné, J.-M. Characterisation of tissue factor-bearing extracellular vesicles with AFM: Comparison of air-tapping-mode AFM and liquid Peak Force AFM. J. Extracell. Vesicles 2013, 2, 21045. [Google Scholar] [CrossRef] [PubMed]
- Pisitkun, T.; Shen, R.-F.; Knepper, M.A. Identification and proteomic profiling of exosomes in human urine. Proc. Natl. Acad. Sci. USA 2004, 101, 13368. [Google Scholar] [CrossRef] [PubMed]
- Colombo, M.; Raposo, G.; Théry, C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 2014, 30, 255–289. [Google Scholar] [CrossRef] [PubMed]
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef] [PubMed]
- Vernon-Parry, K. Scanning electron microscopy: An introduction. III-Vs Rev. 2000, 13, 40–44. [Google Scholar] [CrossRef]
- USHIKI, T.; FUJITA, T. Backscattered electron imaging. Its application to biological specimens stained with heavy metals. Arch. Histol. Jpn. 1986, 49, 139–154. [Google Scholar] [CrossRef] [PubMed]
- Soligo, D.; Lambertenghi-Deliliers, G. Biological applications of backscattered electron imaging of scanning electron microscopy. Scanning 1987, 9, 95–98. [Google Scholar] [CrossRef]
- Rissin, D.M.; Kan, C.W.; Campbell, T.G.; Howes, S.C.; Fournier, D.R.; Song, L.; Piech, T.; Patel, P.P.; Chang, L.; Rivnak, A.J.; et al. Single-molecule enzyme-linked immunosorbent assay detects serum proteins at subfemtomolar concentrations. Nat. Biotechnol. 2010, 28, 595–599. [Google Scholar] [CrossRef]
- Zarovni, N.; Corrado, A.; Guazzi, P.; Zocco, D.; Lari, E.; Radano, G.; Muhhina, J.; Fondelli, C.; Gavrilova, J.; Chiesi, A. Integrated isolation and quantitative analysis of exosome shuttled proteins and nucleic acids using immunocapture approaches. Methods 2015, 87, 46–58. [Google Scholar] [CrossRef] [PubMed]
- Fang, S.; Tian, H.; Li, X.; Jin, D.; Li, X.; Kong, J.; Yang, C.; Yang, X.; Lu, Y.; Luo, Y. Clinical application of a microfluidic chip for immunocapture and quantification of circulating exosomes to assist breast cancer diagnosis and molecular classification. PLoS ONE 2017, 12, e0175050. [Google Scholar] [CrossRef] [PubMed]
- Zou, L.; Liu, X.; Zhou, Y.; Mei, W.; Wang, Q.; Yang, X.; Wang, K. Optical fiber amplifier and thermometer assisted point-of-care biosensor for detection of cancerous exosomes. Sens. Actuators B Chem. 2022, 351, 130893. [Google Scholar] [CrossRef]
- Liang, L.-G.; Kong, M.-Q.; Zhou, S.; Sheng, Y.-F.; Wang, P.; Yu, T.; Inci, F.; Kuo, W.P.; Li, L.-J.; Demirci, U. An integrated double-filtration microfluidic device for isolation, enrichment and quantification of urinary extracellular vesicles for detection of bladder cancer. Sci. Rep. 2017, 7, 46224. [Google Scholar] [CrossRef]
- Vaidyanathan, R.; Naghibosadat, M.; Rauf, S.; Korbie, D.; Carrascosa, L.G.; Shiddiky, M.J.; Trau, M. Detecting exosomes specifically: A multiplexed device based on alternating current electrohydrodynamic induced nanoshearing. Anal. Chem. 2014, 86, 11125–11132. [Google Scholar] [CrossRef]
- Gordon, R.; Sinton, D.; Kavanagh, K.L.; Brolo, A.G. A new generation of sensors based on extraordinary optical transmission. Acc. Chem. Res. 2008, 41, 1049–1057. [Google Scholar] [CrossRef]
- Im, H.; Shao, H.; Park, Y.I.; Peterson, V.M.; Castro, C.M.; Weissleder, R.; Lee, H. Label-free detection and molecular profiling of exosomes with a nano-plasmonic sensor. Nat. Biotechnol. 2014, 32, 490–495. [Google Scholar] [CrossRef]
- Issadore, D.; Min, C.; Liong, M.; Chung, J.; Weissleder, R.; Lee, H. Miniature magnetic resonance system for point-of-care diagnostics. Lab A Chip 2011, 11, 2282–2287. [Google Scholar] [CrossRef] [PubMed]
- Shao, H.; Chung, J.; Balaj, L.; Charest, A.; Bigner, D.D.; Carter, B.S.; Hochberg, F.H.; Breakefield, X.O.; Weissleder, R.; Lee, H. Protein typing of circulating microvesicles allows real-time monitoring of glioblastoma therapy. Nat. Med. 2012, 18, 1835–1840. [Google Scholar] [CrossRef] [PubMed]
- Mehryab, F.; Rabbani, S.; Shahhosseini, S.; Shekari, F.; Fatahi, Y.; Baharvand, H.; Haeri, A. Exosomes as a next-generation drug delivery system: An update on drug loading approaches, characterization, and clinical application challenges. Acta Biomater. 2020, 113, 42–62. [Google Scholar] [CrossRef]
- Mehanny, M.; Lehr, C.-M.; Fuhrmann, G. Extracellular vesicles as antigen carriers for novel vaccination avenues. Adv. Drug Deliv. Rev. 2021, 173, 164–180. [Google Scholar] [CrossRef]
- Dad, H.A.; Gu, T.-W.; Zhu, A.-Q.; Huang, L.-Q.; Peng, L.-H. Plant exosome-like nanovesicles: Emerging therapeutics and drug delivery nanoplatforms. Mol. Ther. 2021, 29, 13–31. [Google Scholar] [CrossRef]
- Kim, G.; Lee, Y.; Ha, J.; Han, S.; Lee, M. Engineering exosomes for pulmonary delivery of peptides and drugs to inflammatory lung cells by inhalation. J. Control. Release 2021, 330, 684–695. [Google Scholar] [CrossRef]
- Chinnappan, M.; Srivastava, A.; Amreddy, N.; Razaq, M.; Pareek, V.; Ahmed, R.; Mehta, M.; Peterson, J.E.; Munshi, A.; Ramesh, R. Exosomes as drug delivery vehicle and contributor of resistance to anticancer drugs. Cancer Lett. 2020, 486, 18–28. [Google Scholar] [CrossRef]
- Dai, J.; Su, Y.; Zhong, S.; Cong, L.; Liu, B.; Yang, J.; Tao, Y.; He, Z.; Chen, C.; Jiang, Y. Exosomes: Key players in cancer and potential therapeutic strategy. Signal Transduct. Target. Ther. 2020, 5, 145. [Google Scholar] [CrossRef]
- Wang, J.; Chen, D.; Ho, E.A. Challenges in the development and establishment of exosome-based drug delivery systems. J. Control. Release 2021, 329, 894–906. [Google Scholar] [CrossRef]
- Crewe, C.; Joffin, N.; Rutkowski, J.M.; Kim, M.; Zhang, F.; Towler, D.A.; Gordillo, R.; Scherer, P.E. An endothelial-to-adipocyte extracellular vesicle axis governed by metabolic state. Cell 2018, 175, 695–708.e613. [Google Scholar] [CrossRef]
- El Andaloussi, S.; Mäger, I.; Breakefield, X.O.; Wood, M.J. Extracellular vesicles: Biology and emerging therapeutic opportunities. Nat. Rev. Drug Discov. 2013, 12, 347–357. [Google Scholar] [CrossRef] [PubMed]
- Mathieu, M.; Martin-Jaular, L.; Lavieu, G.; Théry, C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat. Cell Biol. 2019, 21, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Purushothaman, A.; Bandari, S.K.; Liu, J.; Mobley, J.A.; Brown, E.E.; Sanderson, R.D. Fibronectin on the surface of myeloma cell-derived exosomes mediates exosome-cell interactions. J. Biol. Chem. 2016, 291, 1652–1663. [Google Scholar] [CrossRef] [PubMed]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef]
- Butreddy, A.; Kommineni, N.; Dudhipala, N. Exosomes as Naturally Occurring Vehicles for Delivery of Biopharmaceuticals: Insights from Drug Delivery to Clinical Perspectives. Nanomaterials 2021, 11, 1481. [Google Scholar] [CrossRef]
- Ha, D.; Yang, N.; Nadithe, V. Exosomes as therapeutic drug carriers and delivery vehicles across biological membranes: Current perspectives and future challenges. Acta Pharm. Sin. B 2016, 6, 287–296. [Google Scholar] [CrossRef]
- Lin, J.; Li, J.; Huang, B.; Liu, J.; Chen, X.; Chen, X.-M.; Xu, Y.-M.; Huang, L.-F.; Wang, X.-Z. Exosomes: Novel biomarkers for clinical diagnosis. Sci. World J. 2015, 2015, 657086. [Google Scholar] [CrossRef]
- Jafari, D.; Shajari, S.; Jafari, R.; Mardi, N.; Gomari, H.; Ganji, F.; Moghadam, M.F.; Samadikuchaksaraei, A. Designer Exosomes: A New Platform for Biotechnology Therapeutics. BioDrugs 2020, 34, 567–586. [Google Scholar] [CrossRef]
- Bie, N.; Yong, T.; Wei, Z.; Gan, L.; Yang, X. Extracellular vesicles for improved tumor accumulation and penetration. Adv. Drug Deliv. Rev. 2022, 188, 114450. [Google Scholar] [CrossRef]
- Jiang, X.-C.; Zhang, T.; Gao, J.-Q. The in vivo fate and targeting engineering of crossover vesicle-based gene delivery system. Adv. Drug Deliv. Rev. 2022, 187, 114324. [Google Scholar] [CrossRef]
- Kamerkar, S.; LeBleu, V.S.; Sugimoto, H.; Yang, S.; Ruivo, C.F.; Melo, S.A.; Lee, J.J.; Kalluri, R. Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature 2017, 546, 498–503. [Google Scholar] [CrossRef] [PubMed]
- Lou, G.; Song, X.; Yang, F.; Wu, S.; Wang, J.; Chen, Z.; Liu, Y. Exosomes derived from miR-122-modified adipose tissue-derived MSCs increase chemosensitivity of hepatocellular carcinoma. J. Hematol. Oncol. 2015, 8, 122. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.M.; Yang, Y.; Oh, S.J.; Hong, Y.; Seo, M.; Jang, M. Cancer-derived exosomes as a delivery platform of CRISPR/Cas9 confer cancer cell tropism-dependent targeting. J. Control. Release 2017, 266, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Hoshino, A.; Costa-Silva, B.; Shen, T.; Rodrigues, G.; Hashimoto, A.; Mark, M.T.; Molina, H.; Kohsaka, S.; di Giannatale, A.; Ceder, S.; et al. Tumour exosome integrins determine organotropic metastasis. Nature 2015, 527, 329–335. [Google Scholar] [CrossRef] [PubMed]
- Baglio, S.R.; Lagerweij, T.; Pérez-Lanzón, M.; Ho, X.D.; Léveillé, N.; Melo, S.A.; Cleton-Jansen, A.-M.; Jordanova, E.S.; Roncuzzi, L.; Greco, M. Blocking tumor-educated MSC paracrine activity halts osteosarcoma progression. Clin. Cancer Res. 2017, 23, 3721–3733. [Google Scholar] [CrossRef]
- Ekström, E.J.; Bergenfelz, C.; von Bülow, V.; Serifler, F.; Carlemalm, E.; Jönsson, G.; Andersson, T.; Leandersson, K. WNT5A induces release of exosomes containing pro-angiogenic and immunosuppressive factors from malignant melanoma cells. Mol. Cancer 2014, 13, 88. [Google Scholar] [CrossRef]
- Macia, E.; Ehrlich, M.; Massol, R.; Boucrot, E.; Brunner, C.; Kirchhausen, T. Dynasore, a cell-permeable inhibitor of dynamin. Dev. Cell 2006, 10, 839–850. [Google Scholar] [CrossRef]
- Menck, K.; Scharf, C.; Bleckmann, A.; Dyck, L.; Rost, U.; Wenzel, D.; Dhople, V.M.; Siam, L.; Pukrop, T.; Binder, C. Tumor-derived microvesicles mediate human breast cancer invasion through differentially glycosylated EMMPRIN. J. Mol. Cell Biol. 2015, 7, 143–153. [Google Scholar] [CrossRef]
- Kawamoto, T.; Ohga, N.; Akiyama, K.; Hirata, N.; Kitahara, S.; Maishi, N.; Osawa, T.; Yamamoto, K.; Kondoh, M.; Shindoh, M.; et al. Tumor-derived microvesicles induce proangiogenic phenotype in endothelial cells via endocytosis. PLoS ONE 2012, 7, e34045. [Google Scholar] [CrossRef]
- Wang, C.; Qi, P.; Lu, Y.; Liu, L.; Zhang, Y.; Sheng, Q.; Wang, T.; Zhang, M.; Wang, R.; Song, S. Bicomponent polymeric micelles for pH-controlled delivery of doxorubicin. Drug Deliv. 2020, 27, 344–357. [Google Scholar] [CrossRef]
- Pinnell, J.R.; Cui, M.; Tieu, K. Exosomes in Parkinson disease. J. Neurochem. 2021, 157, 413–428. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Sun, T.; An, J.; Wen, L.; Liu, F.; Bu, Z.; Cui, Y.; Feng, J. Potential roles of exosomes in Parkinson’s disease: From pathogenesis, diagnosis, and treatment to prognosis. Front. Cell Dev. Biol. 2020, 8, 86. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Zheng, T.; Zhang, B. Exosomes in Parkinson’s disease. Neurosci. Bull. 2017, 33, 331–338. [Google Scholar] [CrossRef] [PubMed]
- Porro, C.; Panaro, M.A.; Lofrumento, D.D.; Hasalla, E.; Trotta, T. The multiple roles of exosomes in Parkinson’s disease: An overview. Immunopharmacol. Immunotoxicol. 2019, 41, 469–476. [Google Scholar] [CrossRef]
- Malm, T.; Loppi, S.; Kanninen, K.M. Exosomes in Alzheimer’s disease. Neurochem. Int. 2016, 97, 193–199. [Google Scholar] [CrossRef]
- Zhang, T.; Ma, S.; Lv, J.; Wang, X.; Afewerky, H.K.; Li, H.; Lu, Y. The emerging role of exosomes in Alzheimer’s disease. Ageing Res. Rev. 2021, 68, 101321. [Google Scholar] [CrossRef]
- Xiao, T.; Zhang, W.; Jiao, B.; Pan, C.-Z.; Liu, X.; Shen, L. The role of exosomes in the pathogenesis of Alzheimer’disease. Transl. Neurodegener. 2017, 6, 3. [Google Scholar] [CrossRef]
- Barile, L.; Vassalli, G. Exosomes: Therapy delivery tools and biomarkers of diseases. Pharmacol. Ther. 2017, 174, 63–78. [Google Scholar] [CrossRef]
- Reynolds, J.L.; Mahajan, S.D. Transmigration of Tetraspanin 2 (Tspan2) siRNA Via Microglia Derived Exosomes across the Blood Brain Barrier Modifies the Production of Immune Mediators by Microglia Cells. J. Neuroimmune Pharmacol. Off. J. Soc. NeuroImmune Pharmacol. 2020, 15, 554–563. [Google Scholar] [CrossRef]
- Cukovic, D.; Bagla, S.; Ukasik, D.; Stemmer, P.M.; Jena, B.P.; Naik, A.R.; Sood, S.; Asano, E.; Luat, A.; Chugani, D.C. Exosomes in epilepsy of tuberous sclerosis complex: Carriers of pro-inflammatory microRNAs. Non-Coding RNA 2021, 7, 40. [Google Scholar] [CrossRef]
- Ananbeh, H.; Vodicka, P.; Kupcova Skalnikova, H. Emerging roles of exosomes in Huntington’s disease. Int. J. Mol. Sci. 2021, 22, 4085. [Google Scholar] [CrossRef] [PubMed]
- Beatriz, M.; Vilaça, R.; Lopes, C. Exosomes: Innocent bystanders or critical culprits in neurodegenerative diseases. Front. Cell Dev. Biol. 2021, 9, 1047. [Google Scholar] [CrossRef]
- Kalani, A.; Tyagi, N. Exosomes in neurological disease, neuroprotection, repair and therapeutics: Problems and perspectives. Neural Regen. Res. 2015, 10, 1565–1567. [Google Scholar] [CrossRef] [PubMed]
- Tsilioni, I.; Panagiotidou, S.; Theoharides, T.C. Exosomes in neurologic and psychiatric disorders. Clin. Ther. 2014, 36, 882–888. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Bai, X.; Zhang, A.; Huang, J.; Xu, S.; Zhang, J. Role of exosomes in central nervous system diseases. Front. Mol. Neurosci. 2019, 12, 240. [Google Scholar] [CrossRef] [PubMed]
- Osier, N.; Motamedi, V.; Edwards, K.; Puccio, A.; Diaz-Arrastia, R.; Kenney, K.; Gill, J. Exosomes in acquired neurological disorders: New insights into pathophysiology and treatment. Mol. Neurobiol. 2018, 55, 9280–9293. [Google Scholar] [CrossRef] [PubMed]
- Suh, J.H.; Joo, H.S.; Hong, E.B.; Lee, H.J.; Lee, J.M. Therapeutic application of exosomes in inflammatory diseases. Int. J. Mol. Sci. 2021, 22, 1144. [Google Scholar] [CrossRef] [PubMed]
- Console, L.; Scalise, M.; Indiveri, C. Exosomes in inflammation and role as biomarkers. Clin. Chim. Acta 2019, 488, 165–171. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Peng, H.; Liu, Y. The roles of exosomes in immunoregulation and autoimmune thyroid diseases. Front. Immunol. 2021, 12, 4823. [Google Scholar] [CrossRef]
- Zhu, T.; Wang, Y.; Jin, H.; Li, L. The role of exosome in autoimmune connective tissue disease. Ann. Med. 2019, 51, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Anel, A.; Gallego-Lleyda, A.; de Miguel, D.; Naval, J.; Martínez-Lostao, L. Role of exosomes in the regulation of T-cell mediated immune responses and in autoimmune disease. Cells 2019, 8, 154. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, S.; Suazo, C.; Boltansky, A.; Ursu, M.; Carvajal, D.; Innocenti, G.; Vukusich, A.; Hurtado, M.; Villanueva, S.; Carreño, J.E.; et al. Urinary Exosomes as a Source of Kidney Dysfunction Biomarker in Renal Transplantation. Transplant. Proc. 2013, 45, 3719–3723. [Google Scholar] [CrossRef] [PubMed]
- Saari, H.; Lázaro-Ibáñez, E.; Viitala, T.; Vuorimaa-Laukkanen, E.; Siljander, P.; Yliperttula, M. Microvesicle- and exosome-mediated drug delivery enhances the cytotoxicity of Paclitaxel in autologous prostate cancer cells. J. Control. Release 2015, 220, 727–737. [Google Scholar] [CrossRef] [PubMed]
- Thongboonkerd, V. Roles for exosome in various kidney diseases and disorders. Front. Pharmacol. 2020, 10, 1655. [Google Scholar] [CrossRef]
- Alvarez-Erviti, L.; Seow, Y.; Yin, H.; Betts, C.; Lakhal, S.; Wood, M.J.A. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat. Biotechnol. 2011, 29, 341–345. [Google Scholar] [CrossRef]
- Pascucci, L.; Coccè, V.; Bonomi, A.; Ami, D.; Ceccarelli, P.; Ciusani, E.; Viganò, L.; Locatelli, A.; Sisto, F.; Doglia, S.M.; et al. Paclitaxel is incorporated by mesenchymal stromal cells and released in exosomes that inhibit in vitro tumor growth: A new approach for drug delivery. J. Control. Release 2014, 192, 262–270. [Google Scholar] [CrossRef]
- Sun, D.; Zhuang, X.; Xiang, X.; Liu, Y.; Zhang, S.; Liu, C.; Barnes, S.; Grizzle, W.; Miller, D.; Zhang, H.-G. A novel nanoparticle drug delivery system: The anti-inflammatory activity of curcumin is enhanced when encapsulated in exosomes. Mol. Ther. 2010, 18, 1606–1614. [Google Scholar] [CrossRef]
- Tian, Y.; Li, S.; Song, J.; Ji, T.; Zhu, M.; Anderson, G.J.; Wei, J.; Nie, G. A doxorubicin delivery platform using engineered natural membrane vesicle exosomes for targeted tumor therapy. Biomaterials 2014, 35, 2383–2390. [Google Scholar] [CrossRef]
- Kim, M.S.; Haney, M.J.; Zhao, Y.; Mahajan, V.; Deygen, I.; Klyachko, N.L.; Inskoe, E.; Piroyan, A.; Sokolsky, M.; Okolie, O.; et al. Development of exosome-encapsulated paclitaxel to overcome MDR in cancer cells. Nanomed. Nanotechnol. Biol. Med. 2016, 12, 655–664. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.S.; Haney, M.J.; Zhao, Y.; Yuan, D.; Deygen, I.; Klyachko, N.L.; Kabanov, A.V.; Batrakova, E.V. Engineering macrophage-derived exosomes for targeted paclitaxel delivery to pulmonary metastases: In vitro and in vivo evaluations. Nanomed. Nanotechnol. Biol. Med. 2018, 14, 195–204. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, X.; Xiang, X.; Grizzle, W.; Sun, D.; Zhang, S.; Axtell, R.C.; Ju, S.; Mu, J.; Zhang, L.; Steinman, L.; et al. Treatment of Brain Inflammatory Diseases by Delivering Exosome Encapsulated Anti-inflammatory Drugs From the Nasal Region to the Brain. Mol. Ther. 2011, 19, 1769–1779. [Google Scholar] [CrossRef]
- Qu, M.; Lin, Q.; Huang, L.; Fu, Y.; Wang, L.; He, S.; Fu, Y.; Yang, S.; Zhang, Z.; Zhang, L.; et al. Dopamine-loaded blood exosomes targeted to brain for better treatment of Parkinson’s disease. J. Control. Release 2018, 287, 156–166. [Google Scholar] [CrossRef]
- Skog, J.; Würdinger, T.; van Rijn, S.; Meijer, D.H.; Gainche, L.; Curry, W.T.; Carter, B.S.; Krichevsky, A.M.; Breakefield, X.O. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 2008, 10, 1470–1476. [Google Scholar] [CrossRef] [PubMed]
- Viñas, J.L.; Burger, D.; Zimpelmann, J.; Haneef, R.; Knoll, W.; Campbell, P.; Gutsol, A.; Carter, A.; Allan, D.S.; Burns, K.D. Transfer of microRNA-486-5p from human endothelial colony forming cell-derived exosomes reduces ischemic kidney injury. Kidney Int. 2016, 90, 1238–1250. [Google Scholar] [CrossRef]
- Cho, E.; Nam, G.-H.; Hong, Y.; Kim, Y.K.; Kim, D.-H.; Yang, Y.; Kim, I.-S. Comparison of exosomes and ferritin protein nanocages for the delivery of membrane protein therapeutics. J. Control. Release 2018, 279, 326–335. [Google Scholar] [CrossRef] [PubMed]
- Jia, G.; Han, Y.; An, Y.; Ding, Y.; He, C.; Wang, X.; Tang, Q. NRP-1 targeted and cargo-loaded exosomes facilitate simultaneous imaging and therapy of glioma in vitro and in vivo. Biomaterials 2018, 178, 302–316. [Google Scholar] [CrossRef]
- Alhasan, A.H.; Patel, P.C.; Choi, C.H.J.; Mirkin, C.A. Exosome Encased Spherical Nucleic Acid Gold Nanoparticle Conjugates as Potent MicroRNA Regulation Agents. Small 2014, 10, 186–192. [Google Scholar] [CrossRef] [PubMed]
- Taylor, D.D.; Shah, S. Methods of isolating extracellular vesicles impact down-stream analyses of their cargoes. Methods 2015, 87, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Akuma, P.; Okagu, O.D.; Udenigwe, C.C. Naturally occurring exosome vesicles as potential delivery vehicle for bioactive compounds. Front. Sustain. Food Syst. 2019, 3, 23. [Google Scholar] [CrossRef]
- Smyth, T.; Kullberg, M.; Malik, N.; Smith-Jones, P.; Graner, M.W.; Anchordoquy, T.J. Biodistribution and delivery efficiency of unmodified tumor-derived exosomes. J. Control. Release 2015, 199, 145–155. [Google Scholar] [CrossRef]
- Conlan, R.S.; Pisano, S.; Oliveira, M.I.; Ferrari, M.; Pinto, I.M. Exosomes as reconfigurable therapeutic systems. Trends Mol. Med. 2017, 23, 636–650. [Google Scholar] [CrossRef] [PubMed]
- Imai, T.; Takahashi, Y.; Nishikawa, M.; Kato, K.; Morishita, M.; Yamashita, T.; Matsumoto, A.; Charoenviriyakul, C.; Takakura, Y. Macrophage-dependent clearance of systemically administered B16BL6-derived exosomes from the blood circulation in mice. J. Extracell. Vesicles 2015, 4, 26238. [Google Scholar] [CrossRef] [PubMed]
| Method | Principle | Advantages | Disadvantages | Reference |
|---|---|---|---|---|
| Ultrafiltration | Separation is based on molecular weight and size. | It is faster, requires no special equipment, and is easier to handle than ultracentrifugation. | Exosome clogging and entrapment in filters lead to a poor recovery rate, deformation, and damage to large EVs due to the force required to drive through the filters. | [82,83] |
| Immunological Separation | Exosomes are captured via an antigen-antibody response. | The method saves time by separating bodily fluids immediately, isolation of high purity, and a simple process | Exosome separation on a large scale is impossible due to the high reagent cost, poor capacity, and yield. Non-physiological salt and pH conditions are required. | [84,85] |
| Ultracentrifugation | Sedimentation coefficient of exosomes and other substances in sample | Exosomes may be produced in enormous quantities, with great separation purity. | Time required (>4 h), low recovery rate (5–25%), and poor reproducibility make it unsuitable for clinical diagnosis. | [82,86] |
| Size-Exclusion Chromatography | Utilizes a column of porous polymeric beads to separate exosomes based on size. | High yield and purity | Expensive, time-consuming post-isolation analysis and column contamination. | [87,88] |
| Polymer-based precipitation separation | Hydrophobicity | Simple procedure with a small sample volume | Long run periods and post-separation cleaning are required.Non-exosome contaminants are co-precipitated in the sample. | [89,90,91] |
| Magnetic separation | Magnetic Force | The contactless separation, high specificity, and high throughput | Magnetic labeling | [66,67] |
| Acoustic fluid separation | Separation is based on size | A technique that is label-free, contactless, and quick. | Separation is not extensively used yet. | [88,92] |
| Dielectrophoretic separation | Polarized particle’s size and electric properties | Characteristics of label-free, contactless, fast, and high-throughput | Low resolution, low purity, Joule and electrothermal heating problems | [77,78] |
| Deterministic Lateral Displacement Separation | Critical size for particle separation | Label-free, easy to use | Low separation purity, clogging | [79] |
| Microfluidic devices | Separation based on size, charge, surface properties and interactions | Fast, High precision | Non-scalability on large-scale diagnostics | [80] |
| Sr. No. | Characterization Technique | Principle | Application | References |
|---|---|---|---|---|
| 1. | Nanoparticle Tracking Analysis (NTA) | Particles’ light scattering and Brownian motion | Quantify particle diameter Estimate the presence of antigens on exosomes. | [93,94] |
| 2. | Dynamic Light Scattering (DLS) | Particle light scattering and optical signal | Determine particle size and dispersion. | [95,96] |
| 3. | Atomic Force Microscopy (AFM) | Surface sensing, detection, and imaging | 3D geometry, size, and other biophysical characteristics. Mechanical properties. | [97,98] |
| 4. | Microscopy study Transmission Electron Microscopy (TEM) Scanning Electron Microscopy (SEM) | Accelerated electron beam Low-energy electrons are ejected from only form proximity to the sample surface. | 3D form, size and structure of particles. Surface characteristics comprising size, shape and morphology. | [19,101,102] |
| 5. | Enzyme-linked Immunosorbent Assay (ELISA) | Plate-based enzyme-linked immunosorbent test | Identifies and measures proteins, peptides, hormones, and antibodies; also used to determine exosomes from plasma, serum, and urine using different precise antibodies. | [105,106] |
| 6. | Fluorescence Correlation Microscopy (FCM) | Antibody tagged with a fluorescent dye and measured by a plate reader in microfluidic-dependent FCM. | Immunocapture and quantitative analysis | [107] |
| 7. | Colorimetric detection | Determines the particles in calorimetric detection, quantified using ELISA. | Utilized to detect exosomes from cancer cells. | [26,109,110] |
| 8. | Surface Plasmon Resonance (SPR) detection | Microfluidic-based SPR device. | Improve detection performance by nano-plasmonic exosome (nPLEX) created by modifying a nanosubstrate. Able to functionalize every nanopore depending on the nPLEX chip. | [111,112] |
| 9. | Nuclear Magnetic Resonance (NMR) detection | Micro-NMR technique | Assess the number and presence of proteins in exosomes. Detect exosomes after concentrating microvesicles containing immunogenic nanoparticles via filtering. | [113,114] |
| Kidney Disease/Disorder | Exosome’s Role | Exosome’s Source | Main Findings |
|---|---|---|---|
| Renal cell carcinoma (RCC) | Therapeutics | RCC cell line Pathogenic | CD8 + T-cells activated by exosomes generated from RCC cells combined with GM-CSF and IL-12 showed autologous anti-cancer activity. |
| Biomarker | Urine (human) | Urinary exosomal miR-126-3p in combination with miR-449a or miR-34b-5p might distinguish ccRCC from healthy people. Urinary exosomal miR-126-3p in combination with miR-486-5p in urine might distinguish ccRCC from benign tumors. | |
| Kidney stone disease | Pathogenic | Urine (human) | Urinary exosomes were produced in larger quantities by stone formers. |
| Renal fibrosis | Therapeutics | MSCs (human) | Exosome miR-let7c generated from MSCs has reduced fibrosis in renal tubular epithelial cells. |
| Polycystic kidney disease | Pathogenic | Urine (human) | Multiple PKD-related gene products were excreted into the urine via exosomal secretion. |
| Drug | Type of Drug | Disease Model | Therapeutic Effect | Exosomes Origin | Drug Loading Method | Reference |
|---|---|---|---|---|---|---|
| Paclitaxel | Small molecule drug | Autologous prostate cancer | Enhanced drug cytotoxicity to cancer cells | Prostate cancer cell lines (PC-3 and LNCaP) | Co-incubation | [164] |
| SiRNA | Genetic substances | Alzheimer’s disease | Specific gene knockdown after specific siRNA delivery to the brain | Dendritic cells (gene engineered to express Lamp2b) | Electroporation | [166] |
| Paclitaxel | Small molecule drug | Pancreatic adenocarcinoma | Inhibited growth of human pancreatic adenocarcinoma cell | Mesenchymal stromal cells | Co-incubation | [167] |
| Curcumin | Small molecule drug | Lipopolysaccharide-induced shock | Enhanced anti-inflammatory activity | Mouse lymphoma cell (EL-4) and RAW 264.7 cells | Direct mixing | [168] |
| Doxorubicin | Small molecule drug | Breast cancer | Specific drug delivery to the tumor site and inhibited tumor growth | Immature mouse dendritic cells transfected with the vector expressing iRGD-Lamp2b fusion proteins | Electroporation | [169] |
| Paclitaxel | Small molecule drug | Cancer with multiple drug resistance (MDR) | Overcome MDR cancer in vitro and in vivo | RAW 264.7 cells | Sonication | [170] |
| Paclitaxel | Small molecule drug | Pulmonary metastases | Reduced pulmonary metastases in vitro and in vivo | RAW 264.7 cells | Sonication | [171] |
| Curcumin | Small molecule drug | Brain tumor and autoimmune encephalitis | Inhibited brain inflammation and delayed brain tumor growth | Tumor cells (GL26-Luc, BV2, 3T3L1, 4T1, CT26, A20 and EL-4) | Direct mixing | [172] |
| Dopamine | Small molecule drug | Parkinson’s disease | Enhanced therapeutic effect due to brain-specific drug delivery | Kunming mice blood | Co-incubation | [173] |
| miRNA | Genetic substances | Glioblastoma tumor | Provide diagnostic information | Glioblastoma cells | Transfection | [174] |
| miRNA | Genetic substances | Ischemia kidney injury | Protected kidney function and reduced kidney injury | Human cord blood endothelial colony-forming cells | Transfection | [175] |
| Signal regulatory protein α | Protein | Tumor | Enhanced phagocytosis of tumor cells | HEK293T cells | Transfection | [176] |
| Curcumin | Small molecule drug | Glioma | Improved targeted imaging and therapeutic effect | RAW 264.7 cells | Electroporation | [177] |
| Spherical nucleic acids | Genetic substances | Prostate cancer | 3000-fold enhanced knockdown of miR-21 | PC-3 cells | Naturally encased | [178] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rajput, A.; Varshney, A.; Bajaj, R.; Pokharkar, V. Exosomes as New Generation Vehicles for Drug Delivery: Biomedical Applications and Future Perspectives. Molecules 2022, 27, 7289. https://doi.org/10.3390/molecules27217289
Rajput A, Varshney A, Bajaj R, Pokharkar V. Exosomes as New Generation Vehicles for Drug Delivery: Biomedical Applications and Future Perspectives. Molecules. 2022; 27(21):7289. https://doi.org/10.3390/molecules27217289
Chicago/Turabian StyleRajput, Amarjitsing, Akansh Varshney, Rashi Bajaj, and Varsha Pokharkar. 2022. "Exosomes as New Generation Vehicles for Drug Delivery: Biomedical Applications and Future Perspectives" Molecules 27, no. 21: 7289. https://doi.org/10.3390/molecules27217289
APA StyleRajput, A., Varshney, A., Bajaj, R., & Pokharkar, V. (2022). Exosomes as New Generation Vehicles for Drug Delivery: Biomedical Applications and Future Perspectives. Molecules, 27(21), 7289. https://doi.org/10.3390/molecules27217289




