Kribbellichelins A and B, Two New Antibiotics from Kribbella sp. CA-293567 with Activity against Several Human Pathogens
Abstract
1. Introduction
2. Results and Discussion
2.1. Microbial Isolation and Identification of Actinobacterial Strains
2.2. Actinobacterial Production Screening: Small-Scale Fermentations
2.3. Isolation and Structural Elucidation of Compounds 1–5
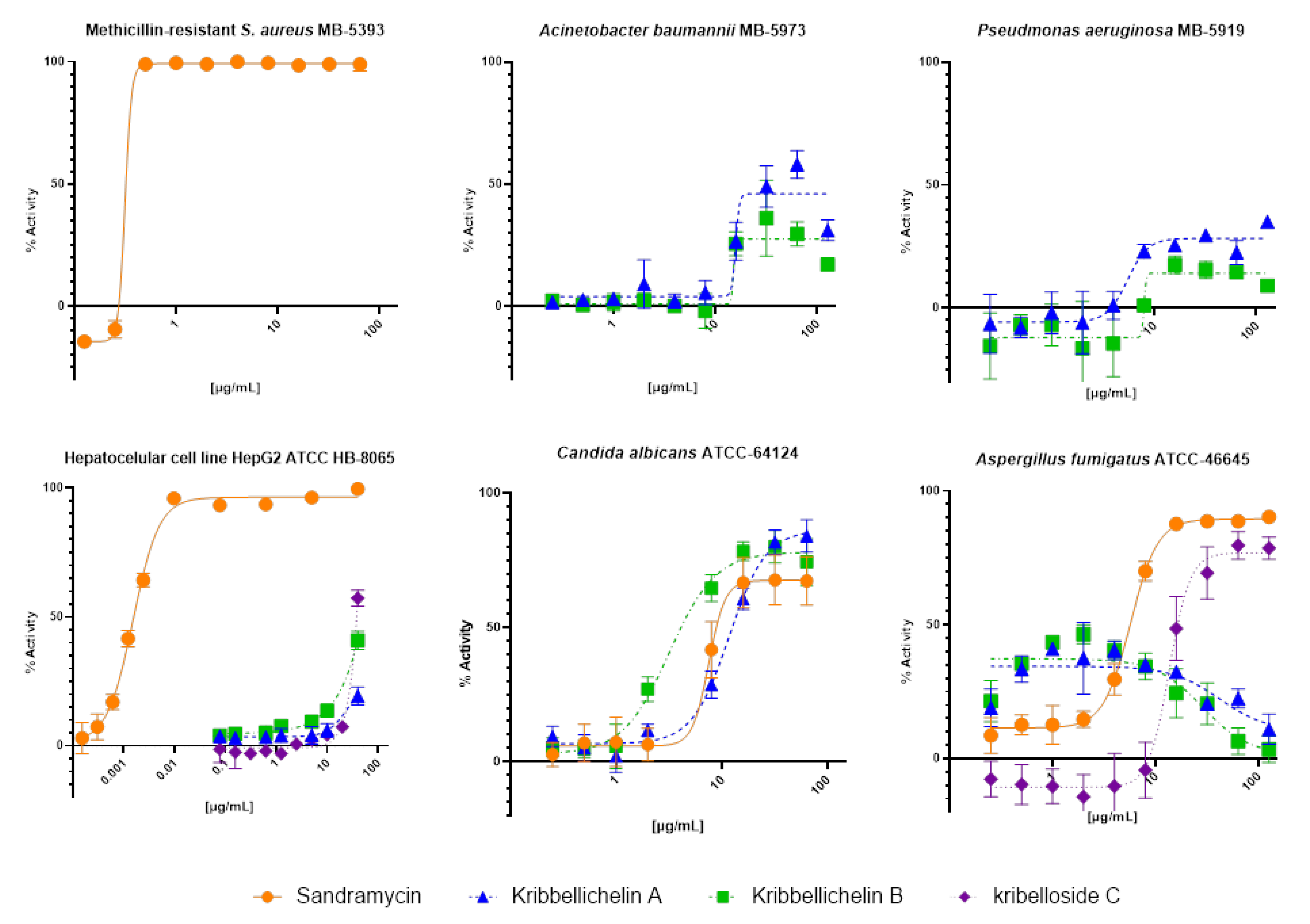
2.4. Biological Activity
3. Materials and Methods
3.1. Microbial Isolation and Identification of Actinobacterial Strains
3.2. General Experimental Procedures
3.3. Small Fermentations and Extraction
3.4. Large-Scale Fermentation, Extraction, and Isolation
3.5. Marfey’s Analysis of Compound 1
3.6. Antimicrobial Assays
3.7. Cytotoxicity Bioassays
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Demain, A.L.; Sanchez, S. Microbial Drug Discovery: 80 Years of Progress. J. Antibiot. 2009, 62, 5–16. [Google Scholar] [CrossRef] [PubMed]
- Murray, C.J.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Robles Aguilar, G.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global Burden of Bacterial Antimicrobial Resistance in 2019: A Systematic Analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- Genilloud, O.; González, I.; Salazar, O.; Martín, J.; Tormo, J.R.; Vicente, F. Current Approaches to Exploit Actinomycetes as a Source of Novel Natural Products. J. Ind. Microbiol. Biotechnol. 2011, 38, 375–389. [Google Scholar] [CrossRef] [PubMed]
- Bode, H.B.; Bethe, B.; Höfs, R.; Zeeck, A. Big Effects from Small Changes: Possible Ways to Explore Nature’s Chemical Diversity. ChemBioChem 2002, 3, 619. [Google Scholar] [CrossRef]
- Chassagne, F.; Cabanac, G.; Hubert, G.; David, B.; Marti, G. The Landscape of Natural Product Diversity and Their Pharmacological Relevance from a Focus on the Dictionary of Natural Products®. Phytochem. Rev. 2019, 18, 601–622. [Google Scholar] [CrossRef]
- Singh, T.A.; Passari, A.K.; Jajoo, A.; Bhasin, S.; Gupta, V.K.; Hashem, A.; Alqarawi, A.A.; Abd Allah, E.F. Tapping Into Actinobacterial Genomes for Natural Product Discovery. Front. Microbiol. 2021, 12, 1662. [Google Scholar] [CrossRef] [PubMed]
- Barka, E.A.; Vatsa, P.; Sanchez, L.; Gaveau-Vaillant, N.; Jacquard, C.; Klenk, H.-P.; Clément, C.; Ouhdouch, Y.; van Wezel, G.P. Taxonomy, Physiology, and Natural Products of Actinobacteria. Microbiol. Mol. Biol. Rev. 2016, 80, 1–43. [Google Scholar] [CrossRef]
- Scherlach, K.; Hertweck, C. Triggering Cryptic Natural Product Biosynthesis in Microorganisms. Org. Biomol. Chem. 2009, 7, 1753. [Google Scholar] [CrossRef]
- Gaiero, J.R.; McCall, C.A.; Thompson, K.A.; Day, N.J.; Best, A.S.; Dunfield, K.E. Inside the Root Microbiome: Bacterial Root Endophytes and Plant Growth Promotion. Am. J. Bot. 2013, 100, 1738–1750. [Google Scholar] [CrossRef]
- Hernández-Bolaños, E.; Montesdeoca-Flores, D.; Abreu-Yanes, E.; Barrios, M.L.; Abreu-Acosta, N. Evaluating Different Methodologies for Bioprospecting Actinomycetes in Canary Islands Soils. Curr. Microbiol. 2020, 77, 2510–2522. [Google Scholar] [CrossRef]
- Sayed, A.M.; Hassan, M.H.A.; Alhadrami, H.A.; Hassan, H.M.; Goodfellow, M.; Rateb, M.E. Extreme Environments: Microbiology Leading to Specialized Metabolites. J. Appl. Microbiol. 2020, 128, 630–657. [Google Scholar] [CrossRef]
- Hui, M.L.-Y.; Tan, L.T.-H.; Letchumanan, V.; He, Y.-W.; Fang, C.-M.; Chan, K.-G.; Law, J.W.-F.; Lee, L.-H. The Extremophilic Actinobacteria: From Microbes to Medicine. Antibiotics 2021, 10, 682. [Google Scholar] [CrossRef]
- Lloyd, A.B. Behaviour of Streptomycetes in Soil. J. Gen. Microbiol. 1969, 56, 165–170. [Google Scholar] [CrossRef]
- Kumar, A.S.; Bais, H.P. Wired to the Roots. Plant Signal. Behav. 2012, 7, 1598–1604. [Google Scholar] [CrossRef]
- Yang, C.-H.; Crowley, D.E.; Borneman, J.; Keen, N.T. Microbial Phyllosphere Populations Are More Complex than Previously Realized. Proc. Natl. Acad. Sci. USA 2001, 98, 3889–3894. [Google Scholar] [CrossRef]
- Bundale, S.; Singh, J.; Begde, D.; Nashikkar, N.; Upadhyay, A. Rare Actinobacteria: A Potential Source of Bioactive Polyketides and Peptides. World J. Microbiol. Biotechnol. 2019, 35, 92. [Google Scholar] [CrossRef]
- Matson, J.A.; Bush, J.A. Sandramycin, a Novel Antitumor Antibiotic Produced by a Nocardioides sp. Production, Isolation, Characterization and Biological Properties. J. Antibiot. 1989, 42, 1763–1767. [Google Scholar] [CrossRef]
- Park, Y.; Yoon, J.-H.; Shin, Y.K.; Suzuki, K.; Kudo, T.; Seino, A.; Kim, H.-J.; Lee, J.-S.; Lee, S.T. Classification of Nocardioides Fulvus IFO 14399 and Nocardioides sp. ATCC 39419 in Kribbella Gen. Nov., as Kribbella flavida sp. Nov. and Kribbella sandramycini sp. Nov. Int. J. Syst. Evol. Microbiol. 1999, 49, 743–752. [Google Scholar] [CrossRef]
- Alitalo, K.; Eriksson, U.; Olofsson, B.; Makinen, T. Novel Neuropilin/Growth Factor Binding and Uses. Thereof. Patent WO2000023565, 27 April 2000. [Google Scholar]
- Li, W.J.; Wang, D.; Zhang, Y.Q.; Schumann, P.; Stackebrandt, E.; Xu, L.H.; Jiang, C.L. Kribbella antibiotica sp. Nov., a Novel Nocardioform Actinomycete Strain Isolated from Soil in Yunnan, China. Syst. Appl. Microbiol. 2004, 27, 160–165. [Google Scholar] [CrossRef]
- Song, J.; Kim, B.Y.; Hong, S.B.; Cho, H.S.; Sohn, K.; Chun, J.; Suh, J.W. Kribbella solani Sp. Nov. and Kribbella jejuensis sp. Nov., Isolated from Potato Tuber and Soil in Jeju, Korea. Int. J. Syst. Evol. Microbiol. 2004, 54, 1345–1348. [Google Scholar] [CrossRef][Green Version]
- Salimi, F.; Hamedi, J.; Motevaseli, E.; Mohammadipanah, F. Isolation and Screening of Rare Actinobacteria, a New Insight for Finding Natural Products with Antivascular Calcification Activity. J. Appl. Microbiol. 2018, 124, 254–266. [Google Scholar] [CrossRef]
- Matson, J.A.; Colson, K.L.; Belofsky, G.N.; Bleiberg, B.B. Sandramycin, a Novel Antitumor Antibiotic Produced by a Nocardioides sp.. II. Structure Determination. J. Antibiot. 1993, 46, 162–166. [Google Scholar] [CrossRef]
- Tormo, J.R.; García, J.B.; DeAntonio, M.; Feliz, J.; Mira, A.; Díez, M.T.; Hernández, P.; Peláez, F. A Method for the Selection of Production Media for Actinomycete Strains Based on Their Metabolite HPLC Profiles. J. Ind. Microbiol. Biotechnol. 2003, 30, 582–588. [Google Scholar] [CrossRef]
- Boger, D.L.; Saionz, K.W. DNA Binding Properties of Key Sandramycin Analogues: Systematic Examination of the Intercalation Chromophore. Bioorg. Med. Chem. 1999, 7, 315–321. [Google Scholar] [CrossRef]
- Boger, D.L.; Chen, J.-H.; Saionz, K.W.; Jin, Q. Synthesis of Key Sandramycin Analogs: Systematic Examination of the Intercalation Chromophore. Bioorg. Med. Chem. 1998, 6, 85–102. [Google Scholar] [CrossRef]
- Jensen, P.R. Natural Products and the Gene Cluster Revolution. Trends Microbiol. 2016, 24, 968–977. [Google Scholar] [CrossRef]
- Pérez-Victoria, I.; Martín, J.; Reyes, F. Combined LC/UV/MS and NMR Strategies for the Dereplication of Marine Natural Products. Planta Med. 2016, 82, 857–871. [Google Scholar] [CrossRef]
- Stojiljkovic, I.; Evavold, B.D.; Kumar, V. Antimicrobial Properties of Porphyrins. Expert Opin. Investig. Drugs 2001, 10, 309–320. [Google Scholar] [CrossRef] [PubMed]
- Bhuiyan, M.N.I.; Takai, R.; Mitsuhashi, S.; Shigetomi, K.; Tanaka, Y.; Kamagata, Y.; Ubukata, M. Zincmethylphyrins and Coproporphyrins, Novel Growth Factors Released by Sphingopyxis sp., Enable Laboratory Cultivation of Previously Uncultured Leucobacter sp. through Interspecies Mutualism. J. Antibiot. 2016, 69, 97–103. [Google Scholar] [CrossRef]
- Igarashi, M.; Sawa, R.; Yamasaki, M.; Hayashi, C.; Umekita, M.; Hatano, M.; Fujiwara, T.; Mizumoto, K.; Nomoto, A. Kribellosides, Novel RNA 5′-Triphosphatase Inhibitors from the Rare Actinomycete Kribbella sp. MI481-42F6. J. Antibiot. 2017, 70, 582–589. [Google Scholar] [CrossRef]
- Dimise, E.J.; Widboom, P.F.; Bruner, S.D. Structure Elucidation and Biosynthesis of Fuscachelins, Peptide Siderophores from the Moderate Thermophile Thermobifida fusca. Proc. Natl. Acad. Sci. USA 2008, 105, 15311–15316. [Google Scholar] [CrossRef] [PubMed]
- Hoeksema, H.; Mizsak, S.A.; Baczynskyj, L.; Pschigoda, L.M. Structure of Rubradirin. J. Am. Chem. Soc. 1982, 104, 5173–5181. [Google Scholar] [CrossRef]
- Carretero-Molina, D.; Ortiz-López, F.J.; Martín, J.; González, I.; Sánchez-Hidalgo, M.; Román-Hurtado, F.; Díaz, C.; de la Cruz, M.; Genilloud, O.; Reyes, F. Pentaminomycins F and G, Nonribosomal Peptides Containing 2-Pyridylalanine. J. Nat. Prod. 2021, 84, 1127–1134. [Google Scholar] [CrossRef] [PubMed]
- Eagle, H.; Musselman, A.D. The Rate of Bactericidal Action of Penicillin in Vitro as a Function of Its Concentration, and Its Paradoxically Reduced Activity at High Concentrations against Certain Organisms. J. Exp. Med. 1948, 88, 99–131. [Google Scholar] [CrossRef]
- Prasetyoputri, A.; Jarrad, A.M.; Cooper, M.A.; Blaskovich, M.A.T. The Eagle Effect and Antibiotic-Induced Persistence: Two Sides of the Same Coin? Trends Microbiol. 2019, 27, 339–354. [Google Scholar] [CrossRef]
- Valero, C.; Colabardini, A.C.; de Castro, P.A.; Amich, J.; Bromley, M.J.; Goldman, G.H. The Caspofungin Paradoxical Effect Is a Tolerant “Eagle Effect” in the Filamentous Fungal Pathogen Aspergillus Fumigatus. MBio 2022, 13, e0044722. [Google Scholar] [CrossRef]
- Innis, M.A.; Gelfand, D.H.; Sninsky, J.J.; White, T.J. PCR Protocols: A Guide to Methods and Application; Academic Press: Cambridge, MA, USA, 1990; Volume 6. [Google Scholar]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S Ribosomal DNA Amplification for Phylogenetic Study. J. Bacteriol. 1991, 173, 697–703. [Google Scholar] [CrossRef]
- Pascual, J.; Macián, M.C.; Arahal, D.R.; Garay, E.; Pujalte, M.J. Multilocus Sequence Analysis of the Central Clade of the Genus Vibrio by Using the 16S RRNA, RecA, PyrH, RpoD, GyrB, RctB and ToxR Genes. Int. J. Syst. Evol. Microbiol. 2010, 60, 154–165. [Google Scholar] [CrossRef]
- Pascual, J.; Blanco, S.; García-López, M.; García-Salamanca, A.; Bursakov, S.A.; Genilloud, O.; Bills, G.F.; Ramos, J.L.; van Dillewijn, P. Assessing Bacterial Diversity in the Rhizosphere of Thymus Zygis Growing in the Sierra Nevada National Park (Spain) through Culture-Dependent and Independent Approaches. PLoS ONE 2016, 11, e0146558. [Google Scholar] [CrossRef]
- Yoon, S.-H.; Ha, S.-M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A Taxonomically United Database of 16S RRNA Gene Sequences and Whole-Genome Assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Thompson, J. The CLUSTAL_X Windows Interface: Flexible Strategies for Multiple Sequence Alignment Aided by Quality Analysis Tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The Neighbor-Joining Method: A New Method for Reconstructing Phylogenetic Trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Jukes, T.H.; Cantor, C.R. Evolution of Protein Molecules. In Mammalian Protein Metabolism; Elsevier: Amsterdam, The Netherlands, 1969; pp. 21–132. [Google Scholar]
- Martín, J.; Crespo, G.; González-Menéndez, V.; Pérez-Moreno, G.; Sánchez-Carrasco, P.; Pérez-Victoria, I.; Ruiz-Pérez, L.M.; González-Pacanowska, D.; Vicente, F.; Genilloud, O.; et al. MDN-0104, an Antiplasmodial Betaine Lipid from Heterospora chenopodii. J. Nat. Prod. 2014, 77, 2118–2123. [Google Scholar] [CrossRef]
- Palomo, S.; González, I.; de la Cruz, M.; Martín, J.; Tormo, J.; Anderson, M.; Hill, R.; Vicente, F.; Reyes, F.; Genilloud, O. Sponge-Derived Kocuria and Micrococcus spp. as Sources of the New Thiazolyl Peptide Antibiotic Kocurin. Mar. Drugs 2013, 11, 1071–1086. [Google Scholar] [CrossRef]
- Pan, R.; Bai, X.; Chen, J.; Zhang, H.; Wang, H. Exploring Structural Diversity of Microbe Secondary Metabolites Using OSMAC Strategy: A Literature Review. Front. Microbiol. 2019, 10, 294. [Google Scholar] [CrossRef]
- Duetz, W.A. Microtiter Plates as Mini-Bioreactors: Miniaturization of Fermentation Methods. Trends Microbiol. 2007, 15, 469–475. [Google Scholar] [CrossRef]
- Minas, W.; Bailey, J.E.; Duetz, W. Streptomycetes in Micro-Cultures: Growth, Production of Secondary Metabolites, and Storage and Retrieval in the 96–Well Format. Antonie Van Leeuwenhoek 2000, 78, 297–305. [Google Scholar] [CrossRef]
- Pérez-Bonilla, M.; Oves-Costales, D.; González, I.; de la Cruz, M.; Martín, J.; Vicente, F.; Genilloud, O.; Reyes, F. Krisynomycins, Imipenem Potentiators against Methicillin-Resistant Staphylococcus aureus, Produced by Streptomyces Canus. J. Nat. Prod. 2020, 83, 2597–2606. [Google Scholar] [CrossRef]
- Audoin, C.; Bonhomme, D.; Ivanisevic, J.; Cruz, M.; Cautain, B.; Monteiro, M.; Reyes, F.; Rios, L.; Perez, T.; Thomas, O. Balibalosides, an Original Family of Glucosylated Sesterterpenes Produced by the Mediterranean Sponge Oscarella balibaloi. Mar. Drugs 2013, 11, 1477–1489. [Google Scholar] [CrossRef]
- Monteiro, M.C.; de la Cruz, M.; Cantizani, J.; Moreno, C.; Tormo, J.R.; Mellado, E.; De Lucas, J.R.; Asensio, F.; Valiante, V.; Brakhage, A.A.; et al. A New Approach to Drug Discovery: High-Throughput Screening of Microbial Natural Extracts against Aspergillus fumigatus Using Resazurin. SLAS Discov. 2012, 17, 542–549. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Ravipati, A.S.; Koyyalamudi, S.R.; Jeong, S.C.; Reddy, N.; Bartlett, J.; Smith, P.T.; de la Cruz, M.; Monteiro, M.C.; Melguizo, Á.; et al. Anti-Fungal and Anti-Bacterial Activities of Ethanol Extracts of Selected Traditional Chinese Medicinal Herbs. Asian Pac. J. Trop. Med. 2013, 6, 673–681. [Google Scholar] [CrossRef]
- Zhang, J.-H.; Chung, T.D.Y.; Oldenburg, K.R. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. SLAS Discov. 1999, 4, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Serrano, R.; González-Menéndez, V.; Mackenzie, T.A.; Ramos, M.C.; Frisvad, J.C.; Larsen, T.O. A Molecular Networking Based Discovery of Diketopiperazine Heterodimers and Aspergillicins from Aspergillus caelatus. J. Nat. Prod. 2022, 85, 25–33. [Google Scholar] [CrossRef]

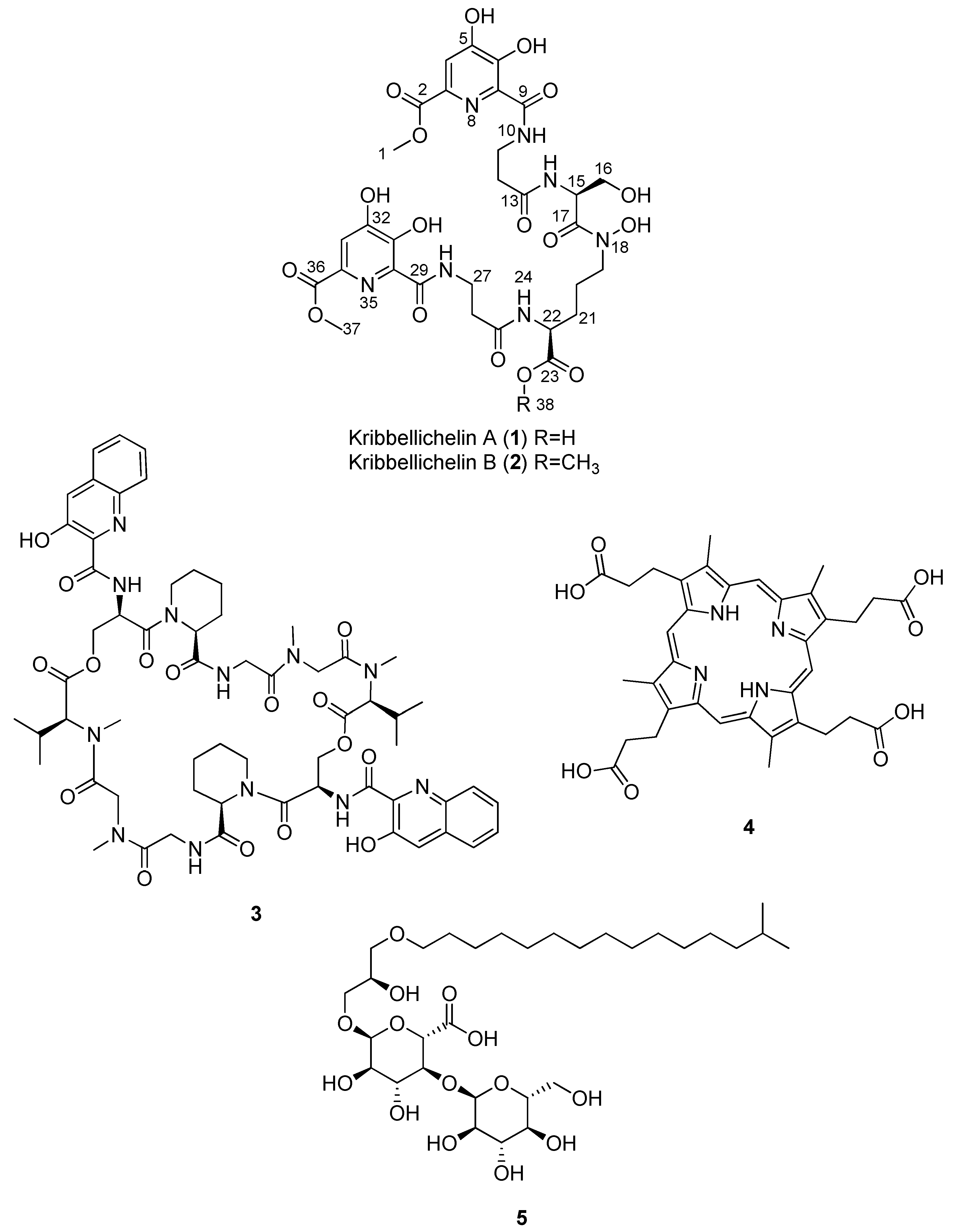

| Compound | MS Signals (m/z) | Exact Mass (Da) | Molecular Formula |
|---|---|---|---|
| Kribbellichelin A (1) | 768 | 767.2246 | C30H37N7O17 |
| Kribbellichelin B (2) | 782 | 781.2402 | C31H39N7O17 |
| Sandramycin (3) | 1221, 1222 | 1220.5502 | C60H76N12O16 |
| Coproporphyrin III (4) | 655 | - | C36H38N4O9 |
| Kribelloside C (5) | 672 | 654.3825 | C31H58O14 |
| 1 | 2 | |||||
|---|---|---|---|---|---|---|
| CD3OH | CD3CN/D2O 1:1 | CD3CN/D2O 1:1 | ||||
| 1H NMR | 13C NMR | 1H NMR | 13C NMR | 1H NMR | 13C NMR | |
| Position | δ in ppm, mult, J in Hz | δ in ppm | δ in ppm (mult, J in Hz) | δ in ppm | δ in ppm (mult, J in Hz) | δ in ppm j |
| 1 | 3.94, s | 53.5 | 3.83, s | 54.0 | 3.85, s | 54.0 |
| 2 | 166.9 | 166.40 c | 166.6 | |||
| 3 | 139.2 | 138.46 d | n.d. | |||
| 4 | 7.54, s | 116.1 | 7.45, s | 115.80 e | 7.52, br s | 115.8 |
| 5 | 155.8 | 155.6 | n.d. | |||
| 6 | 151.9 | 150.9 | n.d. | |||
| 7 | 132.3 | 131.33 f | n.d. | |||
| 8 | ||||||
| 9 | 170.6 | 169.2 | 169.2 | |||
| 10 | 9.34, m | |||||
| 11 | 3.69, m, 2H | 37.0 a | 3.55, m, 2H | 36.52 g | 3.57, m, 2H | 36.5 |
| 12 | 2.64, m, 2H | 36.4 | 2.56, m, 2H | 35.81 h | 2.55, m, 2H | 35.8 |
| 13 | 173.73 b | 173.94 i | 173.8 | |||
| 14 | 8.05, d, 7.5 | |||||
| 15 | 5.12, m | 53.8 | 4.96, m | 53.2 | 4.95, m | 53.2 |
| 16 | 3.81, dd, 11.0, 4.4; 3.73, dd, 11.1, 6.2 | 62.6 | 3.70, dd, 11.3, 4.5; 3.63, dd, 11.3, 6.3 | 61.7 | 3.68, m; 3.62, m | 61.7 |
| 17 | 171.5 | 171.1 | 171.2 | |||
| 18 | ||||||
| 19 | 3.57, m, 2H | 48.9 | 3.51, m; 3.39, m | 48.6 | 3.48, m; 3.37, m | 48.7 |
| 20 | 1.67, m, 2H | 24.3 | 1.53, m, 2H | 23.5 | 1.49, m, 2H | 23.5 |
| 21 | 1.84, m; 1.63, m | 29.7 | 1.69, m; 1.56, m | 28.7 | 1.65, m; 1.51, m | 28.8 |
| 22 | 4.40, m | 53.7 | 4.22, m | 53.3 | 4.24, m | 53.4 |
| 23 | 175.4 | 175.7 | 174.5 | |||
| 24 | 8.30, d, 7.7 | |||||
| 25 | 173.65 b | 173.85 i | 174.0 | |||
| 26 | 2.64, m, 2H | 36.4 | 2.56, m, 2H | 35.65 h | 2.55, m, 2H | 35.8 |
| 27 | 3.69, m, 2H | 36.9 a | 3.55, m, 2H | 36.40 g | 3.57, m, 2H | 36.5 |
| 28 | 9.34, m | |||||
| 29 | 170.6 | 169.2 | 169.2 | |||
| 30 | 132.3 | 131.29 f | n.d. | |||
| 31 | 151.9 | 150.9 | n.d. | |||
| 32 | 155.8 | 155.6 | n.d. | |||
| 33 | 7.54, s | 116.1 | 7.45, s | 115.76 e | 7.52, brs | 115.8 |
| 34 | 139.2 | 138.41 d | n.d. | |||
| 35 | ||||||
| 36 | 166.9 | 166.36 c | 166.6 | |||
| 37 | 3.94, s | 53.5 | 3.83, s | 54.0 | 3.83, s | 54.0 |
| 38 | - | - | 3.57, s | 53.5 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Virués-Segovia, J.R.; Reyes, F.; Ruíz, S.; Martín, J.; Fernández-Pastor, I.; Justicia, C.; de la Cruz, M.; Díaz, C.; Mackenzie, T.A.; Genilloud, O.; et al. Kribbellichelins A and B, Two New Antibiotics from Kribbella sp. CA-293567 with Activity against Several Human Pathogens. Molecules 2022, 27, 6355. https://doi.org/10.3390/molecules27196355
Virués-Segovia JR, Reyes F, Ruíz S, Martín J, Fernández-Pastor I, Justicia C, de la Cruz M, Díaz C, Mackenzie TA, Genilloud O, et al. Kribbellichelins A and B, Two New Antibiotics from Kribbella sp. CA-293567 with Activity against Several Human Pathogens. Molecules. 2022; 27(19):6355. https://doi.org/10.3390/molecules27196355
Chicago/Turabian StyleVirués-Segovia, Jorge R., Fernando Reyes, Sandra Ruíz, Jesús Martín, Ignacio Fernández-Pastor, Carlos Justicia, Mercedes de la Cruz, Caridad Díaz, Thomas A. Mackenzie, Olga Genilloud, and et al. 2022. "Kribbellichelins A and B, Two New Antibiotics from Kribbella sp. CA-293567 with Activity against Several Human Pathogens" Molecules 27, no. 19: 6355. https://doi.org/10.3390/molecules27196355
APA StyleVirués-Segovia, J. R., Reyes, F., Ruíz, S., Martín, J., Fernández-Pastor, I., Justicia, C., de la Cruz, M., Díaz, C., Mackenzie, T. A., Genilloud, O., González, I., & Tormo, J. R. (2022). Kribbellichelins A and B, Two New Antibiotics from Kribbella sp. CA-293567 with Activity against Several Human Pathogens. Molecules, 27(19), 6355. https://doi.org/10.3390/molecules27196355









