Utilizing the DNA Aptamer to Determine Lethal α-Amanitin in Mushroom Samples and Urine by Magnetic Bead-ELISA (MELISA)
Abstract
:1. Introduction
2. Results and Discussion
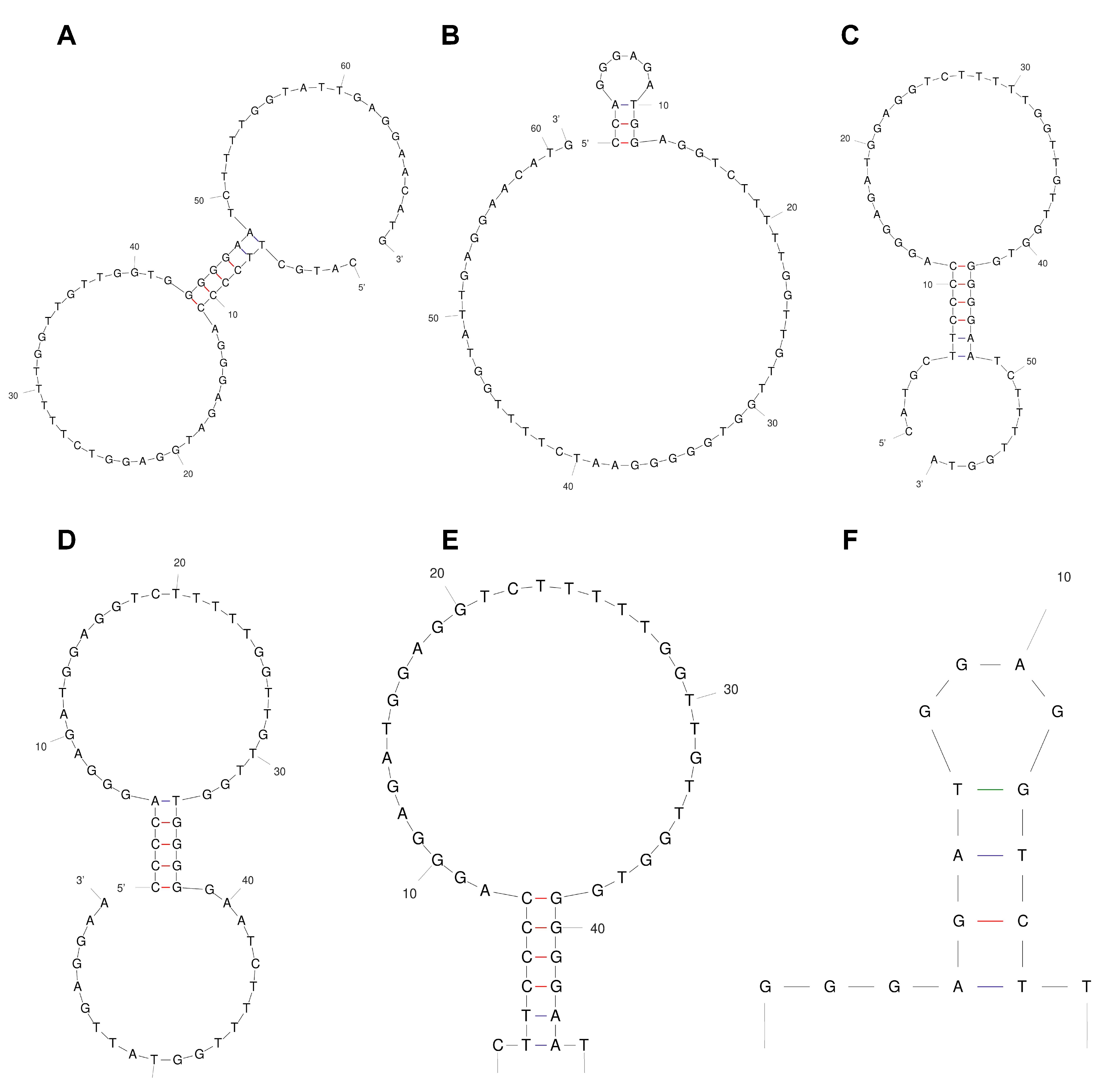
2.1. Truncation and Sequence Design of the Aptamers
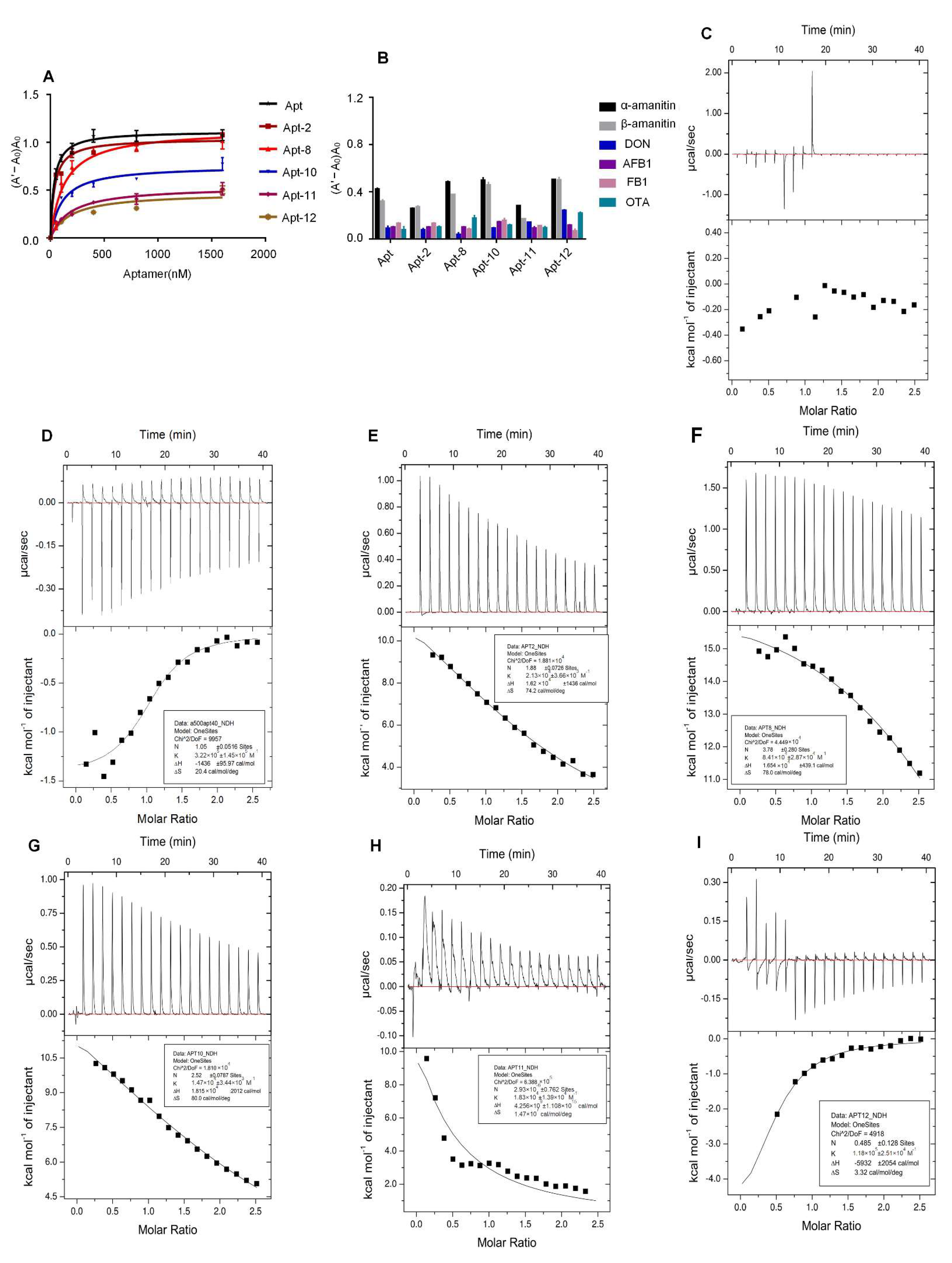
2.2. Specificity and Affinity Testing of the Aptamer
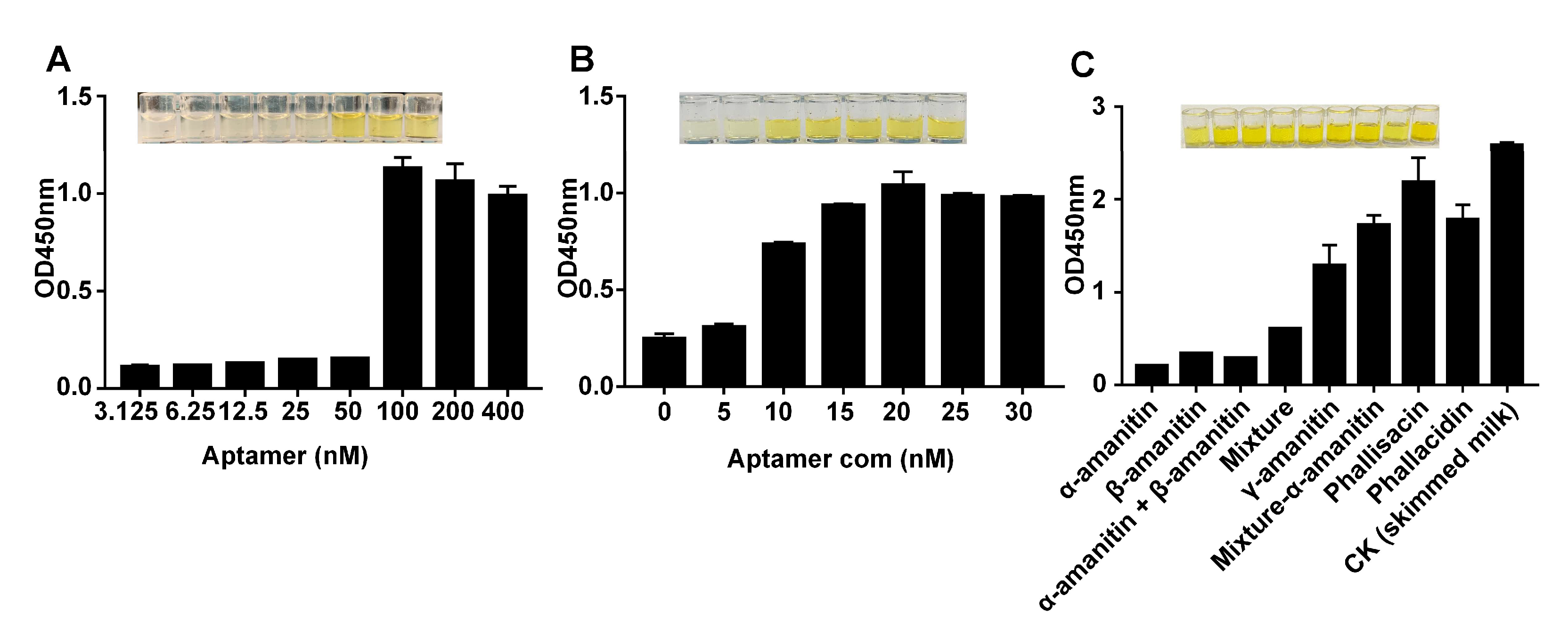
2.3. Optimization of the Experimental Conditions
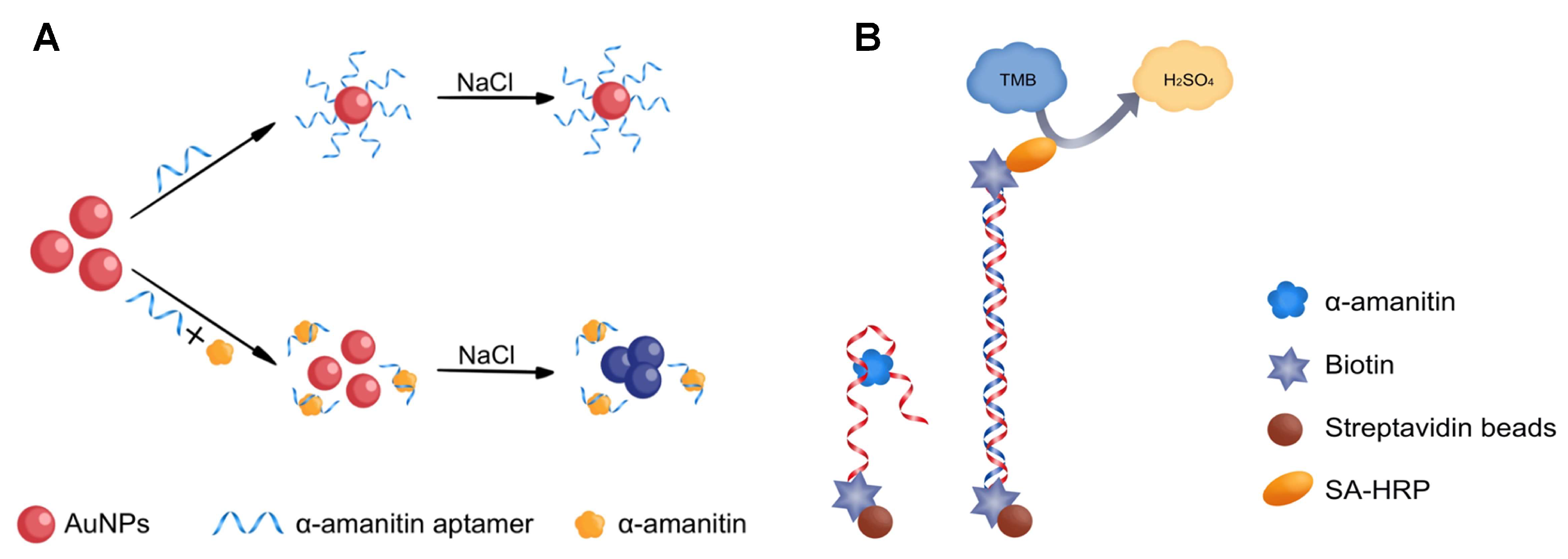
2.4. Detecting α-Amanitin with the Aptamer-Based MELISA
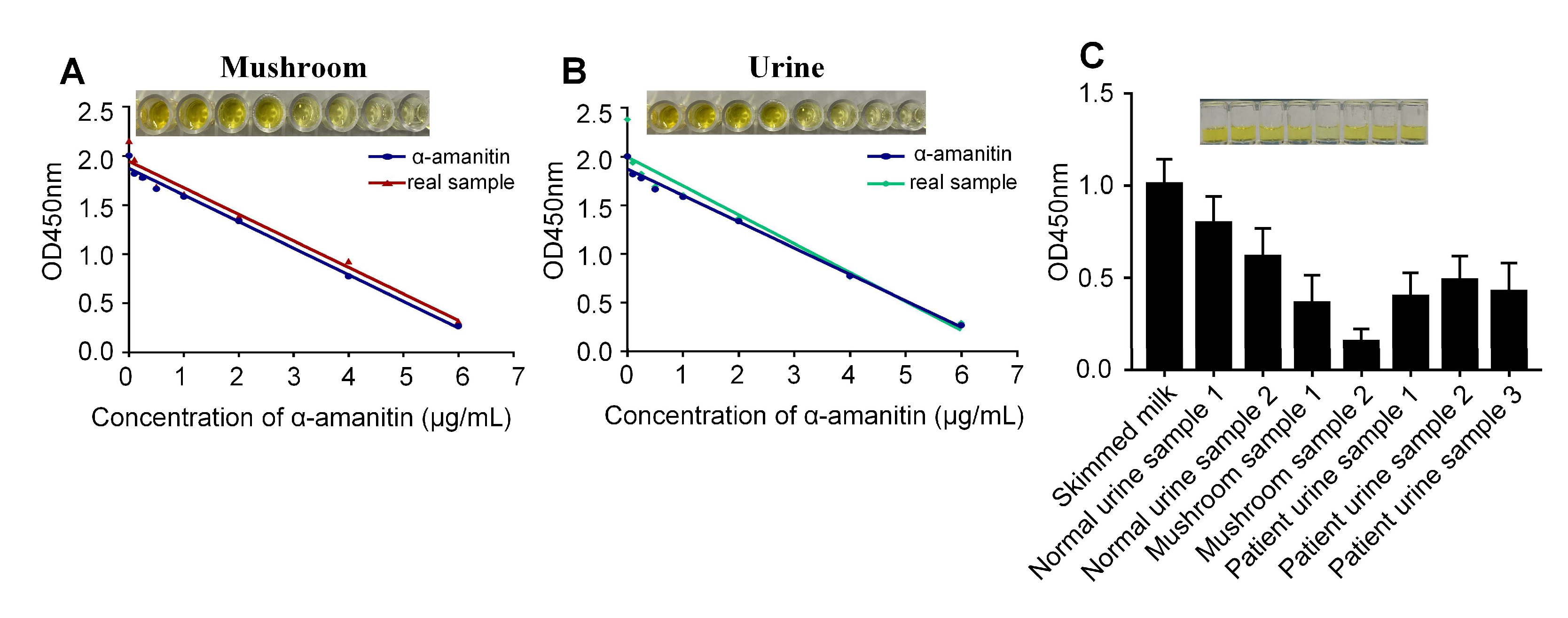
2.5. Detecting α-Amanitin in Actual Samples
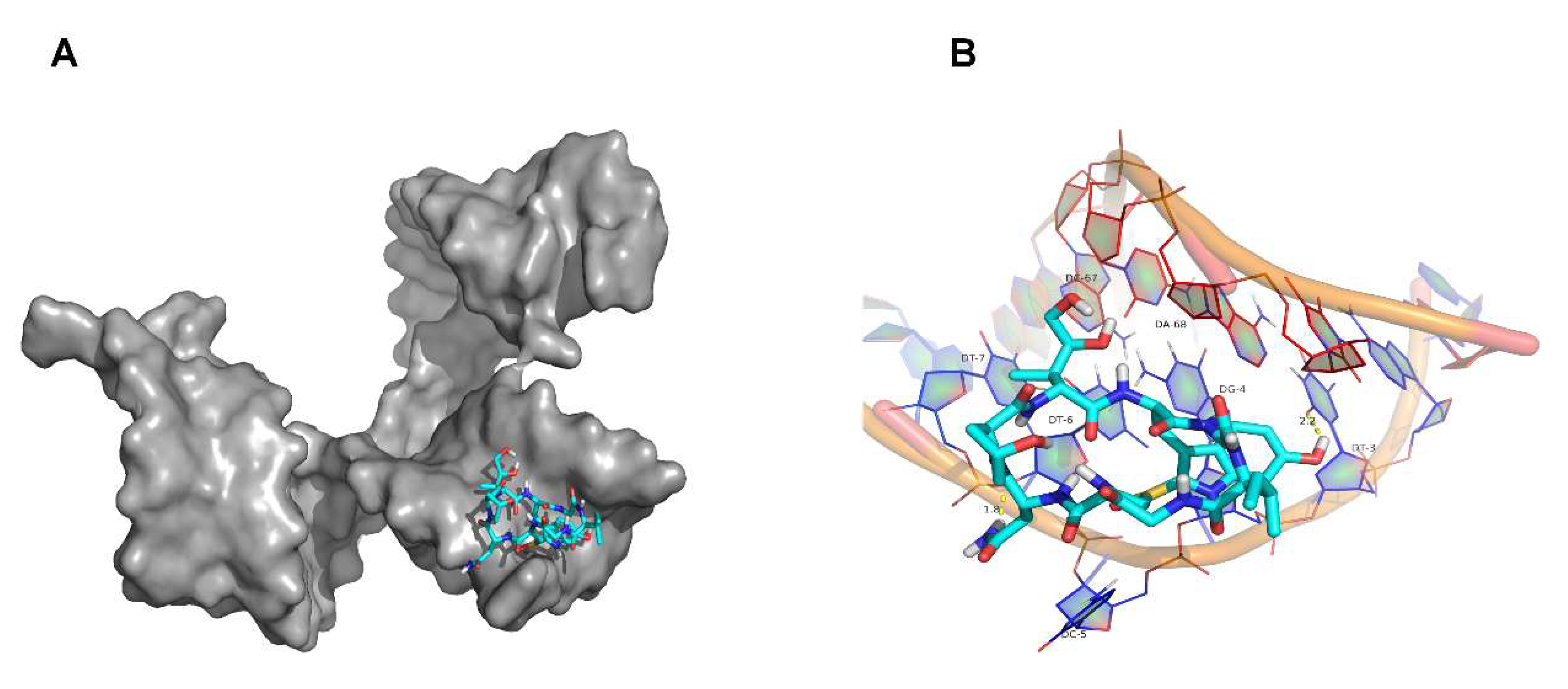
2.6. Molecular Docking Simulation Results
3. Conclusions
4. Materials and Methods
4.1. Reagents
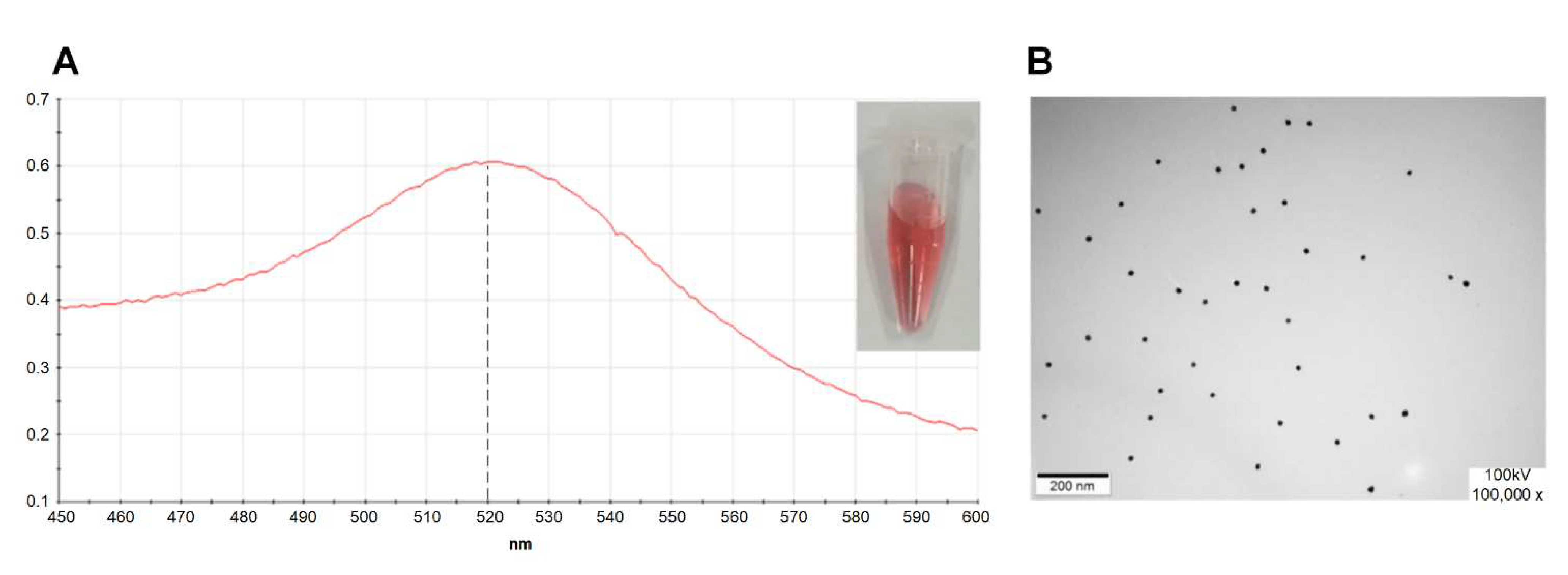
4.2. Preparation and Identification of the AuNPs
4.3. Truncation of the Original α-Amanitin Aptamer Sequence
4.4. Aptamer Specificity and Affinity Tests
4.5. Optimizing the MELISA Experimental Conditions
4.6. Detecting α-Amanitin Using the Aptamer-Based MELISA
4.7. Preparation of the Samples
4.8. Molecular Docking Simulation
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Diaz, J.H. Amatoxin-Containing Mushroom Poisonings: Species, Toxidromes, Treatments, and Outcomes. Wilderness Environ. Med. 2018, 29, 111–118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, C.; Li, H. Expert consensus on clinical diagnosis and treatment of mushroom poisoning with amanita in China (in Chinese). hinese J. Crit. Care Med. (Electron. Ed.) 2020, 13, 20–28. [Google Scholar]
- Santi, L.; Maggioli, C.; Mastroroberto, M.; Tufoni, M.; Napoli, L.; Caraceni, P. Acute Liver Failure Caused by Amanita phalloides Poisoning. Int. J. Hepatol. 2012, 2012, 487480. [Google Scholar] [CrossRef] [PubMed]
- Michelot, D.; Melendez-Howell, L.M. Amanita muscaria: Chemistry, biology, toxicology, and ethnomycology. Mycol. Res. 2003, 107, 131–146. [Google Scholar] [CrossRef]
- Brueckner, F.; Cramer, P. Structural basis of transcription inhibition by alpha-amanitin and implications for RNA polymerase II translocation. Nat. Struct. Mol. Biol. 2008, 15, 811–818. [Google Scholar] [CrossRef] [PubMed]
- Kaya, E.; Yilmaz, I.; Admis, O.; Oktay, M.; Bayram, R.; Bakirci, S.; Yaykasli, K.O.; Kandis, H.; Saritas, A.; Katirci, Y.J.T.R. Effects of erdosteine on alpha amanitin-induced hepatotoxicity in mice. Toxin Rev. 2016, 35, 4–9. [Google Scholar] [CrossRef]
- Garcia, J.; Costa, V.M.; Carvalho, A.T.; Silvestre, R.; Duarte, J.A.; Dourado, D.F.; Arbo, M.D.; Baltazar, T.; Dinis-Oliveira, R.J.; Baptista, P.; et al. A breakthrough on Amanita phalloides poisoning: An effective antidotal effect by polymyxin B. Arch. Toxicol. 2015, 89, 2305–2323. [Google Scholar] [CrossRef]
- Brandenburg, W.E.; Ward, K.J. Mushroom poisoning epidemiology in the United States. Mycologia 2018, 110, 637–641. [Google Scholar] [CrossRef]
- Butera, R.; Locatelli, C.; Coccini, T.; Manzo, L. Diagnostic accuracy of urinary amanitin in suspected mushroom poisoning: A pilot study. J. Toxicol. Clin. Toxicol. 2004, 42, 901–912. [Google Scholar] [CrossRef]
- Sun, J.; Niu, Y.M.; Zhang, Y.T.; Li, H.J.; Yin, Y.; Zhang, Y.Z.; Ma, P.B.; Zhou, J.; Huang, L.; Zhang, H.S.; et al. Toxicity and toxicokinetics of Amanita exitialis in beagle dogs. Toxicon: Off. J. Int. Soc. Toxinol. 2018, 143, 59–67. [Google Scholar] [CrossRef]
- Garcia, J.; Costa, V.M.; Baptista, P.; Bastos Mde, L.; Carvalho, F. Quantification of alpha-amanitin in biological samples by HPLC using simultaneous UV-diode array and electrochemical detection. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2015, 997, 85–95. [Google Scholar] [CrossRef] [Green Version]
- Maurer, H.H.; Kraemer, T.; Ledvinka, O.; Schmitt, C.J.; Weber, A.A. Gas chromatography-mass spectrometry (GC-MS) and liquid chromatography-mass spectrometry (LC-MS) in toxicological analysis. Studies on the detection of clobenzorex and its metabolites within a systematic toxicological analysis procedure by GC-MS and by immunoassay and studies on the detection of alpha- and beta-amanitin in urine by atmospheric pressure ionization electrospray LC-MS. J. Chromatogr. B Biomed. Sci. Appl. 1997, 689, 81–89. [Google Scholar] [PubMed]
- Filigenzi, M.S.; Poppenga, R.H.; Tiwary, A.K.; Puschner, B. Determination of alpha-amanitin in serum and liver by multistage linear ion trap mass spectrometry. J. Agric. Food Chem. 2007, 55, 2784–2790. [Google Scholar] [CrossRef] [PubMed]
- Abbott, N.L.; Hill, K.L.; Garrett, A.; Carter, M.D.; Hamelin, E.I.; Johnson, R.C. Detection of α-, β-, and γ-amanitin in urine by LC-MS/MS using (15)N(10)-α-amanitin as the internal standard. Toxicon: Off. J. Int. Soc. Toxinol. 2018, 152, 71–77. [Google Scholar] [CrossRef] [PubMed]
- Staack, R.; Maurer, H. New Bühlmann ELISA for determination of Amanitins in urine—Are there false positive results due to interferences with ur ine matrix, drugs or their metabolites? Toxichem Krimtech 2000, 68, 68–71. [Google Scholar]
- He, K.; Mao, Q.; Zang, X.; Zhang, Y.; Li, H.; Zhang, D. Production of a broad-specificity monoclonal antibody and application as a receptor to detection amatoxins in mushroom. Biol.: J. Int. Assoc. Biol. Stand. 2017, 49, 57–61. [Google Scholar] [CrossRef]
- Bever, C.S.; Swanson, K.D.; Hamelin, E.I.; Filigenzi, M.; Poppenga, R.H.; Kaae, J.; Cheng, L.W.; Stanker, L.H. Rapid, Sensitive, and Accurate Point-of-Care Detection of Lethal Amatoxins in Urine. Toxins 2020, 12, 123. [Google Scholar] [CrossRef] [Green Version]
- Bever, C.S.; Adams, C.A.; Hnasko, R.M.; Cheng, L.W.; Stanker, L.H. Lateral flow immunoassay (LFIA) for the detection of lethal amatoxins from mushrooms. PLoS ONE 2020, 15, e0231781. [Google Scholar]
- Kinghorn, A.B.; Fraser, L.A.; Lang, S.; Shiu, S.C.C.; Tanner, J.A. Aptamer Bioinformatics. Int. J. Mol. Sci. 2017, 18, 2516. [Google Scholar] [CrossRef] [Green Version]
- Parashar, A. Aptamers in Therapeutics. J. Clin. Diagn. Res. JCDR 2016, 10, 1–6. [Google Scholar] [CrossRef]
- Hermann, T.; Patel, D.J. Adaptive recognition by nucleic acid aptamers. Science 2000, 287, 820–825. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Q.; Yang, Q.; Wu, W. Graphene-Based Steganographic Aptasensor for Information Computing and Monitoring Toxins of Biofilm in Food. Front. Microbiol. 2019, 10, 3139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, P.; Ye, H.; Deng, J.; Khan, I.M.; Yue, L.; Wang, Z. A fluorescence polarization aptasensor coupled with polymerase chain reaction and streptavidin for chloramphenicol detection. Talanta 2019, 205, 120119. [Google Scholar] [CrossRef]
- Wang, C.; Sun, L.; Zhao, Q. A simple aptamer molecular beacon assay for rapid detection of aflatoxin B1. Chin. Chem. Lett. 2019, 30, 1017–1020. [Google Scholar] [CrossRef]
- Trinh, K.H.; Kadam, U.S.; Song, J.; Cho, Y.; Kang, C.H.; Lee, K.O.; Lim, C.O.; Chung, W.S.; Hong, J.C. Novel DNA Aptameric Sensors to Detect the Toxic Insecticide Fenitrothion. Int. J. Mol. Sci. 2021, 22, 846. [Google Scholar] [CrossRef]
- Moutsiopoulou, A.; Broyles, D.; Dikici, E.; Daunert, S.; Deo, S.K. Molecular Aptamer Beacons and Their Applications in Sensing, Imaging, and Diagnostics. Small (Weinh. Der Bergstr. Ger.) 2019, 15, e1902248. [Google Scholar] [CrossRef]
- Gu, C.; Xiang, Y.; Guo, H.; Shi, H. Label-free fluorescence detection of melamine with a truncated aptamer. Analyst 2016, 141, 4511–4517. [Google Scholar] [CrossRef] [Green Version]
- Le, T.T.; Chumphukam, O.; Cass, A.E. Determination of minimal sequence for binding of an aptamer. A comparison of truncation and hybridization inhibition methods. RSC Adv. 2014, 4, 47227–47233. [Google Scholar] [CrossRef]
- Vu, C.Q.; Rotkrua, P.; Tantirungrotechai, Y.; Soontornworajit, B.J.A.C.S. Oligonucleotide Hybridization Combined with Competitive Antibody Binding for the Truncation of a High-Affinity Aptamer. ACS Comb. Sci. 2017, 19, 609–617. [Google Scholar] [CrossRef]
- Feig, A.L. Applications of isothermal titration calorimetry in RNA biochemistry and biophysics. Biopolymers 2007, 87, 293–301. [Google Scholar] [CrossRef] [Green Version]
- Slavkovic, S.; Churcher, Z.R.; Johnson, P.E. Nanomolar binding affinity of quinine-based antimalarial compounds by the cocaine-binding aptamer. Bioorganic Med. Chem. 2018, 26, 5427–5434. [Google Scholar] [CrossRef]
- Li, J.; Hong, D.; Peng, X.; Huang, X.; Xiong, Y.J.R.A. Effect of different-sized spherical gold nanoparticles grown layer by layer on the sensitivity of an immunochromatographic assay. Rsc Adv. 2016, 6, 26178–26185. [Google Scholar] [CrossRef]
- Aldewachi, H.; Chalati, T.; Woodroofe, M.N.; Bricklebank, N.; Sharrack, B.; Gardiner, P. Gold nanoparticle-based colorimetric biosensors. Nanoscale 2017, 10, 18–33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, Q.; Wang, Y.; Jia, J.; Xiang, Y. Colorimetric aptasensors for determination of tobramycin in milk and chicken eggs based on DNA and gold nanoparticles. Food Chem. 2018, 249, 98–103. [Google Scholar] [CrossRef]
- Zhou, X.; Wang, L.; Shen, G.; Zhang, D.; Xie, J.; Mamut, A.; Huang, W.; Zhou, S. Colorimetric determination of ofloxacin using unmodified aptamers and the aggregation of gold nanoparticles. Mikrochim. Acta 2018, 185, 355. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Huang, J.; Tan, S.; Shao, C.; Xiang, J.J.I.J. Preparation of monoclonal antibodies against human NT-Pro BNP and establishment of double-antibody sandwich ELISA. Int. J. Sci. 2020, 7, 152–162. [Google Scholar]
- Deng, Q.; Qiu, M.; Wang, Y.; Lv, P.; Wu, C.; Sun, L.; Ye, R.; Xu, D.; Liu, Y.; Gooneratne, R. A sensitive and validated immunomagnetic-bead based enzyme-linked immunosorbent assay for analyzing total T-2 (free and modified) toxins in shrimp tissues. Ecotoxicol. Environ. Saf. 2017, 142, 441–447. [Google Scholar] [CrossRef] [PubMed]
- Lei, J.H.; Guan, F.; Xu, H.; Chen, L.; Su, B.T.; Zhou, Y.; Wang, T.; Li, Y.L.; Liu, W.Q. Application of an immunomagnetic bead ELISA based on IgY for detection of circulating antigen in urine of mice infected with Schistosoma japonicum. Vet. Parasitol. 2012, 187, 196–202. [Google Scholar] [CrossRef]
- Muszyńska, K.; Ostrowska, D.; Bartnicki, F.; Kowalska, E.; Bodaszewska-Lubaś, M.; Hermanowicz, P.; Faulstich, H.; Strzałka, W. Selection and analysis of a DNA aptamer binding α-amanitin from Amanita phalloides. Acta Biochim. Pol. 2017, 64, 401–406. [Google Scholar] [CrossRef]
- Jia, M.; Sha, J.; Li, Z.; Wang, W.; Zhang, H. High affinity truncated aptamers for ultra-sensitive colorimetric detection of bisphenol A with label-free aptasensor. Food Chem. 2020, 317, 126459. [Google Scholar] [CrossRef]
- Matulakul, P.; Vongpramate, D.; Kulchat, S.; Chompoosor, A.; Thanan, R.; Sithithaworn, P.; Sakonsinsiri, C.; Puangmali, T. Development of Low-Cost AuNP-Based Aptasensors with Truncated Aptamer for Highly Sensitive Detection of 8-Oxo-dG in Urine. ACS Omega 2020, 5, 17423–17430. [Google Scholar] [CrossRef]
- Slavkovic, S.; Johnson, P.E. Isothermal titration calorimetry studies of aptamer-small molecule interactions: Practicalities and pitfalls. Aptamers 2018, 2, 41–51. [Google Scholar]
- Alsager, O.A.; Alotaibi, K.M.; Alswieleh, A.M.; Alyamani, B.J. Colorimetric Aptasensor of Vitamin D3: A Novel Approach to Eliminate Residual Adhesion between Aptamers and Gold Nanoparticles. Sci. Rep. 2018, 8, 12947. [Google Scholar] [CrossRef] [PubMed]
- Ma, P.; Guo, H.; Duan, N.; Ma, X.; Yue, L.; Gu, Q.; Wang, Z. Label free structure-switching fluorescence polarization detection of chloramphenicol with truncated aptamer. Talanta 2021, 230, 122349. [Google Scholar] [CrossRef]
- Trinh, K.H.; Kadam, U.S.; Rampogu, S.; Cho, Y.; Yang, K.A.; Kang, C.H.; Lee, K.W.; Lee, K.O.; Chung, W.S.; Hong, J.C. Development of novel fluorescence-based and label-free noncanonical G4-quadruplex-like DNA biosensor for facile, specific, and ultrasensitive detection of fipronil. J. Hazard. Mater. 2021, 127939. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Zhao, Q. Direct fluorescence anisotropy approach for aflatoxin B1 detection and affinity binding study by using single tetramethylrhodamine labeled aptamer. Talanta 2018, 189, 442–450. [Google Scholar] [CrossRef]
- Yilmaz, I.; Kaya, E.; Sinirlioglu, Z.A.; Bayram, R.; Surmen, M.G.; Colakoglu, S. Clinical importance of toxin concentration in Amanita verna mushroom. Toxicon: Off. J. Int. Soc. Toxinol. 2014, 87, 68–75. [Google Scholar] [CrossRef]
- McKnight, T.A.; McKnight, K.B.; Skeels, M.C. Amatoxin and phallotoxin concentration in amanita bisporigera spores. Mycologia 2010, 102, 763–765. [Google Scholar] [CrossRef]
- Abuknesha, R.A.; Maragkou, A. A highly sensitive and specific enzyme immunoassay for detection of beta-amanitin in biological fluids. Anal. Bioanal. Chem. 2004, 379, 853–860. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Zhao, R.; Wang, L.; Feng, T.; Wei, D.J.M.A. An enzyme-linked immunosorbent assay and a gold-nanoparticle based immunochromatographic test for amatoxins using recombinant antibody. Microchim. Acta 2016, 183, 2211–2219. [Google Scholar] [CrossRef]
- Li, J.; Jing, L.; Song, Y.; Zhang, J.; Chen, Q.; Wang, B.; Xia, X.; Han, Q. Rapid Detection of Rongalite via a Sandwich Lateral Flow Strip Assay Using a Pair of Aptamers. Nanoscale Res. Lett. 2018, 13, 296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soheili, V.; Taghdisi, S.M.; Hassanzadeh Khayyat, M.; Fazly Bazzaz, B.S.; Ramezani, M.; Abnous, K.J.M.A. Colorimetric and ratiometric aggregation assay for streptomycin using gold nanoparticles and a new and highly specific aptamer. Microchimica Acta 2016, 183, 1687–1697. [Google Scholar] [CrossRef]
- Qiao, L.; Wang, H.; He, J.; Yang, S.; Chen, A. Truncated affinity-improved aptamers for 17β-estradiol determination by AuNPs-based colorimetric aptasensor. Food Chem. 2021, 340, 128181. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Lu, Q.; Chen, S.; Wang, F.; Hou, J.; Xu, Z.; Meng, C.; Hu, T.; Hou, Y. Selection and Identification of Novel Aptamers Specific for Clenbuterol Based on ssDNA Library Immobilized SELEX and Gold Nanoparticles Biosensor. Mol. (Basel Switz.) 2018, 23, 2337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fischer, E. Einfluss der Configuration auf die Wirkung der Enzyme. Berichte der Deutschen Chemischen Gesellschaft. 1894, 27, 2985–2993. [Google Scholar] [CrossRef] [Green Version]
- Morris, G.M.; Lim-Wilby, M. Molecular docking. Mol. Modeling Proteins 2008, 443, 365–382. [Google Scholar]
| Name | Sequence (5′-3′) | Kd (Nm) | dG |
|---|---|---|---|
| Apt [39] | CATGCTTCCCCAGGGAGATGGAGGTCTTTTTGGTTGTTGGTGGGGGAATCTTTTGGTATTGAGGAACATG | 33.6 ± 3.928 | −3.13 |
| Apt-2 | CCAGGGAGATGGAGGTCTTTTTGGTTGTTGGTGGGGGAATCTTTTGGTATTGAGGAACATG | 37.9 ± 6.053 | −1.16 |
| Apt-8 | CATGCTTCCCCAGGGAGATGGAGGTCTTTTTGGTTGTTGGTGGGGGAATCTTTTGGTA | 130.9 ± 31.1 | −3.13 |
| Apt-10 | CCCCAGGGAGATGGAGGTCTTTTTGGTTGTTGGTGGGGGAATCTTTTGGTATTGAGGAA | 108.7 ± 18.23 | −2.41 |
| Apt-11 | CTTCCCCAGGGAGATGGAGGTCTTTTTGGTTGTTGGTGGGGGAAT | 184.4 ± 37.08 | −3.13 |
| Apt-12 | GGGAGATGGAGGTCTT | 185.2 ± 60.87 | −0.25 |
| Aptamer | N (Sites) | Kd (μM) | ΔH (kcal/mol) | ΔG (kcal/mol) | −TΔS (kcal/mol) |
|---|---|---|---|---|---|
| Apt | 1.05 | 3.1 | −1.436 | −7.515 | −6.079 |
| Apt-2 | 1.88 | 46.9 | 16.21 | −5.36 | −2.157 |
| Apt-8 | 3.78 | 11.9 | 16.54 | −2.7 | −23.24 |
| Apt-10 | 2.52 | 68 | 18.15 | 5.69 | −23.84 |
| Apt-11 | 2.93 × 104 | 54.6 | 4.256 × 106 | −0.124 × 106 | 4.38 × 106 |
| Apt-12 | 0.485 | 84.7 | −5.932 | −6.921 | 0.989 |
| Samples | Added Concentration (ng/mL) | Kit Measured (ng/mL) | Recovery (%) | MELISA (ng/mL) | Recovery (%) |
|---|---|---|---|---|---|
| Mushroom | 10 | 8.413 | 84.13 | 8.951 | 89.51 |
| 50 | 47.14 | 94.28 | 42.85 | 85.7 | |
| 100 | 90.48 | 90.48 | 88.16 | 88.16 | |
| 200 | 191.3 | 95.65 | 194.8 | 97.4 | |
| Urine | 10 | 8.304 | 83.04 | 8.94 | 89.4 |
| 50 | 46.46 | 92.92 | 43.08 | 86.1 | |
| 100 | 90.21 | 90.21 | 90.16 | 90.16 | |
| 200 | 192.93 | 96.46 | 192.6 | 96.3 |
| Sample | Magnetic Bead-ELISA (n = 2) | HPLC (n = 2) |
|---|---|---|
| Normal urine sample1 | −− | −− |
| Normal urine sample2 | −− | −− |
| Mushroom sample1 | ++ | ++ |
| Mushroom sample2 | ++ | ++ |
| Patient urine sample1 | ++ | ++ |
| Patient urine sample2 | ++ | ++ |
| Patient urine sample3 | ++ | ++ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, J.; Liu, N.; Zhang, X.; Yang, E.; Song, Y.; Zhang, J.; Han, Q. Utilizing the DNA Aptamer to Determine Lethal α-Amanitin in Mushroom Samples and Urine by Magnetic Bead-ELISA (MELISA). Molecules 2022, 27, 538. https://doi.org/10.3390/molecules27020538
Gao J, Liu N, Zhang X, Yang E, Song Y, Zhang J, Han Q. Utilizing the DNA Aptamer to Determine Lethal α-Amanitin in Mushroom Samples and Urine by Magnetic Bead-ELISA (MELISA). Molecules. 2022; 27(2):538. https://doi.org/10.3390/molecules27020538
Chicago/Turabian StyleGao, Jiale, Nuoya Liu, Xiaomeng Zhang, En Yang, Yuzhu Song, Jinyang Zhang, and Qinqin Han. 2022. "Utilizing the DNA Aptamer to Determine Lethal α-Amanitin in Mushroom Samples and Urine by Magnetic Bead-ELISA (MELISA)" Molecules 27, no. 2: 538. https://doi.org/10.3390/molecules27020538
APA StyleGao, J., Liu, N., Zhang, X., Yang, E., Song, Y., Zhang, J., & Han, Q. (2022). Utilizing the DNA Aptamer to Determine Lethal α-Amanitin in Mushroom Samples and Urine by Magnetic Bead-ELISA (MELISA). Molecules, 27(2), 538. https://doi.org/10.3390/molecules27020538






