Derivation, Functionalization of (S)-Goniothalamin from Goniothalamus wightii and Their Derivative Targets SARS-CoV-2 MPro, SPro, and RdRp: A Pharmacological Perspective
Abstract
1. Introduction
2. Results
2.1. Purification of Hexane Extract of G. wightii
2.2. Characterization of Isolated Compound
2.3. Structural Analysis of Compound
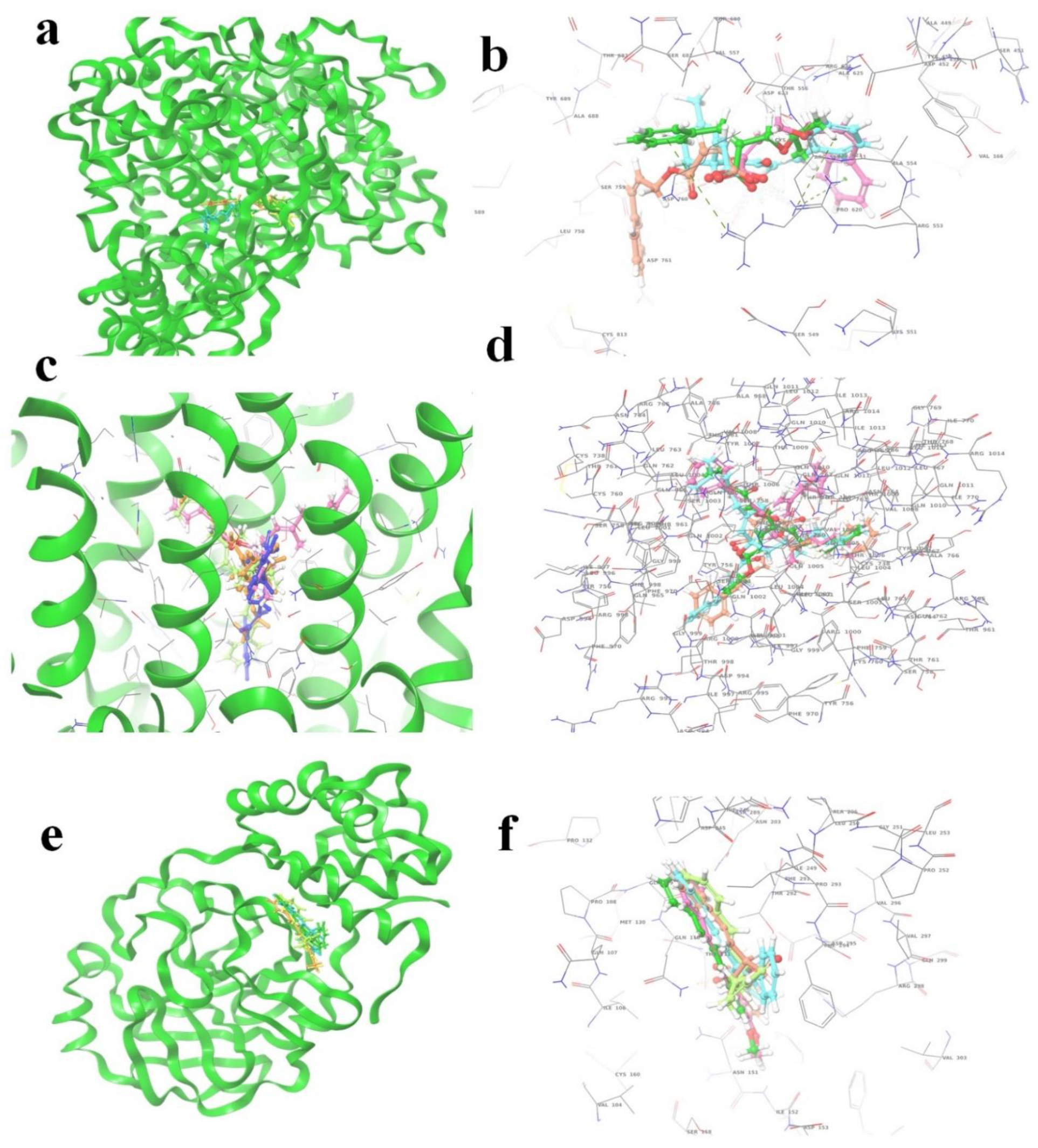
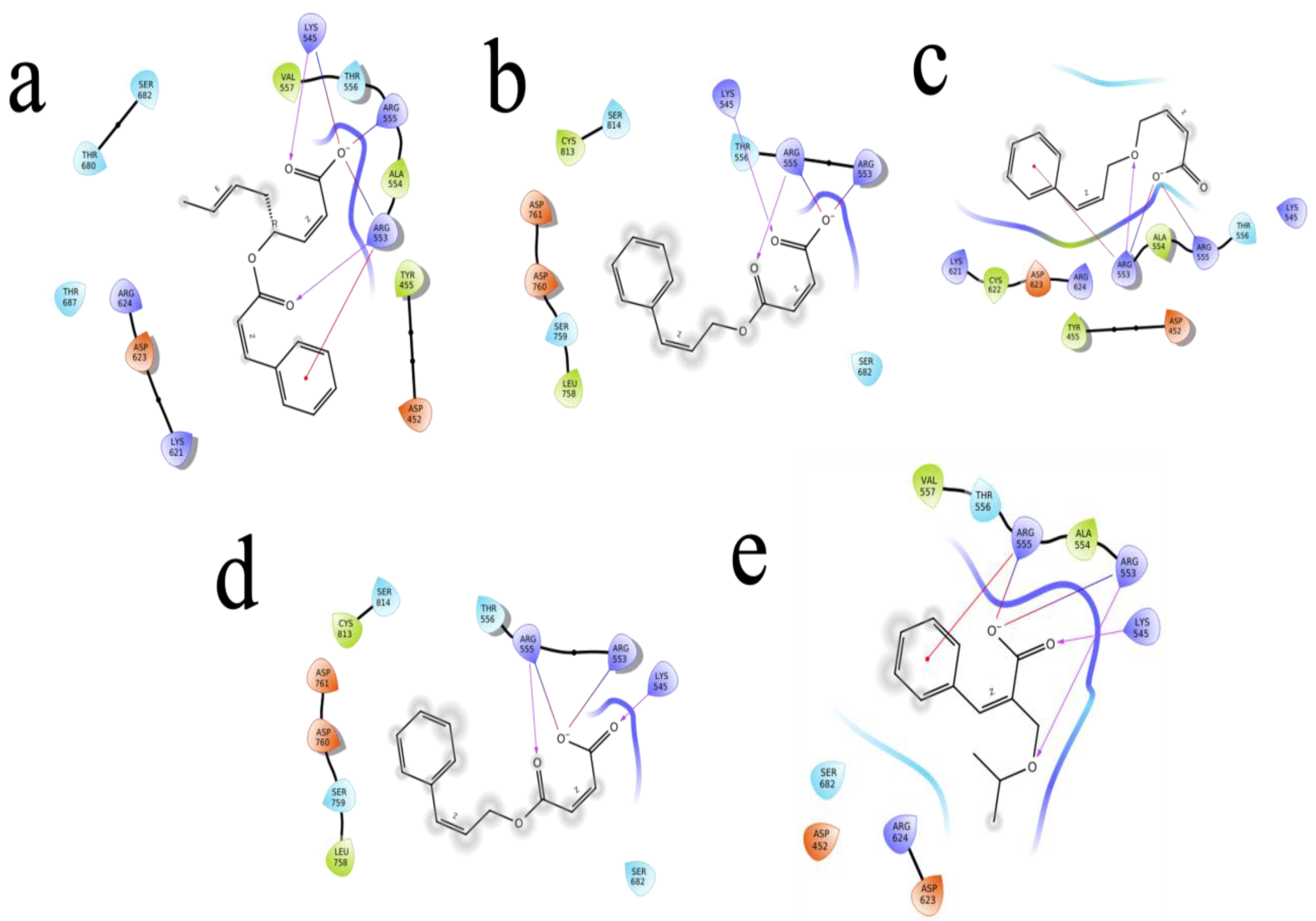
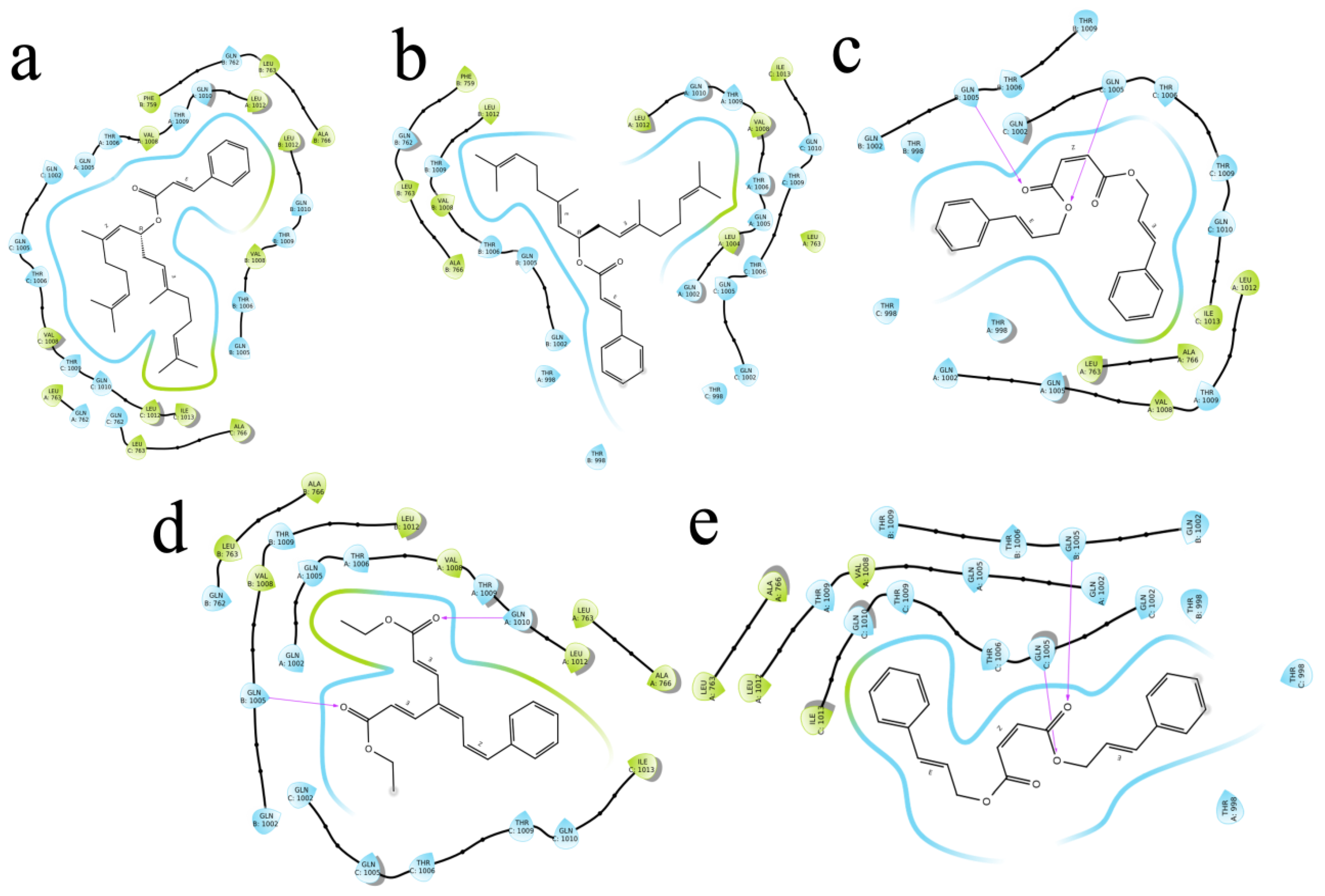
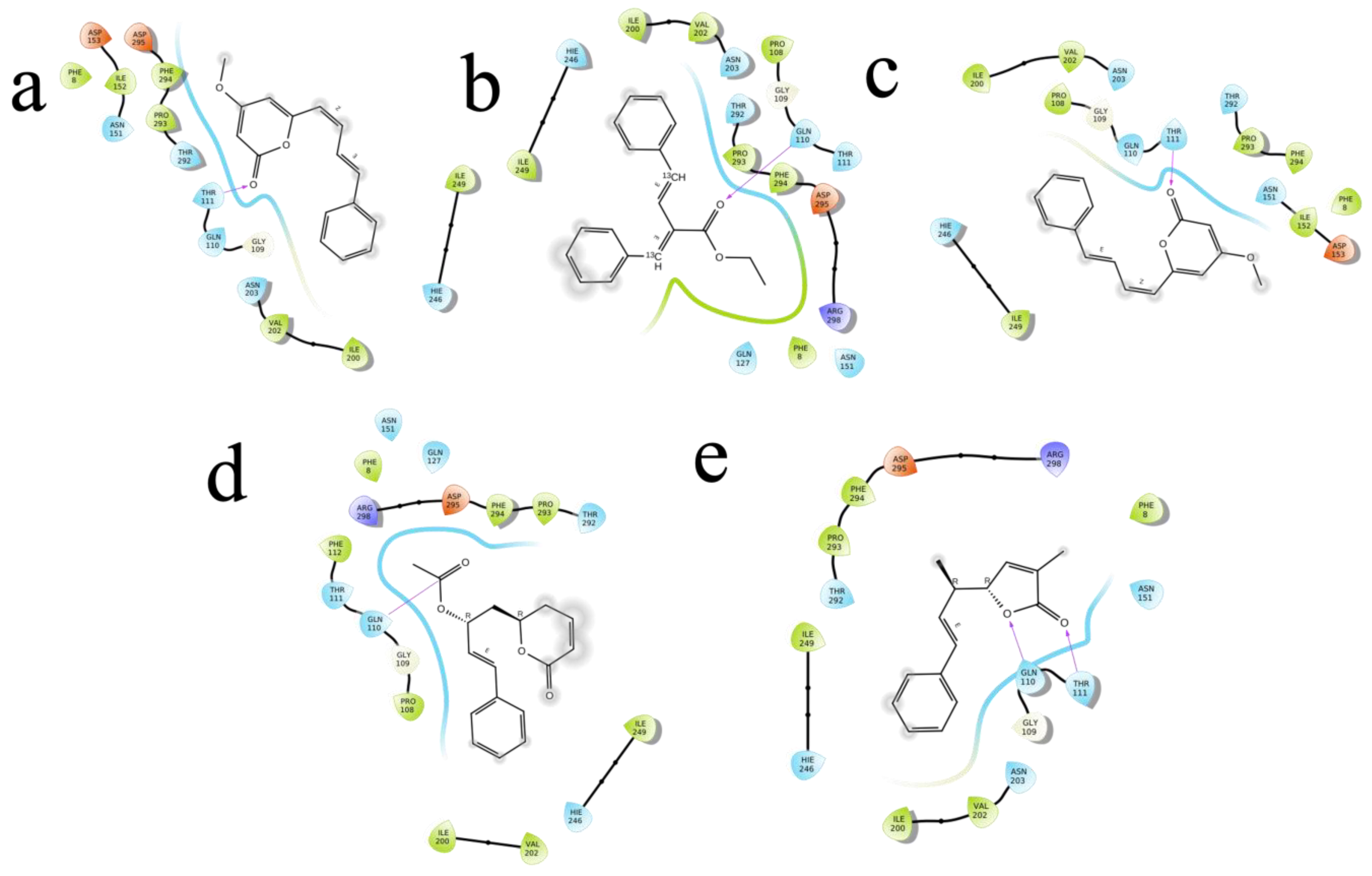
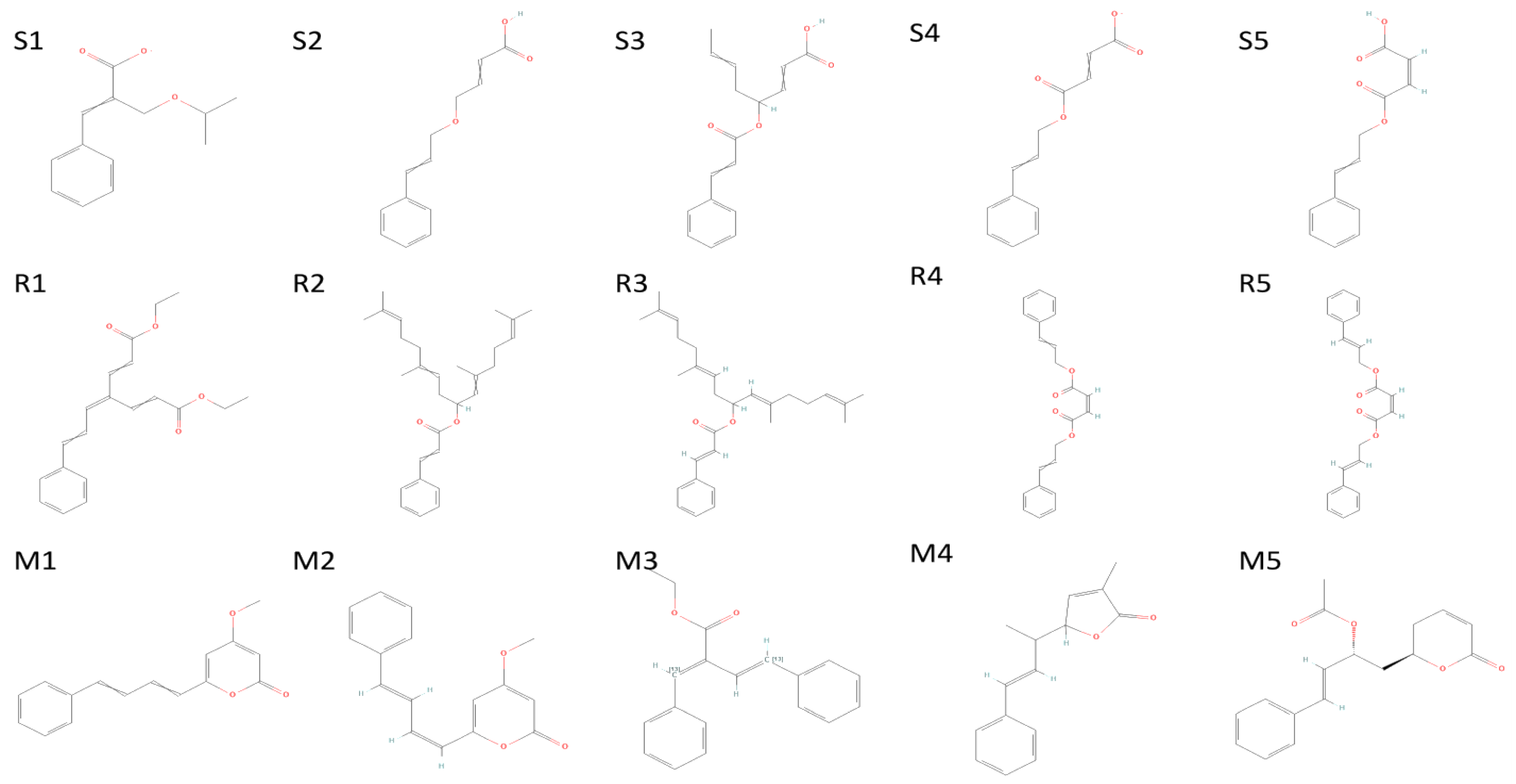
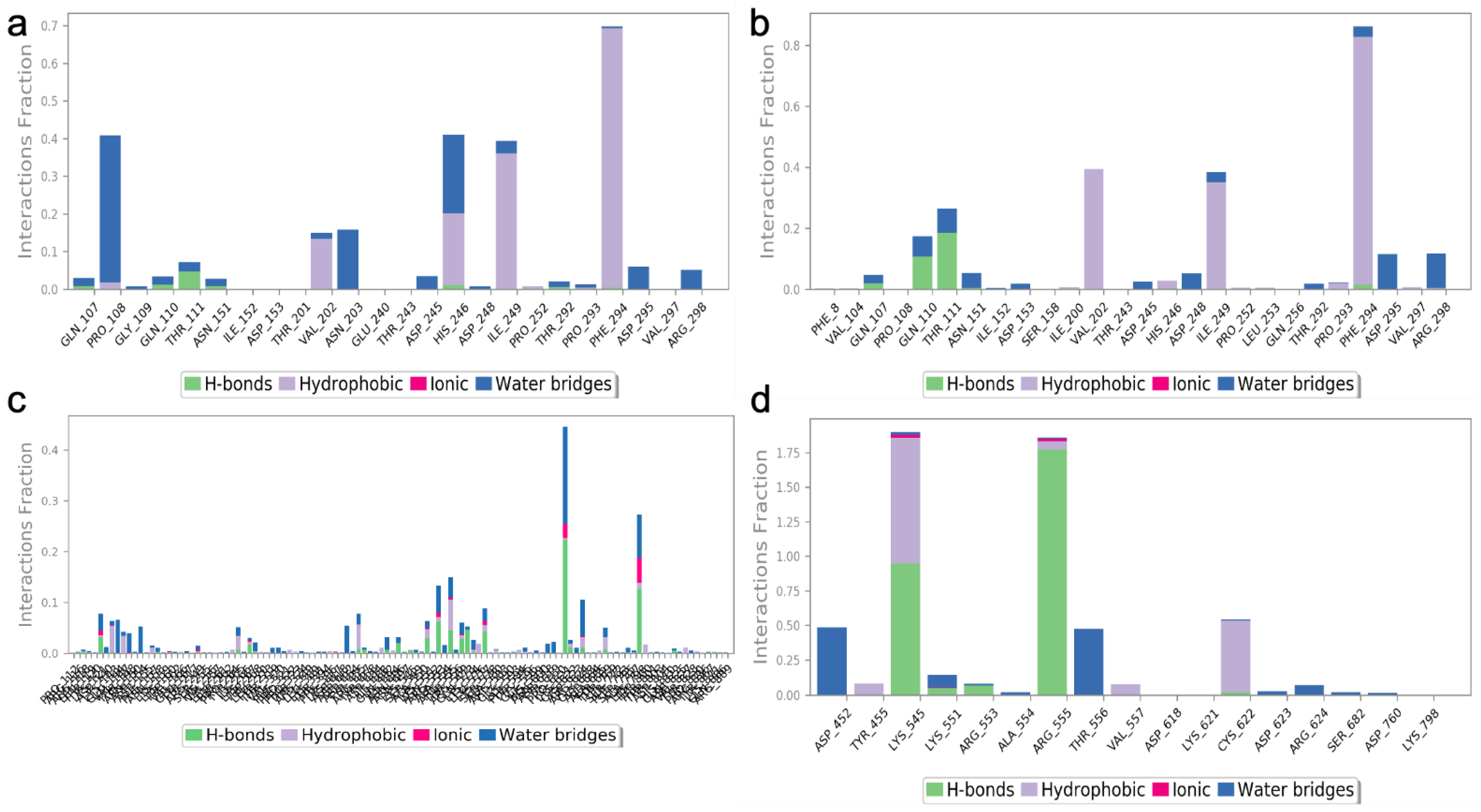
2.4. Molecular Docking
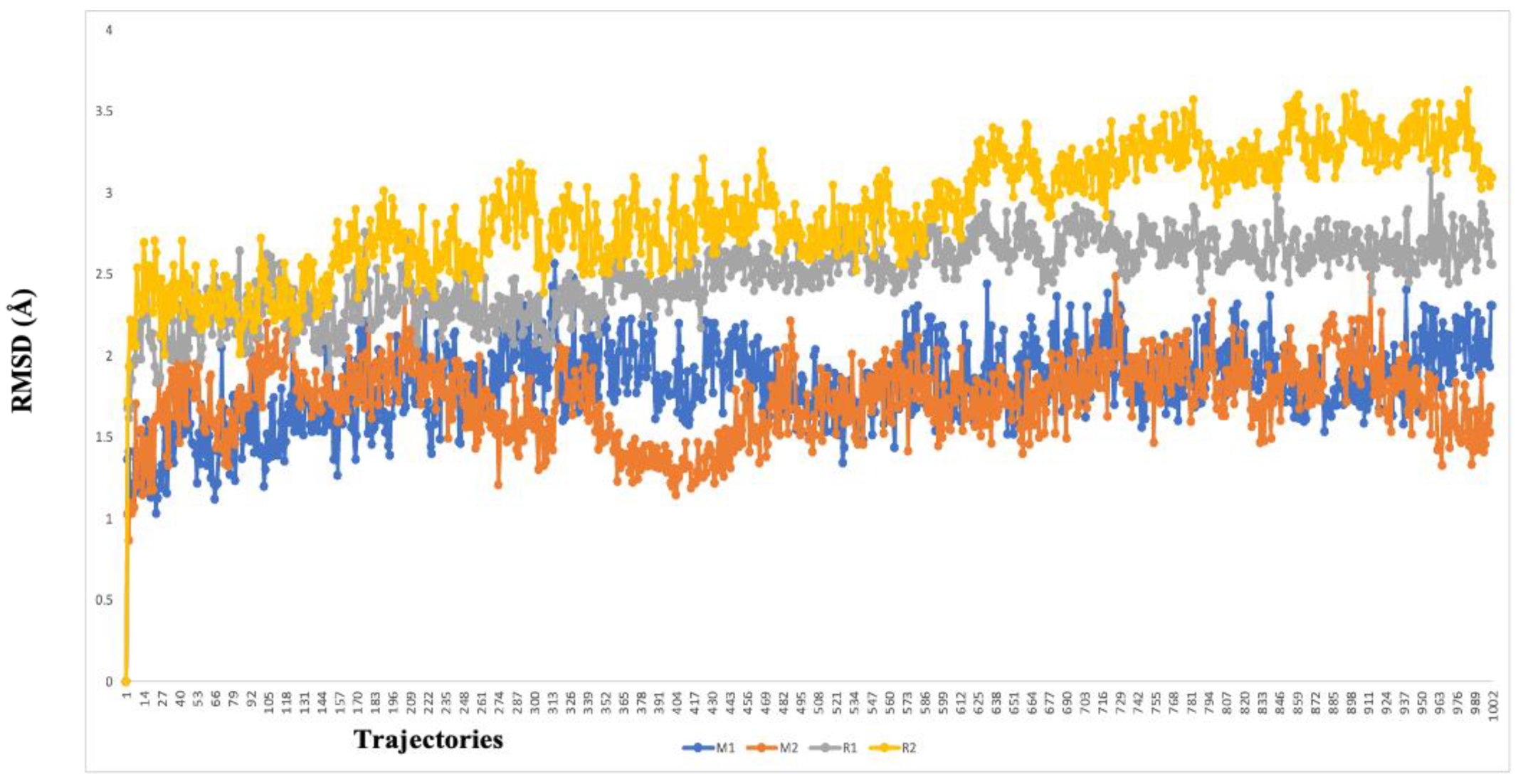
2.5. Molecular-Dynamics Simulation
3. Discussion
4. Materials and Methods
4.1. Chemicals and Reagents
4.2. Collection of Sample and Extraction
4.3. Isolation and Purification of Hexane Extract of G. wightii Leaves
4.4. Characterization of G. wightii Extract
4.5. Structural Analysis
4.6. Computational Analysis
4.6.1. Preprocessing of the Target Protein
4.6.2. Ligand Preparation
4.6.3. Molecular Docking and MD Simulation
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Badraoui, R.; Saoudi, M.; Hamadou, W.S.; Elkahoui, S.; Siddiqui, A.J.; Alam, J.M.; Jamal, A.; Adnan, M.; Suliemen, A.M.E.; Alreshidi, M.M.; et al. Antiviral Effects of Artemisinin and Its Derivatives against SARS-CoV-2 Main Protease: Computational Evidences and Interactions with ACE2 Allelic Variants. Pharmaceuticals 2022, 15, 129. [Google Scholar] [CrossRef] [PubMed]
- Verma, S.; Twilley, D.; Esmear, T.; Oosthuizen, C.B.; Reid, A.-M.; Nel, M.; Lall, N. Anti-SARS-CoV Natural Products with the Potential to Inhibit SARS-CoV-2(COVID-19). Front. Pharmacol. 2020, 11, 561334. [Google Scholar] [CrossRef] [PubMed]
- Biswaranjan, P. Human Health Care against COVID-19 via Environmental Management. Nat. Resour. Hum. Health 2022, 2, 142–149. [Google Scholar] [CrossRef]
- Huang, J.; Tao, G.; Liu, J.; Cai, J.; Huang, Z.; Chen, J.X. Current prevention of COVID-19: Natural products and herbal medicine. Front. Pharmacol. 2020, 11, 588508. [Google Scholar] [CrossRef]
- Shahhamzehei, N.; Abdelfatah, S.; Efferth, T. In Silico and In Vitro Identification of Pan-Coronaviral Main Protease Inhibitors from a Large Natural Product Library. Pharmaceuticals 2022, 15, 308. [Google Scholar] [CrossRef]
- Benarba, B.; Pandiella, A. Medicinal Plants as Sources of Active Molecules against COVID-19. Front. Pharmacol. 2020, 11, 1189. [Google Scholar] [CrossRef]
- Park, S.E. Epidemiology, virology, and clinical features of severe acute respiratory syndrome—Coronavirus-2 (SARS-CoV-2; Coronavirus Disease-19). Korean J. Pediatr. 2020, 63, 119–124. [Google Scholar] [CrossRef]
- Demeke, C.A.; Woldeyohanins, A.E.; Kifle, Z.D. Herbal medicine use for the management of COVID-19: A review article. Metab. Open 2021, 12, 100141. [Google Scholar] [CrossRef]
- Sehailia, M.; Chemat, S. Antimalarial-agent artemisinin and derivatives portray more potent binding to Lys353 and Lys31-binding hotspots of SARS-CoV-2 spike protein than hydroxychloroquine: Potential repurposing of artenimol for COVID-19. J. Biomol. Struct. Dyn. 2020, 39, 6184–6194. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüuger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.-H.; Nitsche, A.; et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020, 181, 271–280. [Google Scholar] [CrossRef]
- Zhou, P.; Yang, X.-L.; Wang, X.-G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.-R.; Zhu, Y.; Li, B.; Huang, C.-L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef]
- Nugraha, R.V.; Ridwansyah, H.; Ghozali, M.; Khairani, A.F.; Atik, N. Traditional Herbal Medicine Candidates as Complementary Treatments for COVID-19: A Review of Their Mechanisms. Pros Cons. Evid. Bas. Com. Alt. Med. 2020, 2020, 2560645. [Google Scholar] [CrossRef]
- Majdi, H.; ZarKalai, F.; Yeddess, W.; Saidani, M. Phenolic Compounds as Antiviral Agents: An In-Silico Investigation against Essential Proteins of SARS-CoV- 2. Nat. Resour. Hum. Health 2022, 2, 62–78. [Google Scholar] [CrossRef]
- Khadka, D.; Dhamala, M.K.; Li, F.; Aryal, P.C.; Magar, P.R.; Bhatta, S.; Thakur, M.S.; Basnet, A.; Cui, D.; Shi, S. The use of medicinal plants to prevent COVID-19 in Nepal. J. Ethnobiol. Ethnomed. 2021, 17, 26. [Google Scholar] [CrossRef]
- Kuchi Bhotla, H.; Balasubramanian, B.; Arumugam, V.A.; Pushparaj, K.; Easwaran, M.; Baskaran, R.; Saravanan, M.; Pappusamy, M.; Meyyazhagan, A. Insinuating Cocktailed Components in Biocompatible-Nanoparticles Could Act as an Impressive Neo-Adjuvant Strategy to Combat COVID-19. Nat. Resour. Hum. Health 2021, 1, 3–7. [Google Scholar] [CrossRef]
- Li, S.; Cheng, C.S.; Zhang, C.; Tang, G.; Tan, H.; Chen, H.Y. Edible and Herbal Plants for the Prevention and Management of COVID-19. Front. Pharm. 2021, 12, 656103. [Google Scholar] [CrossRef]
- Palani, V.; Murugesh, S.; Viji, M.; Santhosh, C.; Wenchao, L.; Balamuralikrishnan, B.; Arumugam, M. Phytoconstituents and their potential antimicrobial, antioxidant and Thomson larvicidal activities of Goniothalamus wightii Hook. F. & Thomson. Arab. J. Sci. Eng. 2020, 45, 4541–4555. [Google Scholar] [CrossRef]
- Khuntia, B.K.; Sharma, V.; Qazi, S.; Das, S.; Sharma, S.; Raza, K.; Sharma, G. Ayurvedic Medicinal Plants against COVID-19: An In Silico Analysis. Nat. Prod. Com. 2021, 16, 1–9. [Google Scholar] [CrossRef]
- Marcelino, R.C.; de Araújo, L.P.; Bueno de Morais Borba, J.; da Silveira, N.J.F. Molecular Docking Study Involving Bioactive Natural Compounds against SARS-CoV-2 Proteins. Nat. Resour. Hum. Health 2022, 2, 366–377. [Google Scholar] [CrossRef]
- Xian, Y.; Zhang, J.; Bian, Z.; Zhou, H.; Zhang, Z.; Lin, Z.; Xu, H. Bioactive natural compounds against human coronaviruses: A review and perspective. Acta Pharm. Sin. B 2020, 10, 1163–1174. [Google Scholar] [CrossRef]
- Ochnik, M.; Franz, D.; Sobczyński, M.; Naporowski, P.; Banach, M.; Orzechowska, B.; Sochocka, M. Inhibition of Human Respiratory Influenza A Virus and Human Betacoronavirus-1 by the Blend of Double-Standardized Extracts of Aronia melanocarpa (Michx.) Elliot and Sambucus nigra L. Pharmaceuticals 2022, 15, 619. [Google Scholar] [CrossRef]
- Shyr, Z.A.; Cheng, Y.-S.; Lo, D.C.; Zheng, W. Drug combination therapy for emerging viral diseases. Drug Discov. Today 2021, 26, 2367–2376. [Google Scholar] [CrossRef]
- Denaro, M.; Smeriglio, A.; Barreca, D.; De Francesco, C.; Occhiuto, C.; Milano, G.; Trombetta, D. Antiviral activity of plants and their isolated bioactive compounds: An update. Phytother. Res. 2020, 34, 742–768. [Google Scholar] [CrossRef]
- Legerská, B.; Chmelová, D.; Ondrejovič, M.; Miertuš, S. The TLC-bioautography as a tool for rapid enzyme inhibitors detection-a review. Crit. Rev. Anal. Chem. 2022, 52, 275–293. [Google Scholar] [CrossRef]
- Hosu, A.; Cimpoiu, C. Evaluation of various biological activities of natural compounds by TLC/HPTLC. J. Liq. Chromatogr. Relat. Technol. 2020, 43, 305–318. [Google Scholar] [CrossRef]
- Hari, N.; Nair, V.P. FTIR spectroscopic analysis of leaf extract in hexane in Jasminum azoricum L. Int. J. Sci. Res. Sci. Technol. 2018, 4, 170–172. [Google Scholar]
- Sayed, A.M.; Basam, S.M.; El-Naggar, E.-M.B.A.; Marzouk, H.S.; El-Hawary, S. LC-MS/MS and GC-MS profiling as well as the antimicrobial effect of leaves of selected Yucca species introduced to Egypt. Sci. Rep. 2020, 10, 1778. [Google Scholar] [CrossRef]
- Gordon, C.J.; Tchesnokov, E.P.; Woolner, E.; Perry, J.K.; Feng, J.Y.; Porter, D.P.; Götte, M. Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J. Biol. Chem. 2020, 295, 6785–6797. [Google Scholar] [CrossRef]
- Jo, S.; Kim, S.; Shin, D.H.; Kim, M.S. Inhibition of SARS-CoV 3CL protease by flavonoids. J. Enzyme Inhib. Med. Chem. 2020, 35, 145–151. [Google Scholar] [CrossRef]
- Remali, J.; Aizat, W.M. A review on plant bioactive compounds and their modes of action against coronavirus infection. Front. Pharmacol. 2021, 11, 589044. [Google Scholar] [CrossRef]
- Abd Wahab, N.Z.; Ibrahim, N. Styrylpyrone Derivative (SPD) Extracted from Goniothalamus umbrosus Binds to Dengue Virus Serotype-2 Envelope Protein and Inhibits Early Stage of Virus Replication. Molecules 2022, 27, 4566. [Google Scholar] [CrossRef] [PubMed]
- Sophonnithiprasert, T.; Aruksakunwong, O.; Tashiro, E.; Kondoh, Y.; Muroi, M.; Osada, H.; Imoto, M.; Watanapokasin, R. Interaction between goniothalamin and peroxisomal multifunctional enzyme type 2 triggering endoplasmic reticulum stress. Heliyon 2020, 6, e05200. [Google Scholar] [CrossRef] [PubMed]
- Semprebon, S.C.; De Fatima, A.; Lepri, S.R.; Sartori, D.; Ribeiro, L.R.; Mantovani, M.S. (S)-Goniothalamin induces DNA damage, apoptosis, and decrease in BIRC5 messenger RNA levels in NCI-H460 cells. Hum. Exp. Toxicol. 2014, 33, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Innajak, S.; Mahabusrakum, W.; Watanapokasin, R. Goniothalamin induces apoptosis associated with autophagy activation through MAPK signaling in SK-BR-3 cells. Oncol. Rep. 2016, 35, 2851–2858. [Google Scholar] [CrossRef]
- Yen, H.K.; Fauzi, A.R.; Din, L.B.; McKelvey-Martin, V.J.; Meng, C.K.; Hussain, S.; Rajab, N.F. Involvement of Seladin-1 in goniothalamin induced apoptosis in urinary bladder cancer cells. BMC Com. Alter. Med. 2014, 14, 2–7. [Google Scholar] [CrossRef]
- Wattanapiromsakul, C.; Wangsintaweekul, B.; Sangprapan, P.; Itharat, A.; Keawpradub, N. Goniothalamin, a cytotoxic compound, isolated from Goniothalamus macrophyllus (Blume) Hook. f. & Thomson var. macrophyllus. Songklanakarin J. Sci. Technol. 2005, 27, 479–487. [Google Scholar]
- Kim, R.P.T.; Bihud, V.; Bin Mohamad, K.; Leong, K.H.; Bin Mohamad, J.; Bin Ahmad, F.; Hazni, H.; Kasim, N.; Halim, S.N.A.; Awang, K. Cytotoxic and antioxidant compounds from the stem bark of Goniothalamus tapisoides Mat Salleh. Molecules 2012, 18, 128–139. [Google Scholar] [CrossRef]
- Olanlokun, J.O.; Olotu, A.F.; David, O.M.; Idowu, T.O.; Soliman, E.M.; Olorunsogo, O.O. A novel compound purified from Alstonia boonei inhibits Plasmodium falciparum lactate dehydrogenase and plasmepsin II. J. Biomol. Struct. Dyn. 2019, 37, 2193–2200. [Google Scholar] [CrossRef]
- Semwal, A.; Kumar, R.; Teotia, U.V.S.; Singh, R. Pharmacognostical evaluation of medicinally important Ficus retusa (Leaves and bark). J. Acute Dis. 2013, 2, 300–303. [Google Scholar] [CrossRef][Green Version]
- Haddad, M.; Zein, S.; Shahrour, H.; Hamadeh, K.; Karaki, N.; Kanaan, H. Antioxidant activity of water-soluble polysaccharide extracted from Eucalyptus cultivated in Lebanon. Asian Pac. J. Trop. Biomed. 2017, 7, 157–160. [Google Scholar] [CrossRef]
- Vignesh, A.; Selvakumar, S.; Vasanth, K. Comparative LC-MS analysis of bioactive compounds, antioxidants and antibacterial activity from leaf and callus extracts of Saraca asoca. Phytomed. Plus 2022, 2, 100167. [Google Scholar] [CrossRef]
- Li, Y.; He, Q.; Du, S.; Guo, S.; Geng, Z.; Deng, Z. Study of methanol extracts from different parts of Peganum harmala L. using 1H-NMR plant metabolomics. J. Anal. Methods Chem. 2018, 2018, 6532789. [Google Scholar] [CrossRef]
- Gunaseelan, S.; Arunkumar, M.; Aravind, M.K.; Gayathri, S.; Rajkeerthana, S.; Mohankumar, V.; Ashokkumar, B.; Varalakshmi, P. Probing marine brown macroalgal phlorotannins as antiviral candidate against SARS-CoV-2: Molecular docking and dynamics simulation approach. Mol. Divers. 2022, 1–20. [Google Scholar] [CrossRef]
- Arunkumar, M.; Gunaseelan, S.; Kubendran Aravind, M.; Mohankumar, V.; Anupam, P.; Harikrishnan, M.; Harikrishnan, M.; Siva, A.; Ashokkumar, B.; Varalakshmi, P. Marine algal antagonists targeting 3CL protease and spike glycoprotein of SARS-CoV-2: A computational approach for anti-COVID-19 drug discovery. J. Biomol. Struct. Dyn. 2021, 1–28. [Google Scholar] [CrossRef]


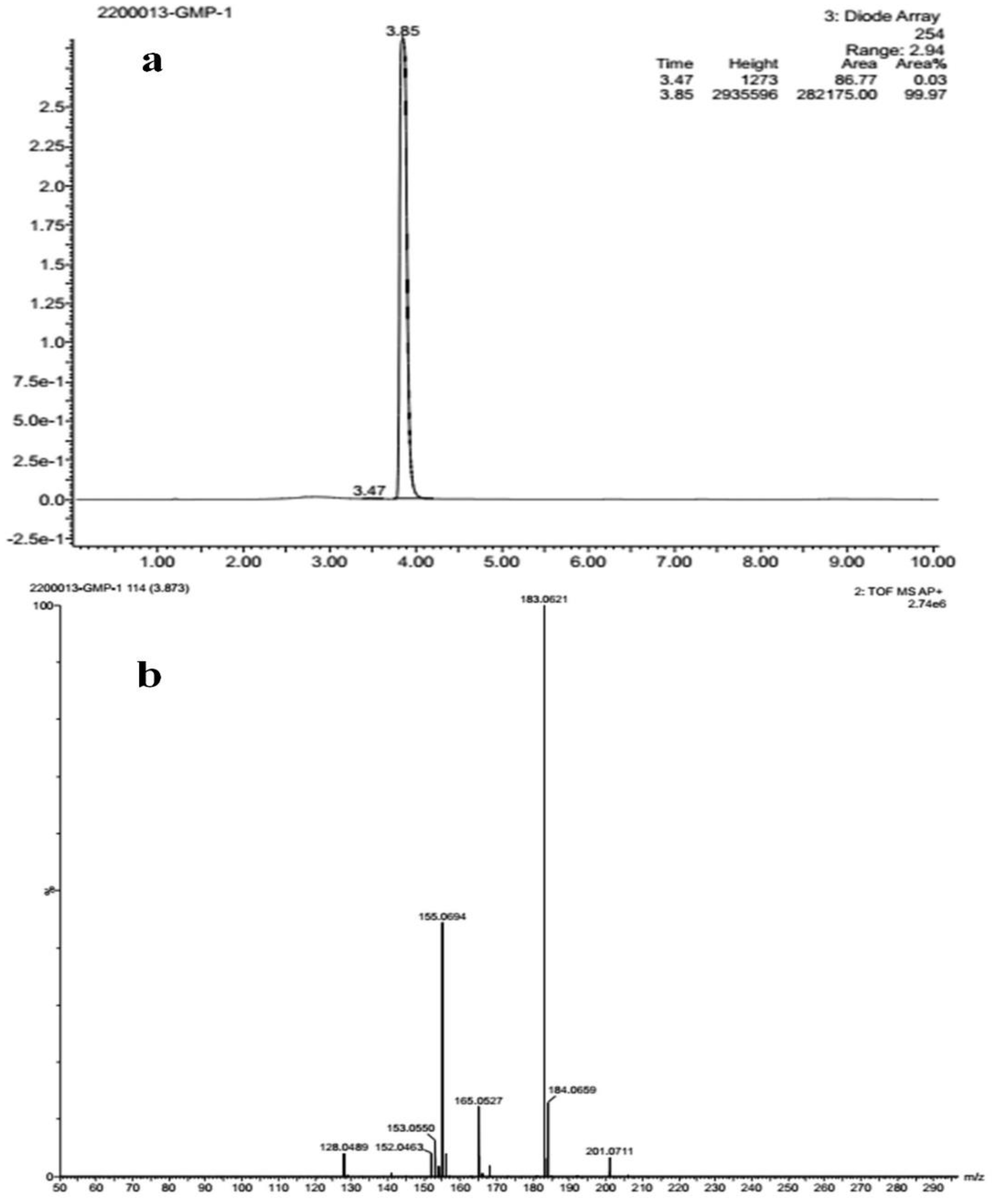
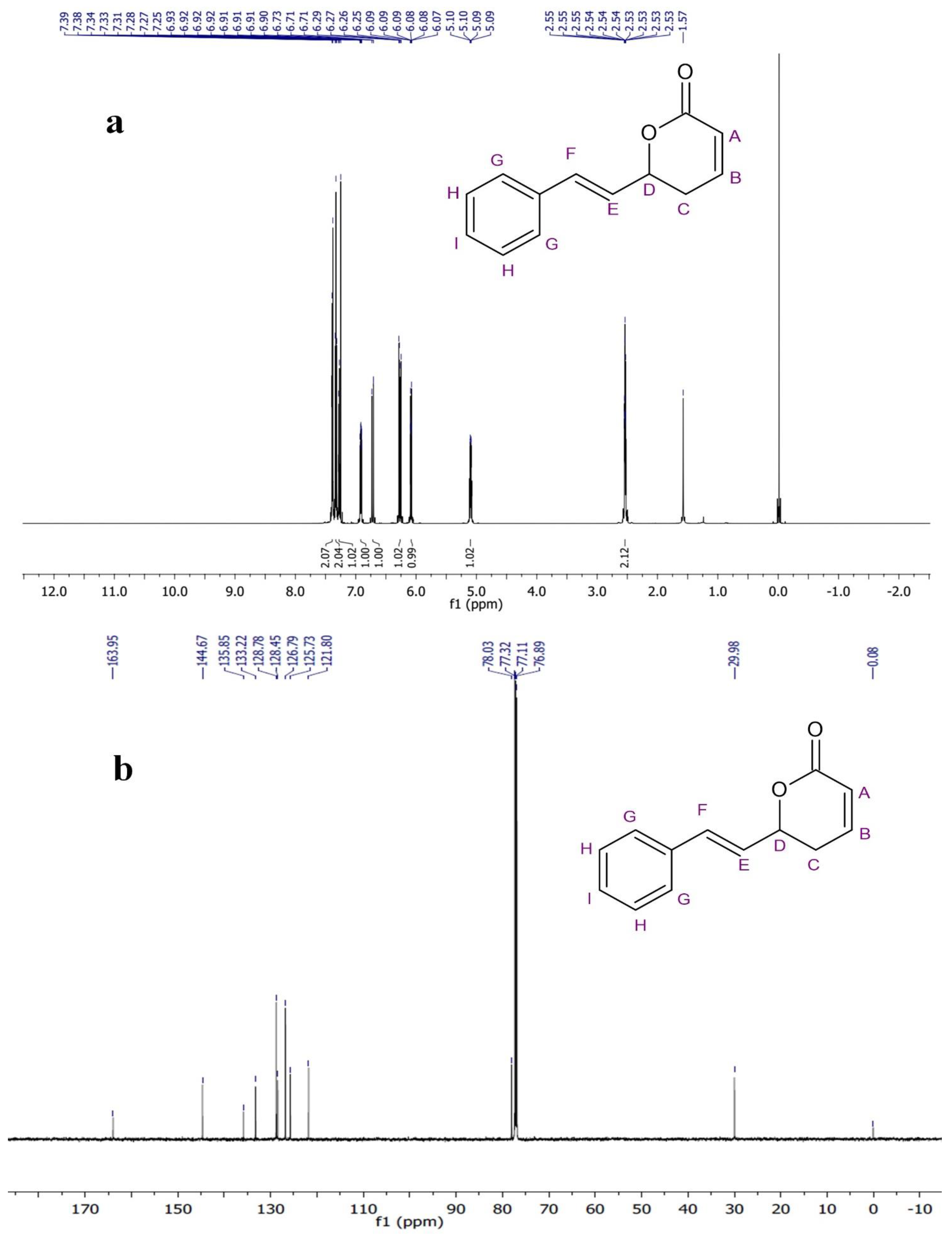

| Wave Number cm−1 | Possible Functional Group | Standard Reference Range |
|---|---|---|
| 3011 | CH stretch (aromatic) | 3000–3150 |
| 2914 | CH stretch (asymmetric) | 3000–2800 |
| 2857 | CH stretch Aliphatic (symmetric) | 3000–2800 |
| 1719 | C=O stretch, lactone | 1720–1750 |
| 1690 | C=C stretch, olefin | 1680–1640 |
| 1455 | CH, bend, aliphatic | 1465–1375 |
| 1393 | C-C stretch, aromatic | 1500–1400 |
| 1250 | C-O stretch | 1320–1000 |
| 1015 | CH bend, Aliphatic (in plane) | 1000–1250 |
| 969 | =C-H bend, olefin | 1000–650 |
| 763 | C-H bend, Aromatic (out-of-plane) | 900–675 |
| ID | Compound Name | PubChem ID | Glide Score | Glide Energy | Glide Emodel | Amino Acid Interaction | Bond Length |
|---|---|---|---|---|---|---|---|
| R1 | 3-Phenyl-2-(propan-2-yloxymethyl)prop-2-enoate | 154129834 | −6.874 | −22.658 | −25.405 | Asp623, Arg624, Arg553, Thr556 | 2.38, (1.88, 2.41), 1.92, (2.36,2.43, 1.53) |
| R2 | 4-(3-Phenylprop-2-enoxy)but-2-enoic acid | 76940553 | −6.786 | −26.902 | −30.473 | Arg624, Thr556 | (2.43,1.67), 1.73 |
| R3 | 4-(3-Phenylprop-2-enoyloxy)octa-2,6-dienoic acid | 67934386 | −6.679 | −26.082 | −31.825 | Arg553, Thr556, Hie572 | 2.47,2.34,2.16 |
| R4 | 4-Oxo-4-(3-phenylprop-2-enoxy)but-2-enoate | 78478352 | −6.569 | −18.008 | −19.319 | Arg555, Thr556 | (3.14,3.15), 2.74 |
| R5 | (Z)-4-oxo-4-(3-phenylprop-2-enoxy)but-2-enoic acid | 70430994 | −6.569 | −18.008 | −19.319 | Arg555, Thr556, Lys545 | 3.73,(2.74,3.68),2.95 |
| S1 | Diethyl 4-(3-phenylprop-2-en-1-ylidene)hepta-2,5-dienedioate | 71430013 | −8.501 | −47.499 | −62.222 | C-A Gln1005, Thr1009, Gln1002, Val1008 C-B Gln1002 C-C Thr1006 | 2.36 (2.16,2.76),2.44,(2.76,2.15),2.56,2.15 2.48 |
| S2 | 2,6,11,15-Tetramethylhexadeca-2,6,10,14-tetraen-8-yl 3-phenylprop-2-enoate | 54538910 | −8.315 | −33.738 | −12.688 | A Gln1005 B Gln1005, Thr1006, Gln1010,Thr1009, Gln762 C Gln1005 | 2.36 (2.16,2.76),2.44,(2.76,2.15),2.56,2.15 2.48 |
| S3 | [(6E,10E)-2,6,11,15-tetramethylhexadeca-2,6,10,14-tetraen-8-yl] (E)-3-phenylprop-2-enoate | 67563707 | −8.286 | −44.473 | −22.029 | Gln1002, Thr1006, Gln1005, Val1008, Thr1009, Gln1005, Gln1002, Thr1009, Thr1006 | (2.19,2.40),(2.26,2.23),2.36,(2.91,2.40),(2.36,2.35) (2.51,2.27,2.38),(2.34,2.31),(2.85,1.88) 2.25 |
| S4 | bis(3-phenylprop-2-enyl) (Z)-but-2-enedioate | 141388166 | −8.127 | −45.087 | −57.403 | Thr1009, Leu763 Gln1002, Gln1005 Thr1009, Thr1006,Gln1002, Gln1010,Gln1005 | 2.39,2.57 2.37,1.87 (2.44,2.66),2.35,(2.01,2.35,2.59),(2.50,2.51),2.28 |
| S5 | bis[(E)-3-phenylprop-2-enyl] (Z)-but-2-enedioate | 70543374 | −8.127 | −45.087 | −57.403 | Leu763, Thr1009 Gln1005 Gln1002, Gln1005,Thr1006, Thr1009, Gln1010 | 2.57,(5.29,2.39) 1.87 (2.37,2.33,2.01,2.59),2.28,2.35,(2.66,2.44),(2.50,2.51) |
| M1 | 4-methoxy-6-[(1Z,3E)-4-phenylbuta-1,3-dienyl]pyran-2-one | 92528557 | −4.755 | −27.663 | −36.773 | Asn203, Gln110, Asn151 | 2.36,(2.31,1.91),2.34 |
| M2 | 4-Methoxy-6-(4-phenylbuta-1,3-dienyl)pyran-2-one | 322722 | −4.723 | −28.379 | −33.967 | Gln110, Asn151 | 2.16,2.30 |
| M3 | ethyl (E,2E)-4-phenyl-2-(phenyl(113C)methylidene)(413C)but-3-enoate | 11482889 | −4.399 | −30.317 | −39.043 | Asn203 | 2.43 |
| M4 | 3-Methyl-5-(1-methyl-3-phenyl-2-propenyl)furan-2(5H)-one | 101575001 | −4.382 | −25.273 | −28.44 | Gln110 | 2.42 |
| M5 | (6R)-5,6-Dihydro-6alpha-[(2R)-2-acetoxy-4-phenyl-3-butenyl]-2H-pyran-2-one | 100927498 | −4.235 | −28.978 | −34.036 | Thr111, Asp295, Gln110 | (2.31,2.25). 2.28,1.76 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Palani, V.; Chinnaraj, S.; Shanmugasundaram, M.; Malaisamy, A.; Maluventhen, V.; Arumugam, V.A.; Rengasamy, K.R.R.; Balasubramanian, B.; Liu, W.-C.; Arumugam, M. Derivation, Functionalization of (S)-Goniothalamin from Goniothalamus wightii and Their Derivative Targets SARS-CoV-2 MPro, SPro, and RdRp: A Pharmacological Perspective. Molecules 2022, 27, 6962. https://doi.org/10.3390/molecules27206962
Palani V, Chinnaraj S, Shanmugasundaram M, Malaisamy A, Maluventhen V, Arumugam VA, Rengasamy KRR, Balasubramanian B, Liu W-C, Arumugam M. Derivation, Functionalization of (S)-Goniothalamin from Goniothalamus wightii and Their Derivative Targets SARS-CoV-2 MPro, SPro, and RdRp: A Pharmacological Perspective. Molecules. 2022; 27(20):6962. https://doi.org/10.3390/molecules27206962
Chicago/Turabian StylePalani, Vino, Santhosh Chinnaraj, Murugesh Shanmugasundaram, Arunkumar Malaisamy, Viji Maluventhen, Vijaya Anand Arumugam, Kannan R. R. Rengasamy, Balamuralikrishnan Balasubramanian, Wen-Chao Liu, and Maruthupandian Arumugam. 2022. "Derivation, Functionalization of (S)-Goniothalamin from Goniothalamus wightii and Their Derivative Targets SARS-CoV-2 MPro, SPro, and RdRp: A Pharmacological Perspective" Molecules 27, no. 20: 6962. https://doi.org/10.3390/molecules27206962
APA StylePalani, V., Chinnaraj, S., Shanmugasundaram, M., Malaisamy, A., Maluventhen, V., Arumugam, V. A., Rengasamy, K. R. R., Balasubramanian, B., Liu, W.-C., & Arumugam, M. (2022). Derivation, Functionalization of (S)-Goniothalamin from Goniothalamus wightii and Their Derivative Targets SARS-CoV-2 MPro, SPro, and RdRp: A Pharmacological Perspective. Molecules, 27(20), 6962. https://doi.org/10.3390/molecules27206962









