Melatonin Attenuates RANKL-Induced Osteoclastogenesis via Inhibition of Atp6v0d2 and DC-STAMP through MAPK and NFATc1 Signaling Pathways
Abstract
:1. Introduction
2. Results
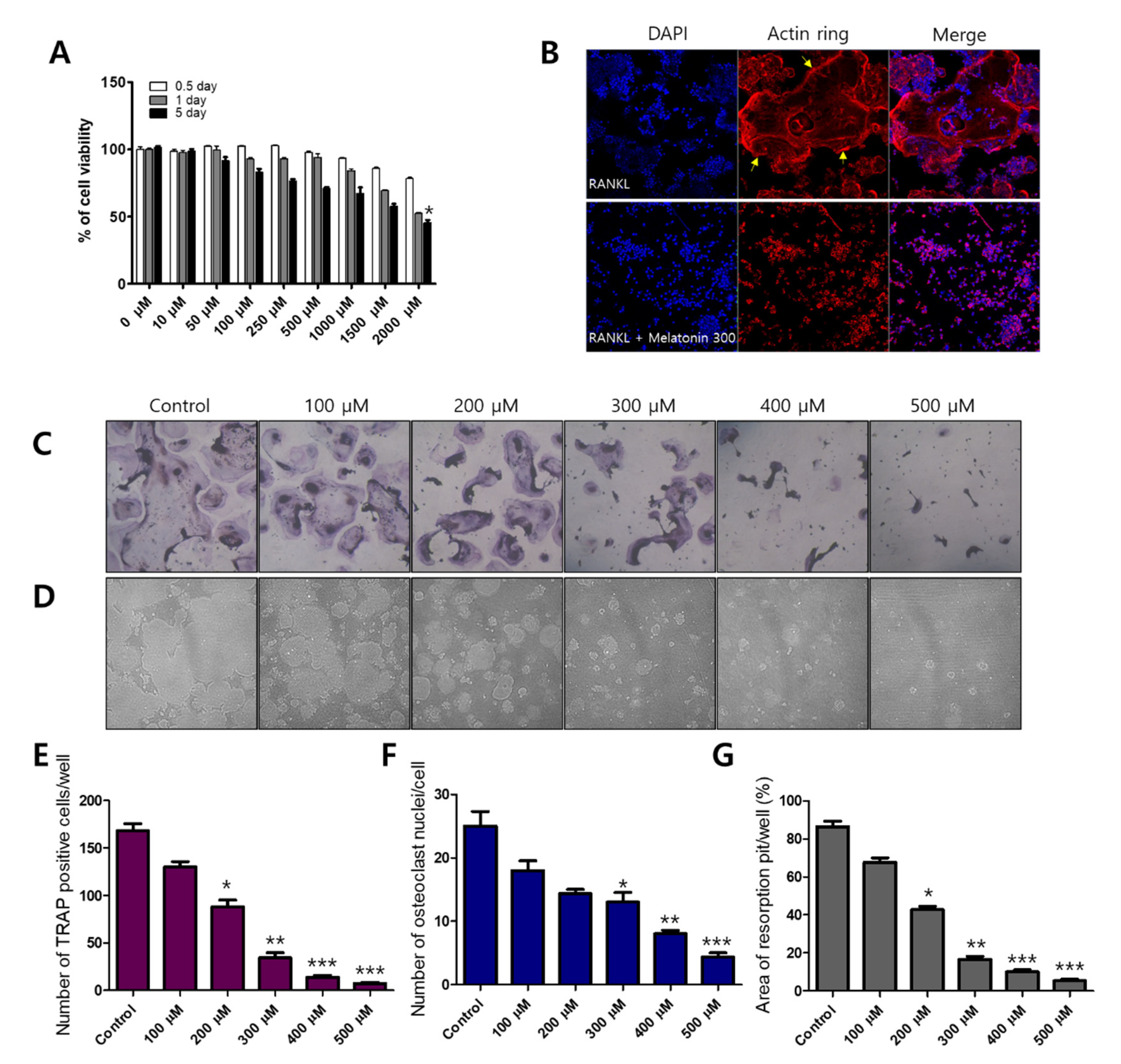
2.1. Melatonin Inhibits RANKL-Induced Osteoclast Formation and Resorption Pit Formation in RAW 264.7 Cells
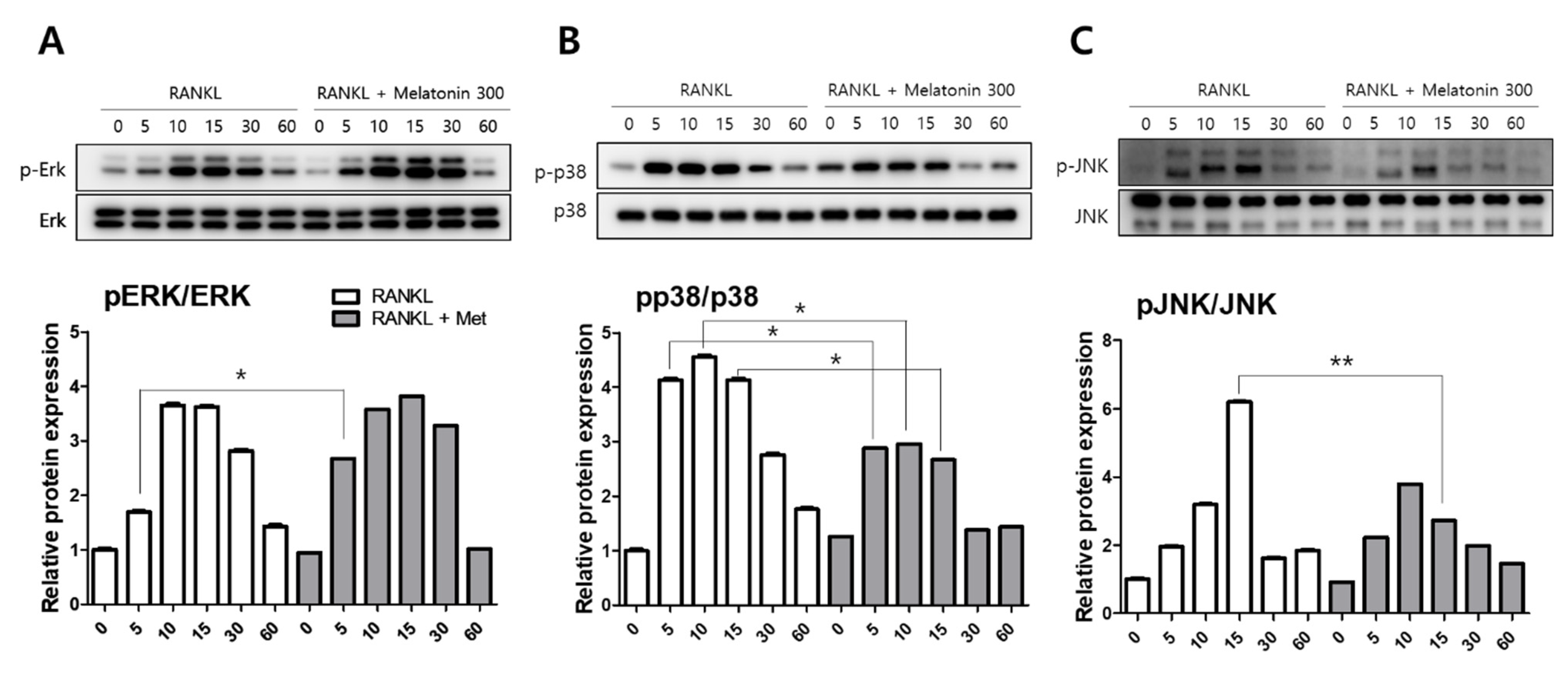
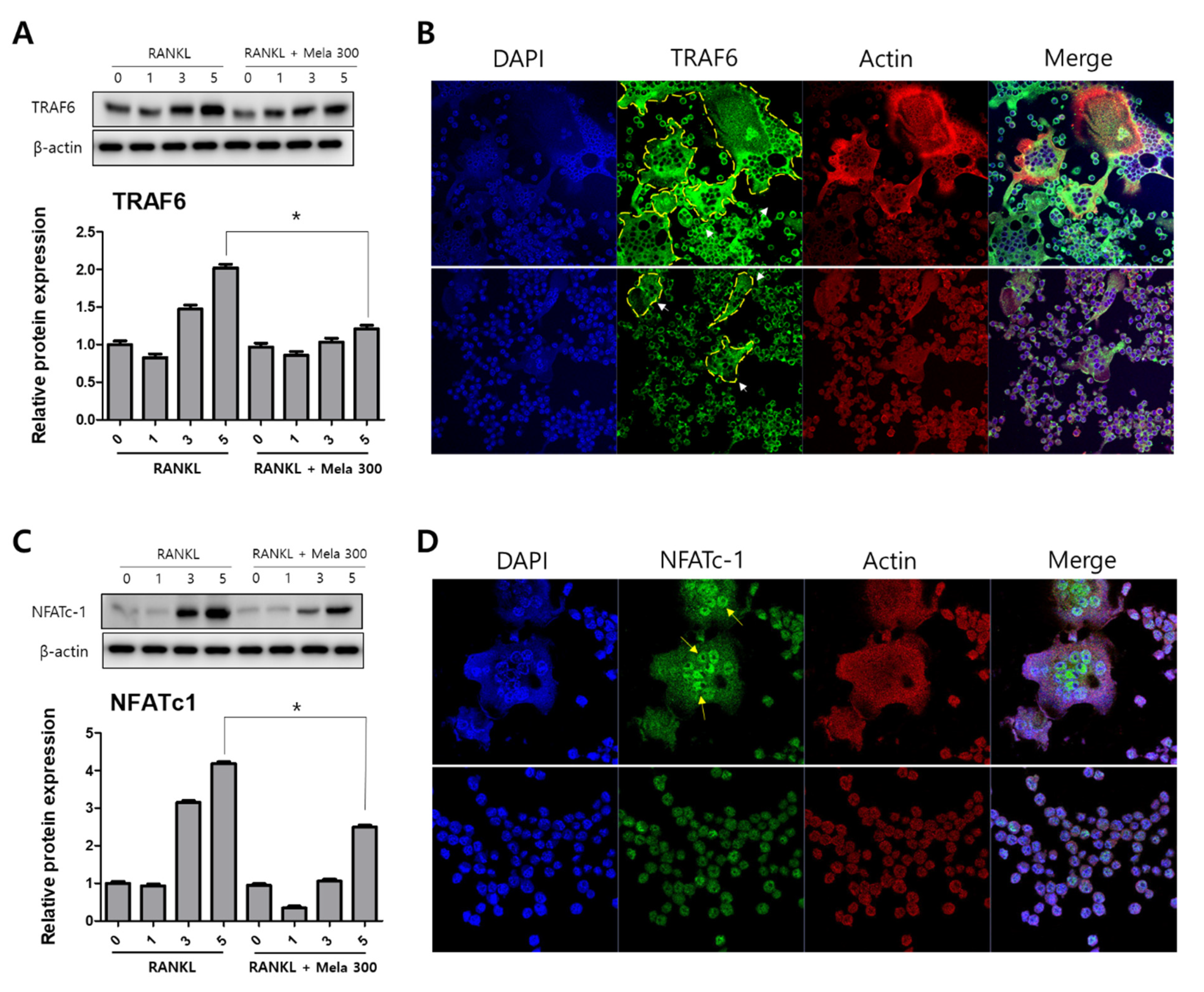
2.2. Melatonin Inhibits MAPK and NFATc1 Signaling Pathway on RANKL-Induced Osteoclastogenesis
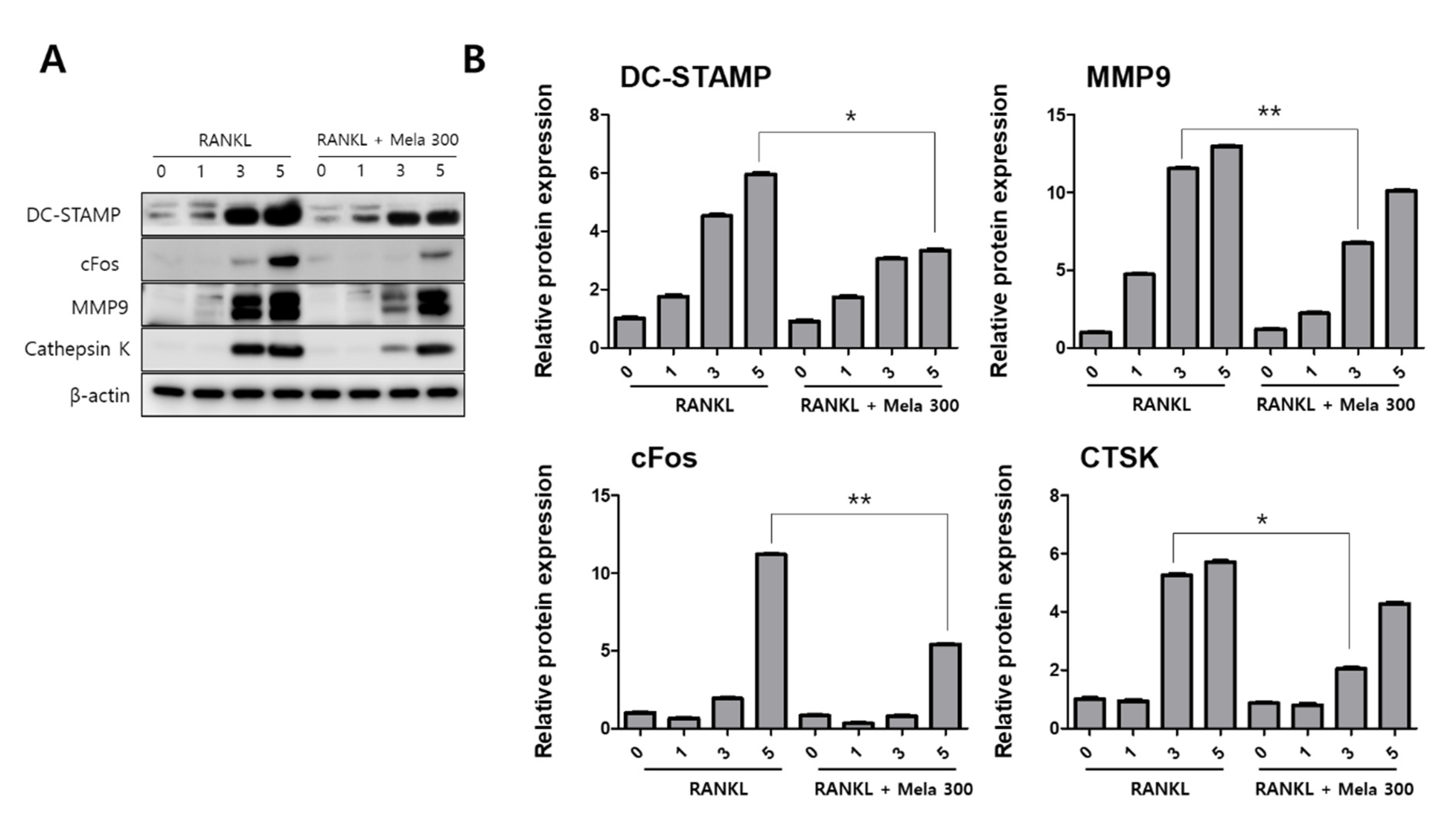
2.3. Melatonin Inhibits V-ATPase Vo Domain (Atp6v0d2) and DC-STAMP Expression
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Cell Viability Assay
4.3. Tartrate-Resistant Acid Phosphatase (TRAP) Staining
4.4. Pit Formation Assay
4.5. Western Blot Analysis
4.6. Immunofluorescence Analysis
4.7. ATPase Activity
4.8. Real-Time PCR
4.9. Small Interfering RNA (siRNA) Transfection
4.10. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Satué, M.; Ramis, J.M.; del Mar Arriero, M.; Monjo, M. A new role for 5-methoxytryptophol on bone cells function in vitro. J. Cell. Biochem. 2015, 116, 551–558. [Google Scholar] [CrossRef]
- Lerner, A.B.; Case, J.D.; Takahashi, Y.; Lee, T.H.; Mori, W. Isolation of melatonin, the pineal gland factor that lightens melanocyteS1. J. Am. Chem. Soc. 1958, 80, 2587. [Google Scholar] [CrossRef]
- Ghareghani, M.; Scavo, L.; Arnoult, D.; Zibara, K.; Farhadi, N. Melatonin therapy reduces the risk of osteoporosis and normalizes bone formation in multiple sclerosis. Fundam Clin Pharm. 2018, 32, 181–187. [Google Scholar] [CrossRef]
- Kotlarczyk, M.P.; Lassila, H.C.; O’Neil, C.K.; D’Amico, F.; Enderby, L.T.; Witt-Enderby, P.A.; Balk, J.L. Melatonin osteoporosis prevention study (MOPS): A randomized, double-blind, placebo-controlled study examining the effects of melatonin on bone health and quality of life in perimenopausal women. J. Pineal Res. 2012, 52, 414–426. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Barceló, E.J.; Mediavilla, M.; Tan, D.; Reiter, R. Scientific basis for the potential use of melatonin in bone diseases: Osteoporosis and adolescent idiopathic scoliosis. J. Osteoporos. 2010, 2010, 830231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, J.-H.; Jang, A.-R.; Park, M.-J.; Kim, D.-I.; Park, J.-H. Melatonin Inhibits Osteoclastogenesis and Bone Loss in Ovariectomized Mice by Regulating PRMT1-Mediated Signaling. Endocrinology 2021, 162, bqab057. [Google Scholar] [CrossRef]
- Maria, S.; Samsonraj, R.M.; Munmun, F.; Glas, J.; Silvestros, M.; Kotlarczyk, M.P.; Rylands, R.; Dudakovic, A.; van Wijnen, A.J.; Enderby, L.T.; et al. Biological effects of melatonin on osteoblast/osteoclast cocultures, bone, and quality of life: Implications of a role for MT2 melatonin receptors, MEK1/2, and MEK5 in melatonin-mediated osteoblastogenesis. J. Pineal. Res. 2018, 64. [Google Scholar] [CrossRef] [PubMed]
- Ping, Z.; Hu, X.; Wang, L.; Shi, J.; Tao, Y.; Wu, X.; Hou, Z.; Guo, X.; Zhang, W.; Yang, H.; et al. Melatonin attenuates titanium particle-induced osteolysis via activation of Wnt/beta-catenin signaling pathway. Acta Biomater. 2017, 51, 513–525. [Google Scholar] [CrossRef]
- Ping, Z.; Wang, Z.; Shi, J.; Wang, L.; Guo, X.; Zhou, W.; Hu, X.; Wu, X.; Liu, Y.; Zhang, W.; et al. Inhibitory effects of melatonin on titanium particle-induced inflammatory bone resorption and osteoclastogenesis via suppression of NF-kappaB signaling. Acta Biomater. 2017, 62, 362–371. [Google Scholar] [CrossRef]
- Maria, S.; Witt-Enderby, P.A. Melatonin effects on bone: Potential use for the prevention and treatment for osteopenia, osteoporosis, and periodontal disease and for use in bone-grafting procedures. J. Pineal. Res. 2014, 56, 115–125. [Google Scholar] [CrossRef]
- Hadjidakis, D.J.; Androulakis, I.I. Bone Remodeling. Ann. N. Y. Acad. Sci. 2006, 1092, 385–396. [Google Scholar] [CrossRef]
- Wu, Q.-Y.; Wang, J.; Tong, X.; Chen, J.; Wang, B.; Miao, Z.-N.; Li, X.; Ye, J.-X.; Yuan, F.-L. Emerging role of circadian rhythm in bone remodeling. J. Mol. Med. 2019, 97, 19–24. [Google Scholar] [CrossRef] [PubMed]
- Martin, T.J.; Sims, N.A. RANKL/OPG.; Critical role in bone physiology. Rev. Endocr. Metab. Disord. 2015, 16, 131–139. [Google Scholar] [CrossRef]
- Boyce, B.F.; Zuscik, M.J.; Xing, L. Biology of bone and cartilage. In Genetics of Bone Biology and Skeletal Disease; Elsevier: Berlin/Heidelberg, Germany, 2018; pp. 173–195. [Google Scholar]
- Canalis, E.; McCarthy, T.; Centrella, M. Growth factors and the regulation of bone remodeling. J. Clin. Investig. 1988, 81, 277–281. [Google Scholar] [CrossRef] [Green Version]
- Eriksen, E.F. Cellular mechanisms of bone remodeling. Rev. Endocr. Metab. Disord. 2010, 11, 219–227. [Google Scholar] [CrossRef] [Green Version]
- Raisz, L.G. Physiology and pathophysiology of bone remodeling. Clin. Chem. 1999, 45, 1353–1358. [Google Scholar]
- O’Brien, C.A.; Nakashima, T.; Takayanagi, H. Osteocyte control of osteoclastogenesis. Bone 2013, 54, 258–263. [Google Scholar] [CrossRef] [Green Version]
- Rody, W.J.; King, G.J.; Gu, G. Osteoclast recruitment to sites of compression in orthodontic tooth movement. Am. J. Orthod. Dentofac. Orthop. 2001, 120, 477–489. [Google Scholar] [CrossRef]
- Udagawa, N.; Takahashi, N.; Akatsu, T.; Tanaka, H.; Sasaki, T.; Nishihara, T.; Koga, T.; Martin, T.J.; Suda, T. Origin of osteoclasts: Mature monocytes and macrophages are capable of differentiating into osteoclasts under a suitable microenvironment prepared by bone marrow-derived stromal cells. Proc. Natl. Acad. Sci. USA 1990, 87, 7260–7264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boyle, W.J.; Simonet, W.S.; Lacey, D.L. Osteoclast differentiation and activation. Nature 2003, 423, 337–342. [Google Scholar] [CrossRef] [PubMed]
- Takayanagi, H. Osteoimmunology: Shared mechanisms and crosstalk between the immune and bone systems. Nat. Rev. Immunol. 2007, 7, 292–304. [Google Scholar] [CrossRef]
- Matsuo, K.; Galson, D.L.; Zhao, C.; Peng, L.; Laplace, C.; Wang, K.Z.; Bachler, M.A.; Amano, H.; Aburatani, H.; Ishikawa, H. Nuclear factor of activated T-cells (NFAT) rescues osteoclastogenesis in precursors lacking c-Fos. J. Biol. Chem. 2004, 279, 26475–26480. [Google Scholar] [CrossRef] [Green Version]
- Lacey, D.L.; Timms, E.; Tan, H.L.; Kelley, M.J.; Dunstan, C.R.; Burgess, T.; Elliott, R.; Colombero, A.; Elliott, G.; Scully, S.; et al. Osteoprotegerin Ligand Is a Cytokine that Regulates Osteoclast Differentiation and Activation. Cell 1998, 93, 165–176. [Google Scholar] [CrossRef] [Green Version]
- Xu, J.; Feng, H.T.; Wang, C.; Yip, K.H.; Pavlos, N.; Papadimitriou, J.M.; Wood, D.; Zheng, M.H. Effects of Bafilomycin A1: An inhibitor of vacuolar H (+)-ATPases on endocytosis and apoptosis in RAW cells and RAW cell-derived osteoclasts. J. Cell. Biochem. 2003, 88, 1256–1264. [Google Scholar] [CrossRef] [PubMed]
- Väänänen, H.K.; Karhukorpi, E.K.; Sundquist, K.; Wallmark, B.; Roininen, I.; Hentunen, T.; Tuukkanen, J.; Lakkakorpi, P. Evidence for the presence of a proton pump of the vacuolar H(+)-ATPase type in the ruffled borders of osteoclasts. J. Cell Biol. 1990, 111, 1305–1311. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Kim, H.J.; Bae, M.K.; Kim, Y.D. Suppression of Osteoclastogenesis by Melatonin: A Melatonin Receptor-Independent Action. Int. J. Mol. Sci. 2017, 18, 1142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, P.I.; Chang, A.C.; Lai, J.L.; Lin, T.H.; Tsai, C.H.; Chen, P.C.; Jiang, Y.J.; Lin, L.W.; Huang, W.C.; Yang, S.F.; et al. Melatonin interrupts osteoclast functioning and suppresses tumor-secreted RANKL expression: Implications for bone metastases. Oncogene 2021, 40, 1503–1515. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Rao, L.; Jia, H.; Chen, J.; Lu, X.; Yang, G.; Li, Q.; Lee, K.K.H.; Yang, L. Baicalin positively regulates osteoclast function by activating MAPK/Mitf signalling. J. Cell. Mol. Med. 2017, 21, 1361–1372. [Google Scholar] [CrossRef]
- Oh, J.H.; Lee, J.Y.; Park, J.H.; No, J.H.; Lee, N.K. Obatoclax regulates the proliferation and fusion of osteoclast precursors through the inhibition of ERK activation by RANKL. Mol. Cells 2015, 38, 279–284. [Google Scholar] [CrossRef] [Green Version]
- Wu, H.; Xu, G.; Li, Y.P. Atp6v0d2 is an essential component of the osteoclast-specific proton pump that mediates extracellular acidification in bone resorption. J. Bone Miner. Res. 2009, 24, 871–885. [Google Scholar] [CrossRef] [Green Version]
- Robling, A.G.; Castillo, A.B.; Turner, C.H. Biomechanical and molecular regulation of bone remodeling. Annu. Rev. Biomed. Eng. 2006, 8, 455–498. [Google Scholar] [CrossRef] [PubMed]
- Orwoll, E.; Scheele, W.; Paul, S.; Adami, S.; Syversen, U.; Diez-Perez, A.; Kaufman, J.-M.; Clancy, A.; Gaich, G. The Effect of Teriparatide [Human Parathyroid Hormone (1–34)] Therapy on Bone Density in Men with Osteoporosis. J. Bone Miner. Res. 2003, 18, 9–17. [Google Scholar] [CrossRef]
- Ladizesky, M.G.; Cutrera, R.A.; Boggio, V.; Somoza, J.; Centrella, J.M.; Mautalen, C.; Cardinali, D.P. Effect of melatonin on bone metabolism in ovariectomized rats. Life Sci. 2001, 70, 557–565. [Google Scholar] [CrossRef]
- Cardinali, D.P.; Ladizesky, M.G.; Boggio, V.; Cutrera, R.A.; Mautalen, C. Melatonin effects on bone: Experimental facts and clinical perspectives. J. Pineal Res. 2003, 34, 81–87. [Google Scholar] [CrossRef]
- Kim, S.-H.; Kim, M.O.; Kim, H.J.; Neupane, S.; Kim, H.J.; Lee, J.H.; Kim, H.-H.; Kim, J.-Y.; Lee, Y. Bortezomib prevents ovariectomy-induced osteoporosis in mice by inhibiting osteoclast differentiation. J. Bone Miner. Metab. 2018, 36, 537–546. [Google Scholar] [CrossRef]
- Roodman, G.D. Mechanisms of bone metastasis. N. Engl. J. Med. 2004, 350, 1655–1664. [Google Scholar] [CrossRef]
- Munmun, F.A. The Role of MEK1/2 and MEK5 in Melatonin-Mediated Actions on Osteoblasts and Osteoclasts Differentiation, Bone Formation, Bone Microarchitecture, and Bone Biomechanics. 2021. Available online: https://dsc.duq.edu/etd/2020/ (accessed on 20 November 2021).
- Saftig, P.; Hunziker, E.; Wehmeyer, O.; Jones, S.; Boyde, A.; Rommerskirch, W.; Moritz, J.D.; Schu, P.; von Figura, K. Impaired osteoclastic bone resorption leads to osteopetrosis in cathepsin-K-deficient mice. Proc. Natl. Acad. Sci. USA 1998, 95, 13453–13458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hayman, A.R. Tartrate-resistant acid phosphatase (TRAP) and the osteoclast/immune cell dichotomy. Autoimmunity 2008, 41, 218–223. [Google Scholar] [CrossRef]
- Boyce, B.F.; Xing, L. The RANKL/RANK/OPG pathway. Curr. Osteoporos. Rep. 2007, 5, 98–104. [Google Scholar] [CrossRef]
- Lee, K.; Seo, I.; Choi, M.H.; Jeong, D. Roles of Mitogen-Activated Protein Kinases in Osteoclast Biology. Int. J. Mol. Sci. 2018, 19, 3004. [Google Scholar] [CrossRef] [Green Version]
- Asagiri, M.; Sato, K.; Usami, T.; Ochi, S.; Nishina, H.; Yoshida, H.; Morita, I.; Wagner, E.F.; Mak, T.W.; Serfling, E.; et al. Autoamplification of NFATc1 expression determines its essential role in bone homeostasis. J. Exp. Med. 2005, 202, 1261–1269. [Google Scholar] [CrossRef] [Green Version]
- Yagi, M.; Miyamoto, T.; Sawatani, Y.; Iwamoto, K.; Hosogane, N.; Fujita, N.; Morita, K.; Ninomiya, K.; Suzuki, T.; Miyamoto, K.; et al. DC-STAMP is essential for cell–cell fusion in osteoclasts and foreign body giant cells. J. Exp. Med. 2005, 202, 345. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.-H.; Rho, J.; Jeong, D.; Sul, J.-Y.; Kim, T.; Kim, N.; Kang, J.-S.; Miyamoto, T.; Suda, T.; Lee, S.-K. v-ATPase V 0 subunit d2–deficient mice exhibit impaired osteoclast fusion and increased bone formation. Nat. Med. 2006, 12, 1403. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Wang, Z.; Duan, N.; Zhu, G.; Schwarz, E.M.; Xie, C. Osteoblast–osteoclast interactions. Connect. Tissue Res. 2018, 59, 99–107. [Google Scholar] [CrossRef] [PubMed]
- Bae, W.-J.; Park, J.S.; Kang, S.-K.; Kwon, I.-K.; Kim, E.-C. Effects of Melatonin and Its Underlying Mechanism on Ethanol-Stimulated Senescence and Osteoclastic Differentiation in Human Periodontal Ligament Cells and Cementoblasts. Int. J. Mol. Sci. 2018, 19, 1742. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, S.-S.; Jeong, S.-P.; Park, B.-S.; Kim, I.-R. Melatonin Attenuates RANKL-Induced Osteoclastogenesis via Inhibition of Atp6v0d2 and DC-STAMP through MAPK and NFATc1 Signaling Pathways. Molecules 2022, 27, 501. https://doi.org/10.3390/molecules27020501
Kim S-S, Jeong S-P, Park B-S, Kim I-R. Melatonin Attenuates RANKL-Induced Osteoclastogenesis via Inhibition of Atp6v0d2 and DC-STAMP through MAPK and NFATc1 Signaling Pathways. Molecules. 2022; 27(2):501. https://doi.org/10.3390/molecules27020501
Chicago/Turabian StyleKim, Seong-Sik, Soon-Pill Jeong, Bong-Soo Park, and In-Ryoung Kim. 2022. "Melatonin Attenuates RANKL-Induced Osteoclastogenesis via Inhibition of Atp6v0d2 and DC-STAMP through MAPK and NFATc1 Signaling Pathways" Molecules 27, no. 2: 501. https://doi.org/10.3390/molecules27020501
APA StyleKim, S.-S., Jeong, S.-P., Park, B.-S., & Kim, I.-R. (2022). Melatonin Attenuates RANKL-Induced Osteoclastogenesis via Inhibition of Atp6v0d2 and DC-STAMP through MAPK and NFATc1 Signaling Pathways. Molecules, 27(2), 501. https://doi.org/10.3390/molecules27020501





