Simple Enzyme Immobilization for Flow Chemistry? An Assessment of Available Strategies for an Acetaldehyde-Dependent Aldolase
Abstract
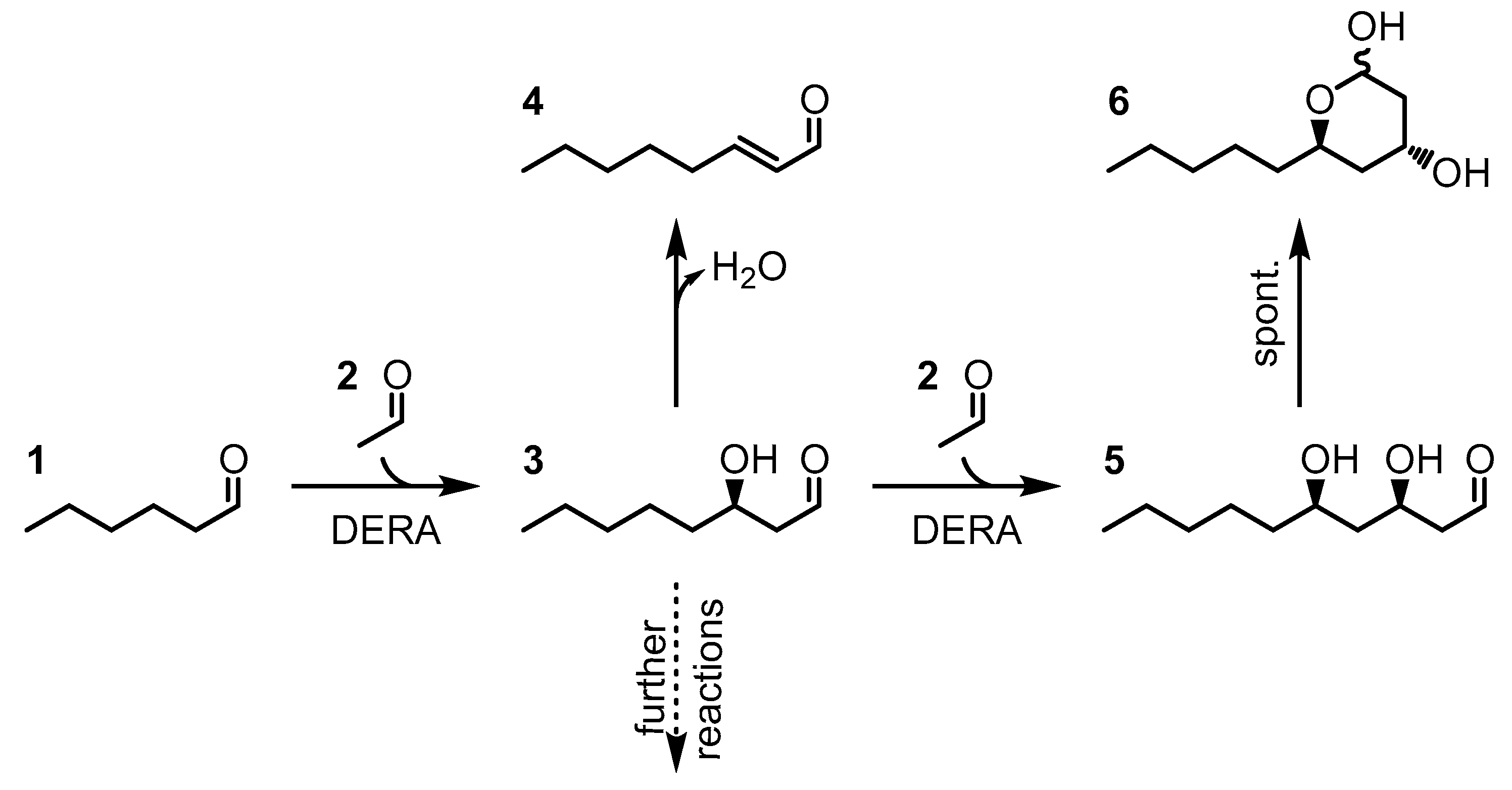
1. Introduction
2. Results
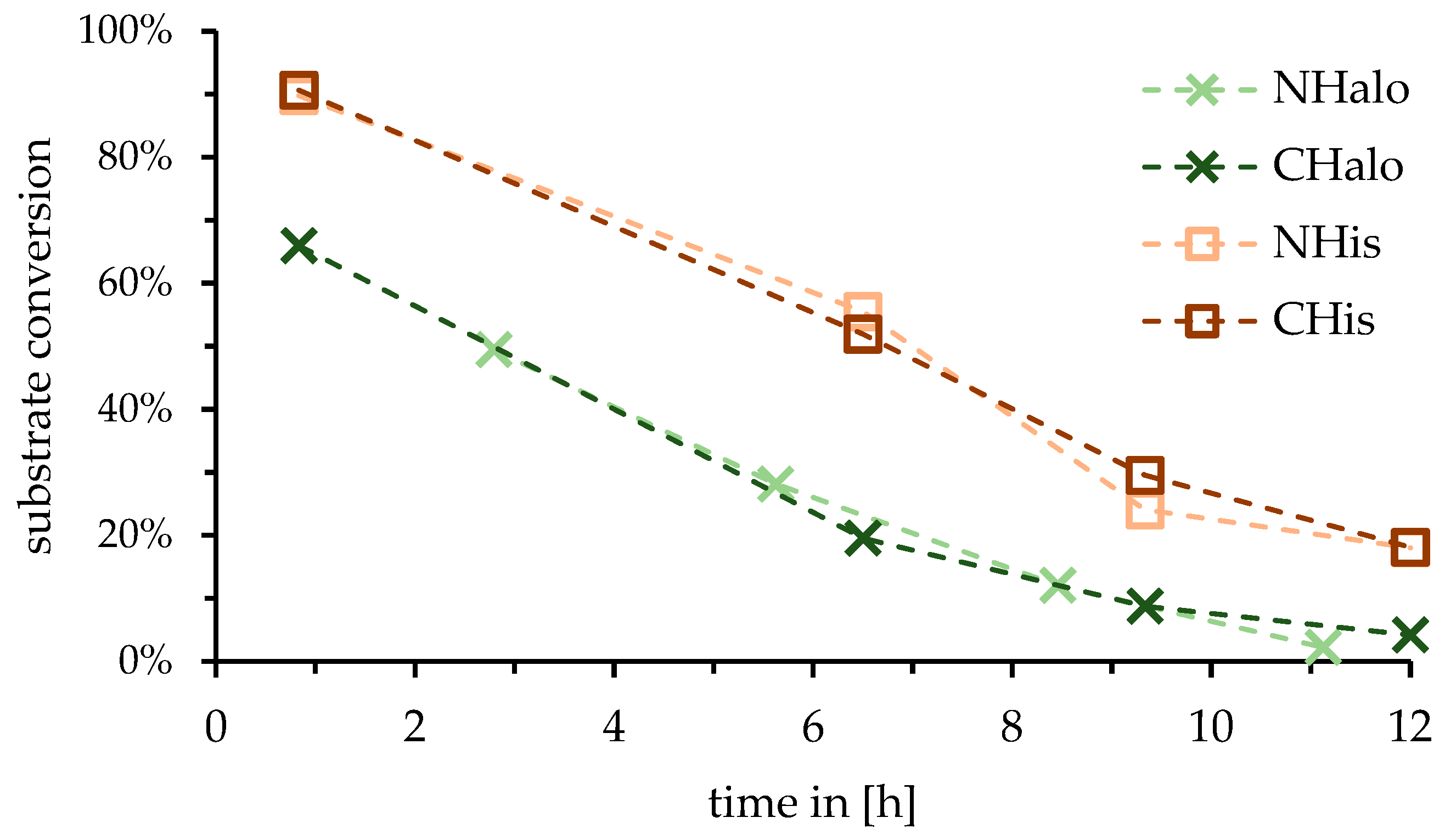
2.1. Influence of Tag Position of Enzyme for Reactor
2.2. Initial Performance and Stability of Reactors
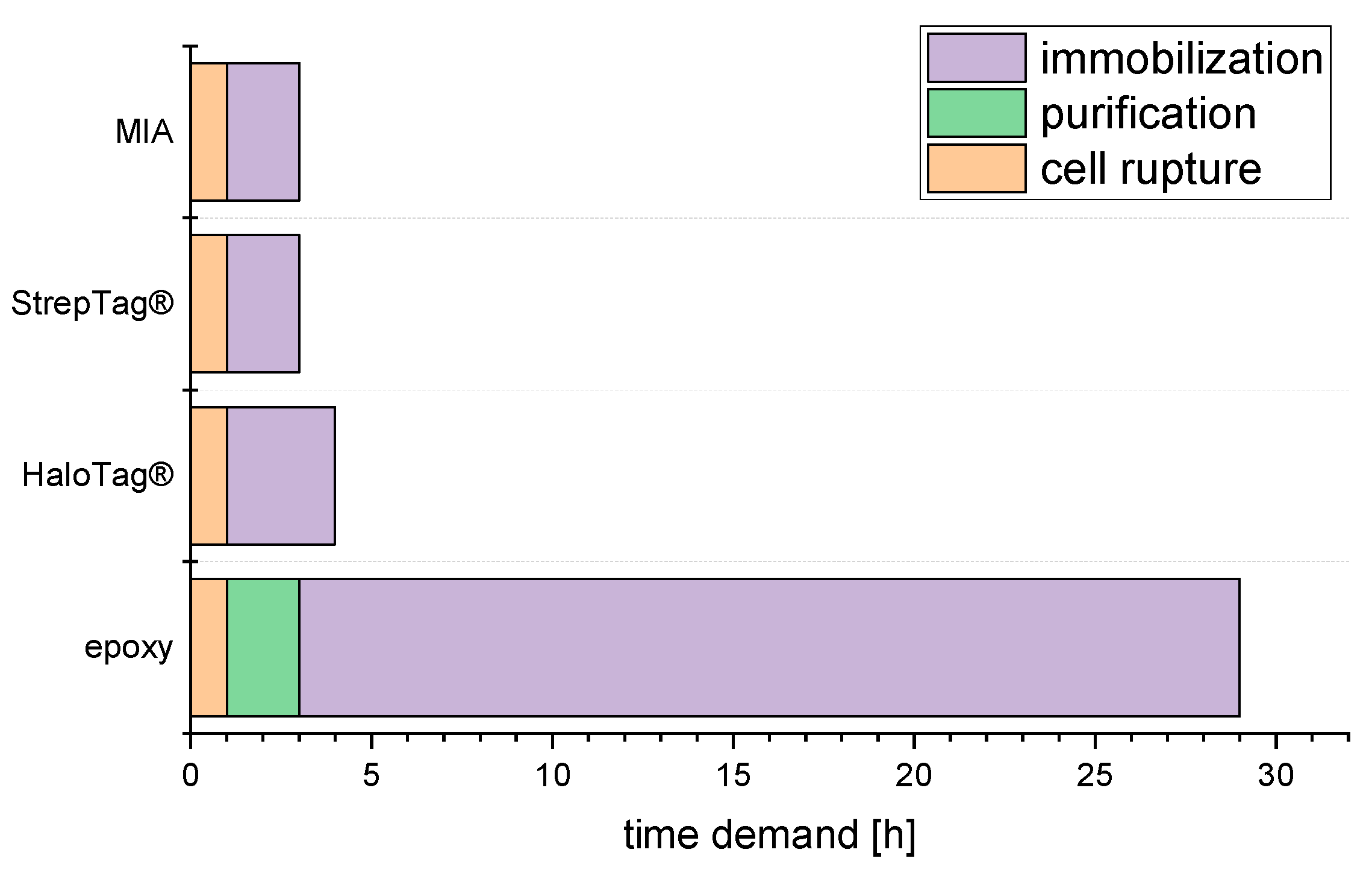
2.3. Time Demand from Harvested Culture to Ready Reactor
3. Discussion
3.1. Influence of Tag Position of Enzyme for Reactor
3.2. Initial Performance and Stability of Reactors
3.3. Time demand from Harvested Culture to Ready Reactor
4. Materials and Methods
4.1. Chemicals
4.2. Software
4.3. Methods
4.3.1. Cloning of DERA Mutants
4.3.2. Protein Production
4.3.3. Immobilization of DERA Variants
4.3.4. DERA Activity Assay
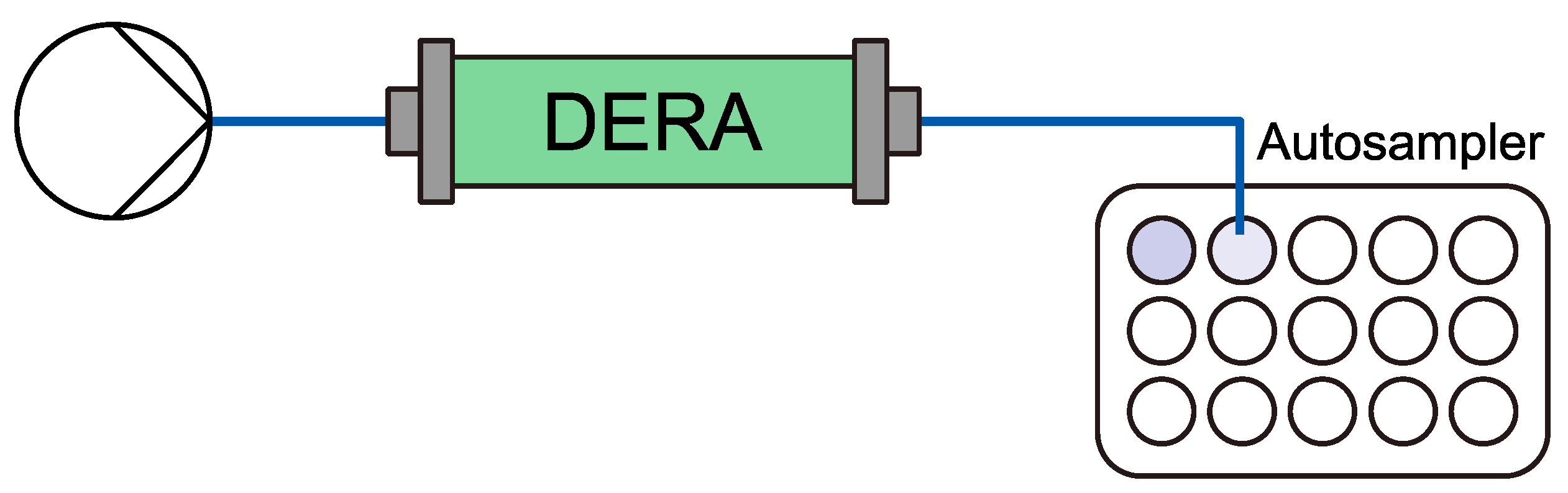
4.3.5. Initial performance Flow Experiment Setup
4.3.6. Tag Position Dependent Performance
4.3.7. Data Evaluation
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Plutschack, M.B.; Pieber, B.; Gilmore, K.; Seeberger, P.H. The Hitchhiker’s Guide to Flow Chemistry. Chem. Rev. 2017, 117, 11796–11893. [Google Scholar] [CrossRef]
- Sheldon, R.A.; Brady, D.; Bode, M.L. The Hitchhiker’s guide to biocatalysis: Recent advances in the use of enzymes in organic synthesis. Chem. Sci. 2020, 11, 2587–2605. [Google Scholar] [CrossRef] [PubMed]
- Sheldon, R.A. Enzyme Immobilization: The Quest for Optimum Performance. Adv. Synth. Catal. 2007, 349, 1289–1307. [Google Scholar] [CrossRef]
- Homaei, A.A.; Sariri, R.; Vianello, F.; Stevanato, R. Enzyme immobilization: An update. J. Chem. Biol. 2013, 6, 185–205. [Google Scholar] [CrossRef] [PubMed]
- Sheldon, R.A.; van Pelt, S. Enzyme immobilisation in biocatalysis: Why, what and how. Chem. Soc. Rev. 2013, 42, 6223–6235. [Google Scholar] [CrossRef] [PubMed]
- Racker, E. Enzymatic Synthesis and Breakdown of Desoxyribose Phosphate. J. Biol. Chem. 1952, 196, 347–365. [Google Scholar] [CrossRef]
- Tozzi, M.G.; Camici, M.; Mascia, L.; Sgarrella, F.; Ipata, P.L. Pentose phosphates in nucleoside interconversion and catabolism. FEBS J. 2006, 273, 1089–1101. [Google Scholar] [CrossRef]
- Gijsen, H.J.M.; Wong, C.-H. Unprecedented Asymmetric Aldol Reactions with Three Aldehyde Substrates Catalyzed by 2-Deoxyribose-5-phosphate Aldolase. J. Am. Chem. Soc. 1994, 116, 8422–8423. [Google Scholar] [CrossRef]
- Hindges, J.; Döbber, J.; Hayes, M.R.; Classen, T.; Pohl, M.; Pietruszka, J. Covalently Immobilized 2-Deoxyribose-5-phosphate Aldolase (DERA) for Biocatalysis in Flow: Utilization of the 3-Hydroxyaldehyde Intermediate in Reaction Cascades. ChemCatChem 2022, 14, e202200390. [Google Scholar] [CrossRef]
- Bramski, J.; Dick, M.; Pietruszka, J.; Classen, T. Probing the acetaldehyde-sensitivity of 2-deoxy-ribose-5-phosphate aldolase (DERA) leads to resistant variants. J. Biotechnol. 2017, 258, 56–58. [Google Scholar] [CrossRef]
- Dick, M.; Weiergräber, O.H.; Classen, T.; Bisterfeld, C.; Bramski, J.; Gohlke, H.; Pietruszka, J. Trading off stability against activity in extremophilic aldolases. Sci. Rep. 2016, 6, 17908. [Google Scholar] [CrossRef] [PubMed]
- Nara, T.Y.; Togashi, H.; Ono, S.; Egami, M.; Sekikawa, C.; Suzuki, Y.-h.; Masuda, I.; Ogawa, J.; Horinouchi, N.; Shimizu, S.; et al. Improvement of aldehyde tolerance and sequential aldol condensation activity of deoxyriboaldolase via immobilization on interparticle pore type mesoporous silica. J. Mol. Catal. B Enzym. 2011, 68, 181–186. [Google Scholar] [CrossRef]
- Wang, A.; Wang, M.; Wang, Q.; Chen, F.; Zhang, F.; Li, H.; Zeng, Z.; Xie, T. Stable and efficient immobilization technique of aldolase under consecutive microwave irradiation at low temperature. Bioresour. Technol. 2011, 102, 469–474. [Google Scholar] [CrossRef]
- Wang, A.; Gao, W.; Zhang, F.; Chen, F.; Du, F.; Yin, X. Amino acid-mediated aldolase immobilisation for enhanced catalysis and thermostability. Bioprocess Biosyst. Eng. 2012, 35, 857–863. [Google Scholar] [CrossRef] [PubMed]
- Subrizi, F.; Crucianelli, M.; Grossi, V.; Passacantando, M.; Botta, G.; Antiochia, R.; Saladino, R. Versatile and Efficient Immobilization of 2-Deoxyribose-5-phosphate Aldolase (DERA) on Multiwalled Carbon Nanotubes. ACS Catal. 2014, 4, 3059–3068. [Google Scholar] [CrossRef]
- Reinicke, S.; Rees, H.C.; Espeel, P.; Vanparijs, N.; Bisterfeld, C.; Dick, M.; Rosencrantz, R.R.; Brezesinski, G.; de Geest, B.G.; Du Prez, F.E.; et al. Immobilization of 2-Deoxy-d-ribose-5-phosphate Aldolase in Polymeric Thin Films via the Langmuir–Schaefer Technique. ACS Appl. Mater. Interfaces 2017, 9, 8317–8326. [Google Scholar] [CrossRef]
- Zhang, S.; Bisterfeld, C.; Bramski, J.; Vanparijs, N.; De Geest, B.G.; Pietruszka, J.; Böker, A.; Reinicke, S. Biocatalytically Active Thin Films via Self-Assembly of 2-Deoxy-d-ribose-5-phosphate Aldolase–Poly(N-isopropylacrylamide) Conjugates. Bioconjug. Chem. 2018, 29, 104–116. [Google Scholar] [CrossRef]
- Zhang, S.; Bramski, J.; Tutus, M.; Pietruszka, J.; Böker, A.; Reinicke, S. A Biocatalytically Active Membrane Obtained from Immobilization of 2-Deoxy-d-ribose-5-phosphate Aldolase on a Porous Support. ACS Appl. Mater. Interfaces 2019, 11, 34441–34453. [Google Scholar] [CrossRef]
- Grabner, B.; Pokhilchuk, Y.; Gruber-Woelfler, H. DERA in Flow: Synthesis of a Statin Side Chain Precursor in Continuous Flow Employing Deoxyribose-5-Phosphate Aldolase Immobilized in Alginate-Luffa Matrix. Catalysts 2020, 10, 137. [Google Scholar] [CrossRef]
- Fei, H.; Xu, G.; Wu, J.-P.; Yang, L.-R. Improvement of the thermal stability and aldehyde tolerance of deoxyriboaldolase via immobilization on nano-magnet material. J. Mol. Catal. B Enzym. 2014, 101, 87–91. [Google Scholar] [CrossRef]
- Reinicke, S.; Fischer, T.; Bramski, J.; Pietruszka, J.; Böker, A. Biocatalytically active microgels by precipitation polymerization of N-isopropyl acrylamide in the presence of an enzyme. RSC Adv. 2019, 9, 28377–28386. [Google Scholar] [CrossRef] [PubMed]
- Los, G.V.; Encell, L.P.; McDougall, M.G.; Hartzell, D.D.; Karassina, N.; Zimprich, C.; Wood, M.G.; Learish, R.; Ohana, R.F.; Urh, M.; et al. HaloTag: A Novel Protein Labeling Technology for Cell Imaging and Protein Analysis. ACS Chem. Biol. 2008, 3, 373–382. [Google Scholar] [CrossRef]
- Encell, L.P.; Friedman Ohana, R.; Zimmerman, K.; Otto, P.; Vidugiris, G.; Wood, M.G.; Los, G.V.; McDougall, M.G.; Zimprich, C.; Karassina, N.; et al. Development of a dehalogenase-based protein fusion tag capable of rapid, selective and covalent attachment to customizable ligands. Curr. Chem. Genom. 2012, 6, 55–71. [Google Scholar] [CrossRef] [PubMed]
- Döbber, J.; Pohl, M. HaloTag™: Evaluation of a covalent one-step immobilization for biocatalysis. J. Biotechnol. 2017, 241, 170–174. [Google Scholar] [CrossRef] [PubMed]
- Porath, J.; Carlsson, J.A.N.; Olsson, I.; Belfrage, G. Metal chelate affinity chromatography, a new approach to protein fractionation. Nature 1975, 258, 598–599. [Google Scholar] [CrossRef]
- Hochuli, E.; Bannwarth, W.; Döbeli, H.; Gentz, R.; Stüber, D. Genetic Approach to Facilitate Purification of Recombinant Proteins with a Novel Metal Chelate Adsorbent. Bio/Technology 1988, 6, 1321–1325. [Google Scholar] [CrossRef]
- Schmidt, T.G.M.; Koepke, J.; Frank, R.; Skerra, A. Molecular Interaction Between the Strep-tag Affinity Peptide and its Cognate Target, Streptavidin. J. Mol. Biol. 1996, 255, 753–766. [Google Scholar] [CrossRef]
- Skerra, A.; Schmidt, T.G.M. Applications of a peptide ligand for streptavidin: The Strep-tag. Biomol. Eng. 1999, 16, 79–86. [Google Scholar] [CrossRef]
- Gennari, A.; Mobayed, F.H.; da Silva Rafael, R.; Rodrigues, R.C.; Sperotto, R.A.; Volpato, G.; Volken de Souza, C.F. Modification of Immobead 150 support for protein immobilization: Effects on the properties of immobilized Aspergillus oryzae β-galactosidase. Biotechnol. Prog. 2018, 34, 934–943. [Google Scholar] [CrossRef]
- Döbber, J.; Pohl, M.; Ley, S.V.; Musio, B. Rapid, selective and stable HaloTag-LbADH immobilization directly from crude cell extract for the continuous biocatalytic production of chiral alcohols and epoxides. React. Chem Eng. 2018, 3, 8–12. [Google Scholar] [CrossRef]
- Schulte, M.; Petrović, D.; Neudecker, P.; Hartmann, R.; Pietruszka, J.; Willbold, S.; Willbold, D.; Panwalkar, V. Conformational Sampling of the Intrinsically Disordered C-Terminal Tail of DERA Is Important for Enzyme Catalysis. ACS Catal. 2018, 8, 3971–3984. [Google Scholar] [CrossRef]
- O’Keefe, S.F.; Wilson, L.A.; Resurreccion, A.P.; Murphy, P.A. Determination of the binding of hexanal to soy glycinin and. beta.-conglycinin in an aqueous model system using a headspace technique. J. Agric. Food Chem. 1991, 39, 1022–1028. [Google Scholar] [CrossRef]
- Wang, K.; Arntfield, S.D. Binding of selected volatile flavour mixture to salt-extracted canola and pea proteins and effect of heat treatment on flavour binding. Food Hydrocoll. 2015, 43, 410–417. [Google Scholar] [CrossRef]
- Daubert, T.E.; Danner, R.P. Physical and Thermodynamic Properties of Pure Chemicals: Data Compilation; Hemisphere Publish Corp.: New York, NY, USA, 1989. [Google Scholar]
- Horton, R.M.; Hunt, H.D.; Ho, S.N.; Pullen, J.K.; Pease, L.R. Engineering hybrid genes without the use of restriction enzymes: Gene splicing by overlap extension. Gene 1989, 77, 61–68. [Google Scholar] [CrossRef]
- Gibson, D.G.; Young, L.; Chuang, R.-Y.; Venter, J.C.; Hutchison, C.A.; Smith, H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Meth. 2009, 6, 343–345. [Google Scholar] [CrossRef]
- Bertani, G. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 1951, 62, 293–300. [Google Scholar] [CrossRef]
- Tartoff, K.D.; Hobbs, C.A. Improved Media for Growing Plasmid and Cosmid Clones. Bethesda Res. Lab. Focus 1987, 9, 12. [Google Scholar]
- Racker, E. Spectrophotometric measurement of hexokinase and phosphohexokinase activity. J. biol. Chem 1947, 167, 843–853. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Bisterfeld, C. Rationales Design der Acetaldehyd-abhängigen Aldolasen und deren Anwendung in der organischen Synthese. Inaugural-Dissertation, Heinrich-Heine-Universität Düsseldorf, Düsseldorf, Germany, 2016. [Google Scholar]
- de Saint Laumer, J.-Y.; Leocata, S.; Tissot, E.; Baroux, L.; Kampf, D.M.; Merle, P.; Boschung, A.; Seyfried, M.; Chaintreau, A. Prediction of response factors for gas chromatography with flame ionization detection: Algorithm improvement, extension to silylated compounds, and application to the quantification of metabolites. J. Sep. Sci. 2015, 38, 3209–3217. [Google Scholar] [CrossRef]
- Švarc, A.; Findrik Blažević, Z.; Vasić-Rački, Đ.; Charnock, S.J.; Vrsalović Presečki, A. A multi-enzyme strategy for the production of a highly valuable lactonized statin side-chain precursor. Chem. Eng. Res. Des. 2020, 164, 35–45. [Google Scholar] [CrossRef]
- Schägger, H.; von Jagow, G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 1987, 166, 368–379. [Google Scholar] [CrossRef]
- Kang, D.; Gho, Y.S.; Suh, M.-K.; Kang, C. Highly Sensitive and Fast Protein Detection with Coomassie Brilliant Blue in Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis. Bull. Korean Chem. Soc. 2002, 23, 1511–1512. [Google Scholar] [CrossRef]
- Details of R square. Available online: https://www.originlab.com/doc/en/Origin-Help/Details_of_R_square (accessed on 14 July 2022).
- Entry P0A6L0 · DEOC_ECOLI. Available online: https://www.uniprot.org/unipro]tkb/P0A6L0 (accessed on 20 July 2022).

| Peak Yield | Half-Life | Exp. Yield | Av. Yield | |
|---|---|---|---|---|
| HPY/t | τ1/2 | HPY | HPY/t | |
| µmol/min | min | µmol | µmol/min | |
| DERA-CHalo-Link | 2.92 | 30 | 61.5 | 2.07 |
| DERA-CStrep-Tactin | 1.18 | 7 | 5.7 | 0.85 |
| DERA-CHis-Ni-S6FF | 4.48 | 339 | 1050 | 3.09 |
| DERA-CHisNu-Ni-S6FF | 3.36 | 538 | 1300 | 2.43 |
| DERA-CHis-Immobead | 0.28 | 80 | 15.9 | 0.20 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wäscher, M.; Classen, T.; Pietruszka, J. Simple Enzyme Immobilization for Flow Chemistry? An Assessment of Available Strategies for an Acetaldehyde-Dependent Aldolase. Molecules 2022, 27, 6483. https://doi.org/10.3390/molecules27196483
Wäscher M, Classen T, Pietruszka J. Simple Enzyme Immobilization for Flow Chemistry? An Assessment of Available Strategies for an Acetaldehyde-Dependent Aldolase. Molecules. 2022; 27(19):6483. https://doi.org/10.3390/molecules27196483
Chicago/Turabian StyleWäscher, Martin, Thomas Classen, and Jörg Pietruszka. 2022. "Simple Enzyme Immobilization for Flow Chemistry? An Assessment of Available Strategies for an Acetaldehyde-Dependent Aldolase" Molecules 27, no. 19: 6483. https://doi.org/10.3390/molecules27196483
APA StyleWäscher, M., Classen, T., & Pietruszka, J. (2022). Simple Enzyme Immobilization for Flow Chemistry? An Assessment of Available Strategies for an Acetaldehyde-Dependent Aldolase. Molecules, 27(19), 6483. https://doi.org/10.3390/molecules27196483






