Mechanistic Aspects for the Modulation of Enzyme Reactions on the DNA Scaffold
Abstract
1. Introduction
2. Construction of the Artificial Enzymatic Reaction Systems Inspired by Nature
2.1. The Spatial Organization of Enzymes in Nature
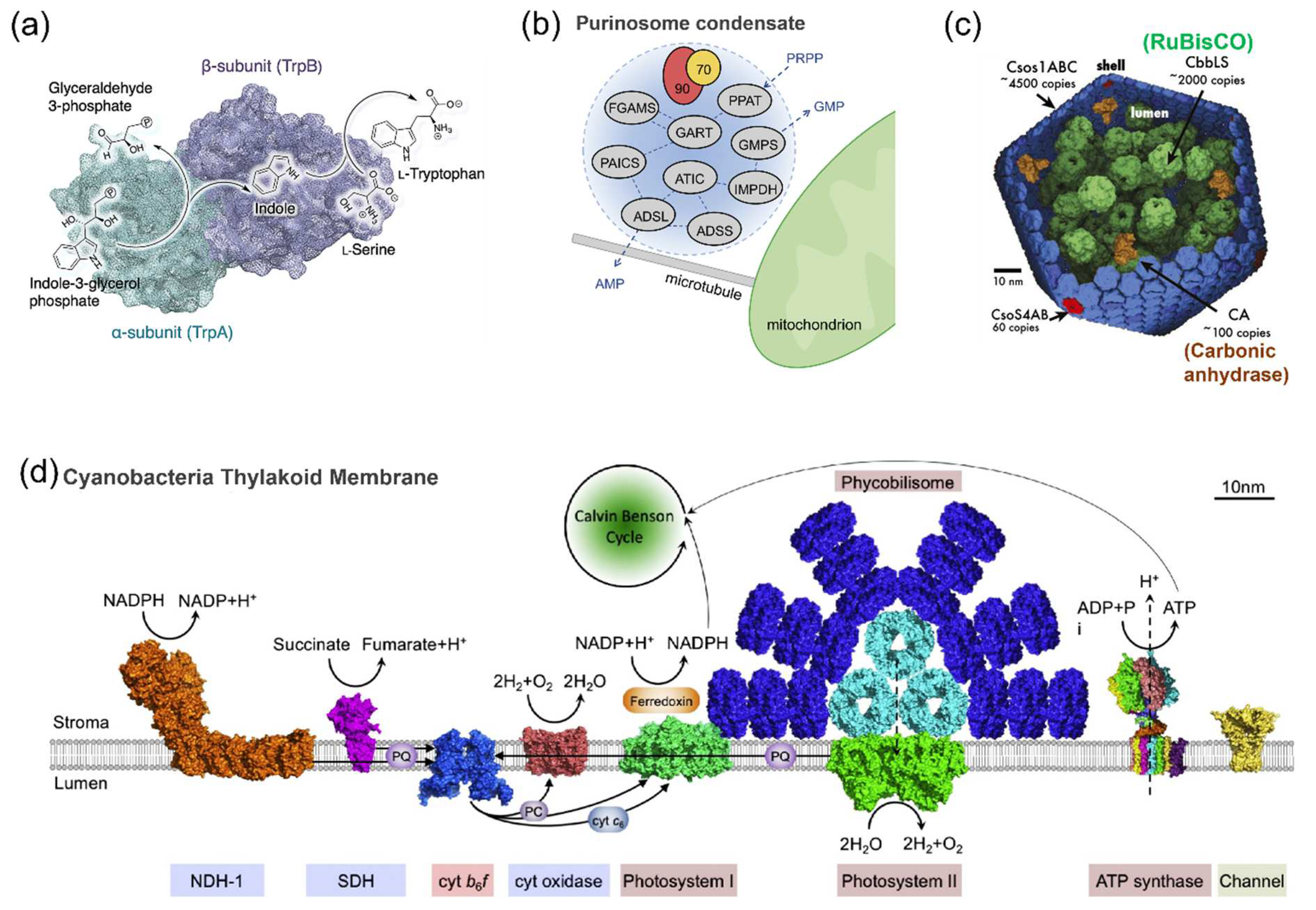
2.2. Enzymatic Reactions on Various Carriers
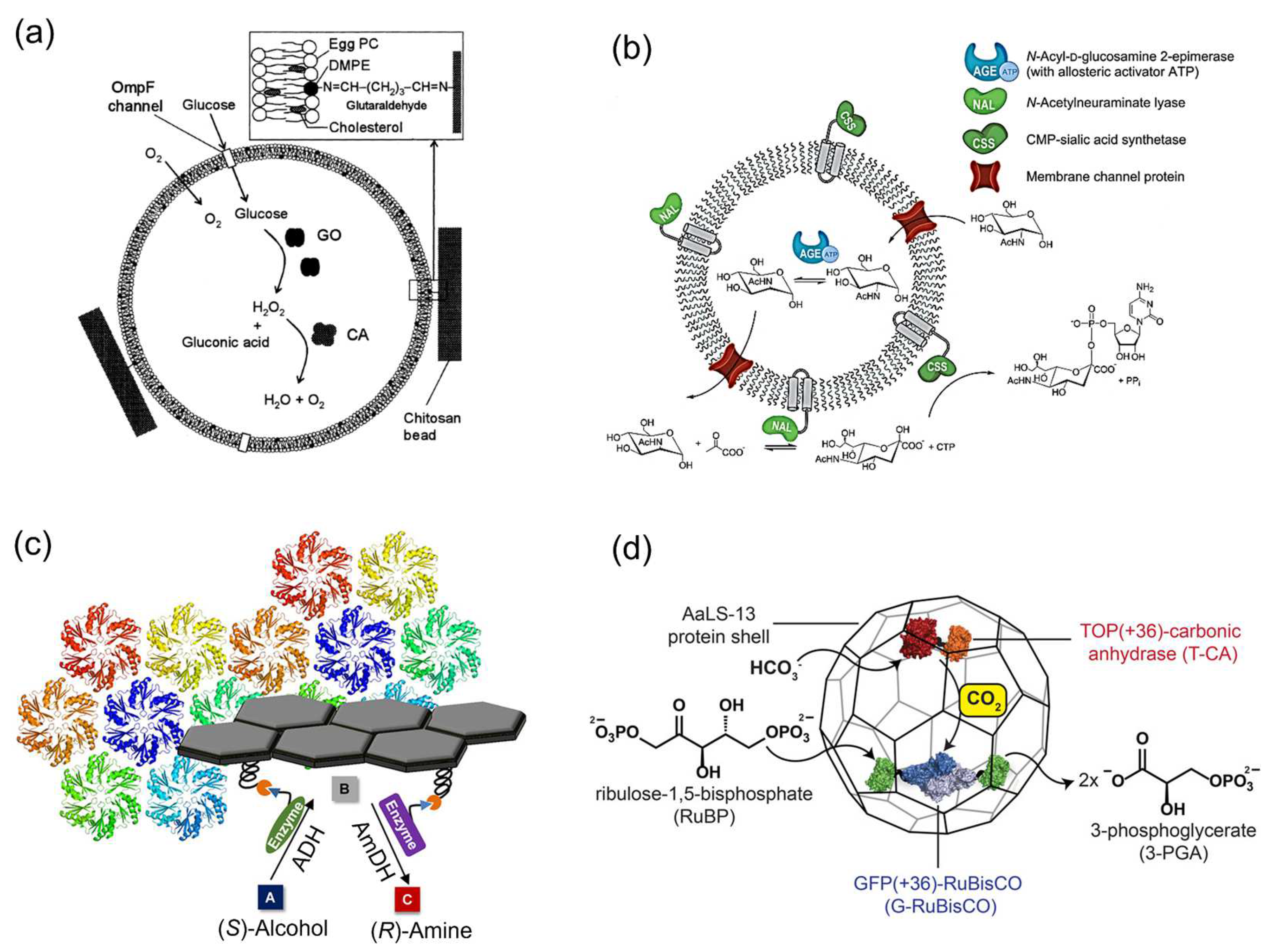
3. Catalytic Enhancement of Single Type of Enzyme Assembled on the DNA Scaffold
3.1. DNA Origami Scaffold
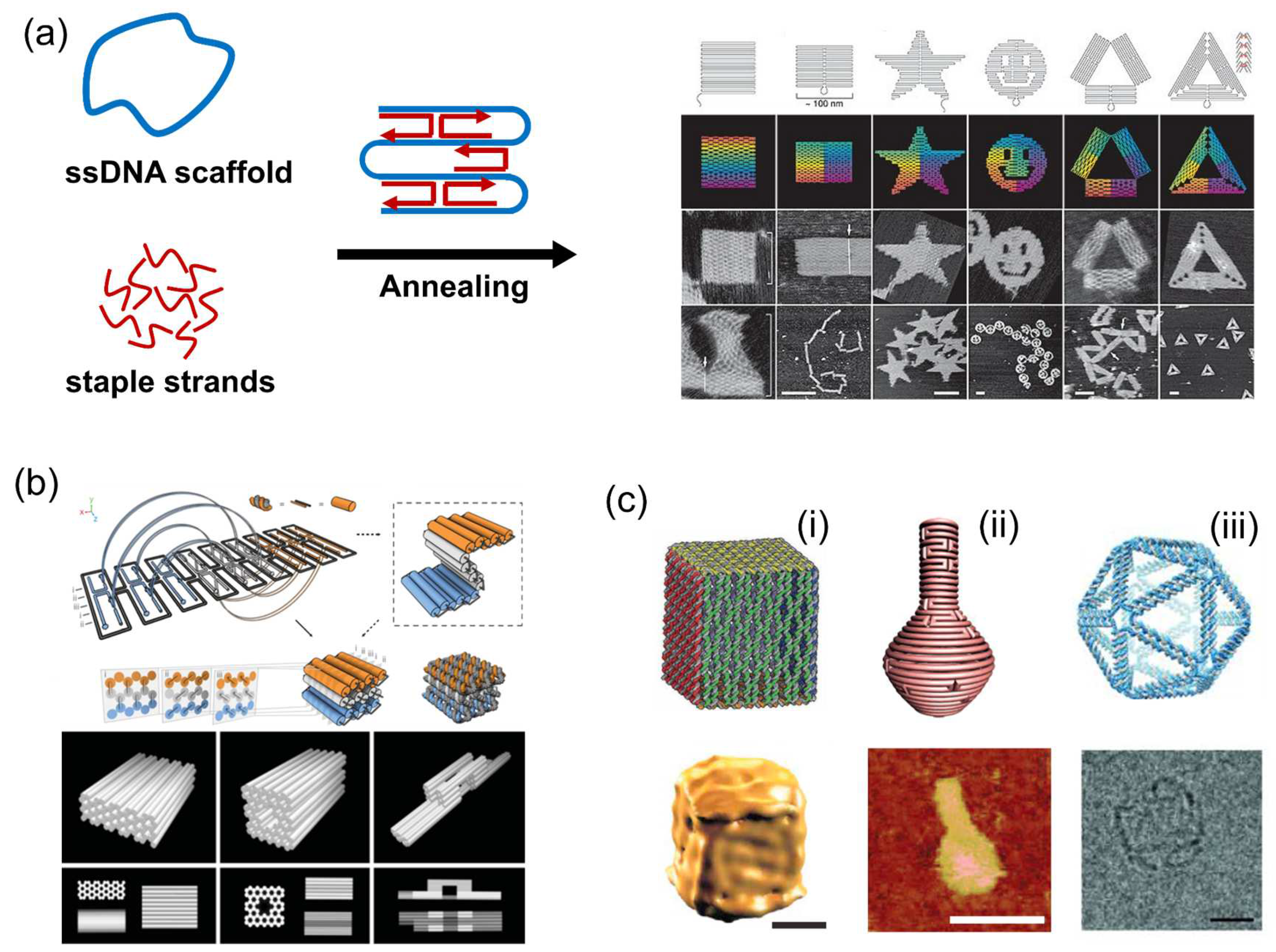
3.2. “Favorable Microenvironment” Provided by DNA
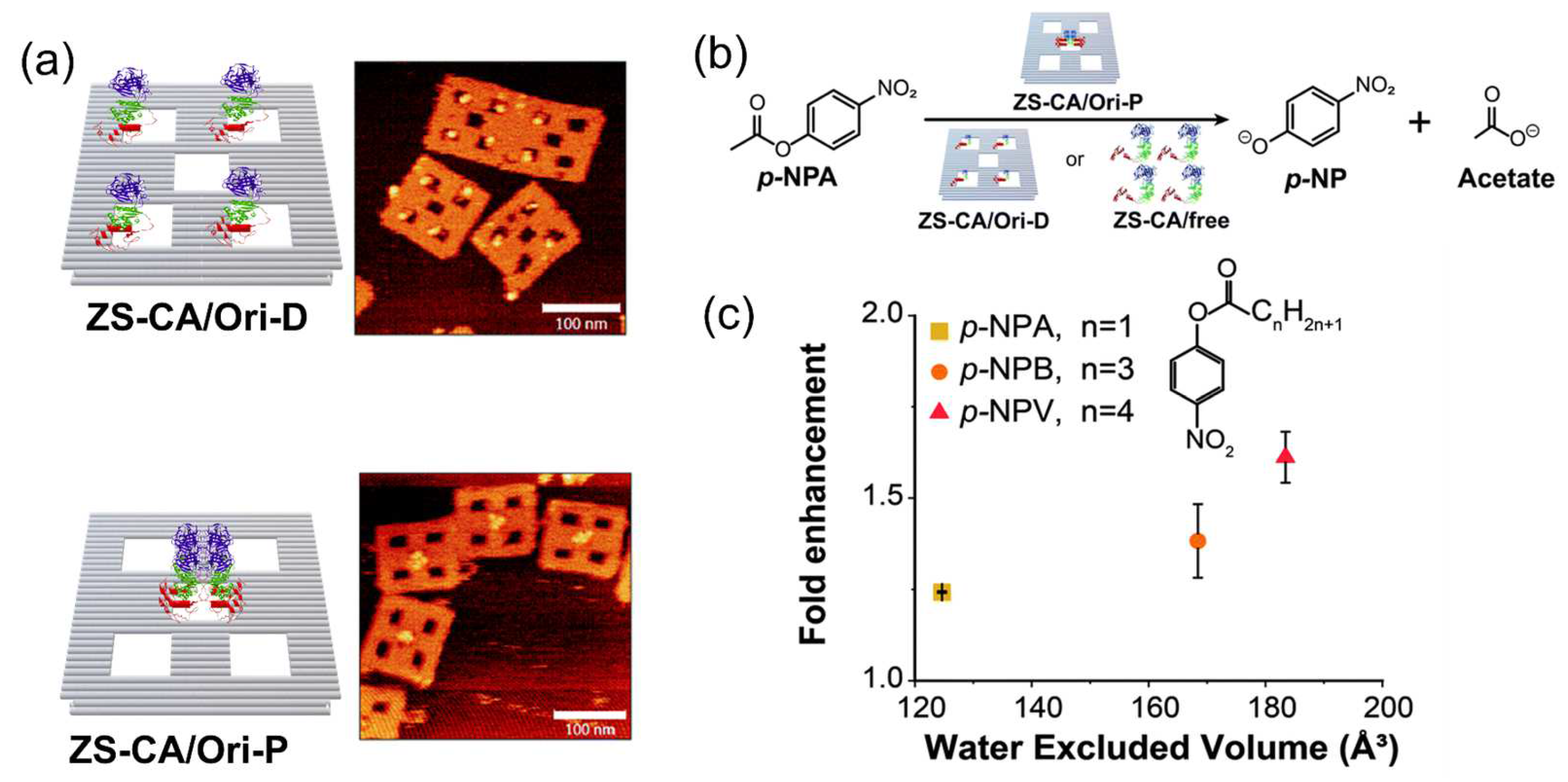
3.3. Protection Effect Derived from DNA Scaffold
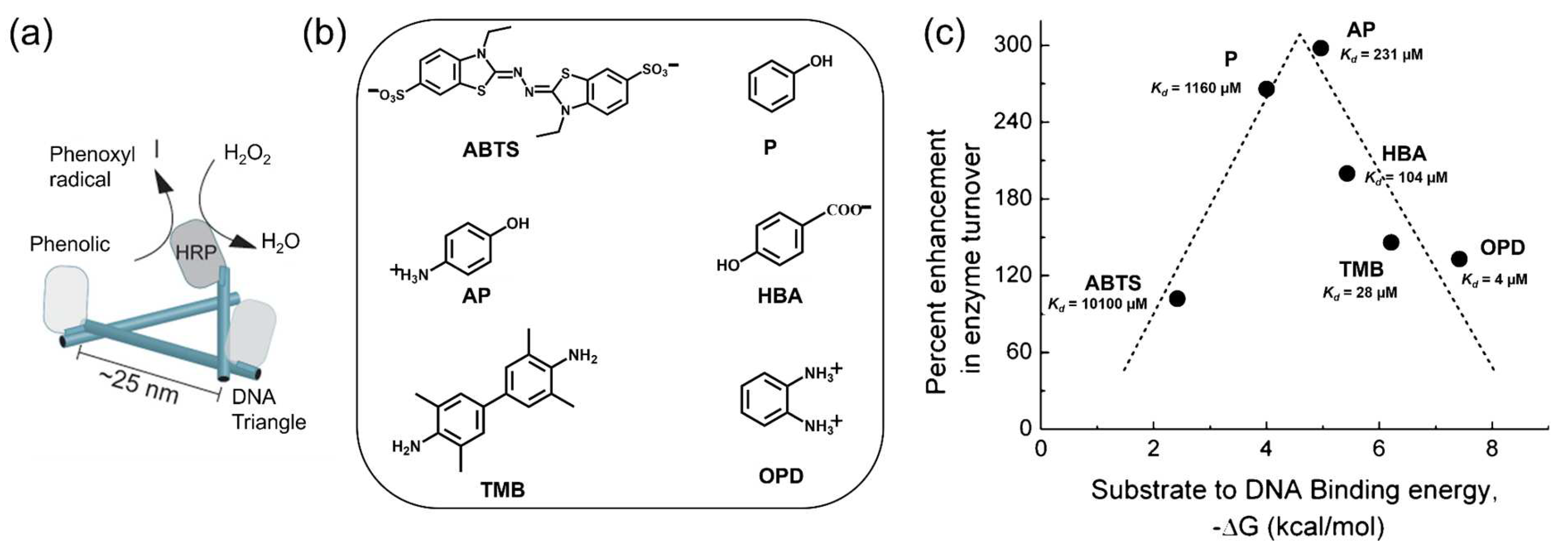
3.4. Substrate Affinity to the DNA Scaffold
3.5. Ordered Hydration Layer on the DNA Scaffold Surface
3.6. Local pH Environment
3.7. General Factors for the Catalytic Enhancement of DNA-Scaffolded Enzymes

3.8. Packed State of Enzymes on the DNA Scaffold
4. Enhanced Efficiency of Enzyme Cascade Reactions on the DNA Scaffold
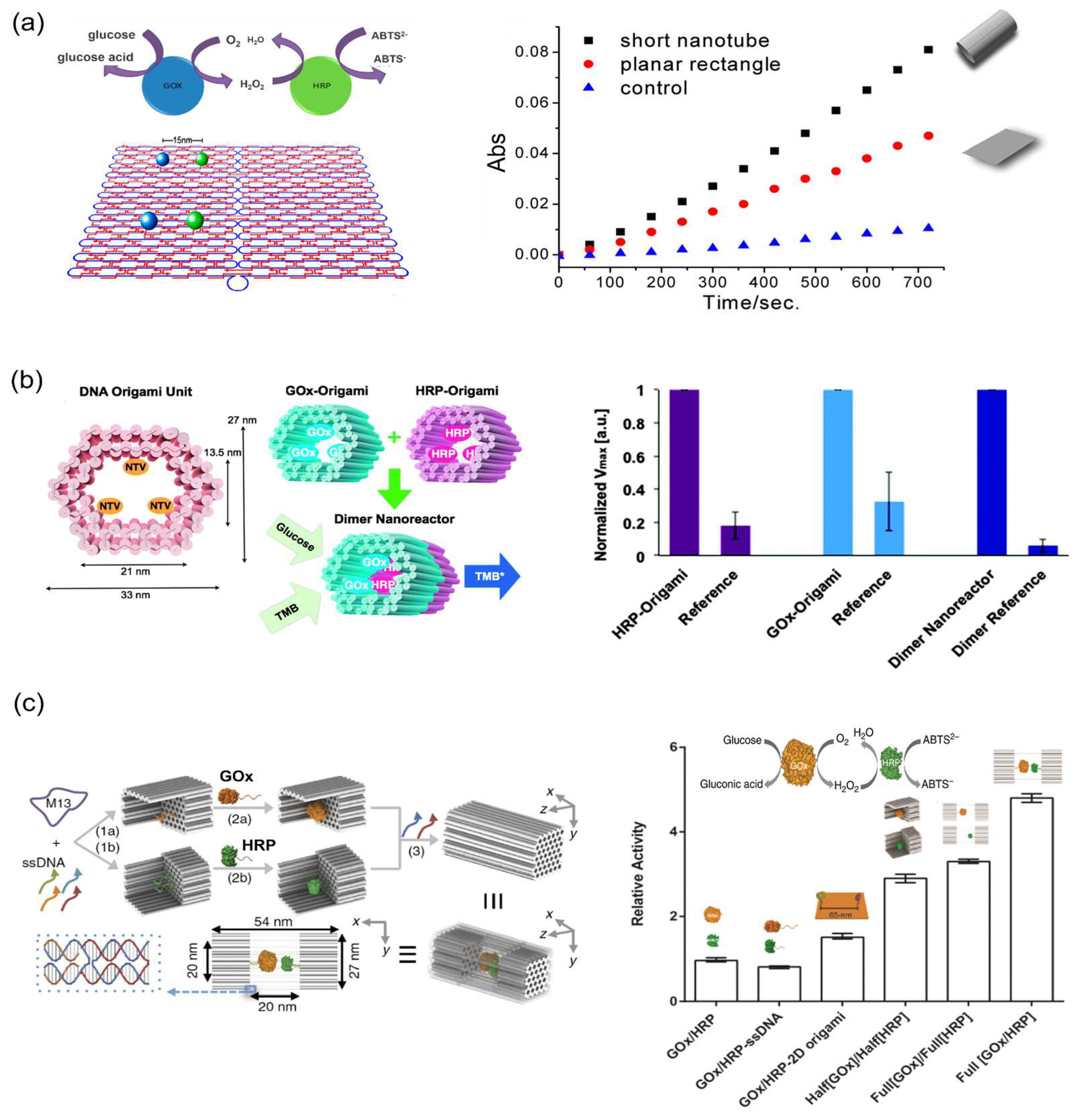
4.1. Activity Enhancement through the DNA Scaffold

4.2. Swinging Arms Facilitating the Substrate Channeling
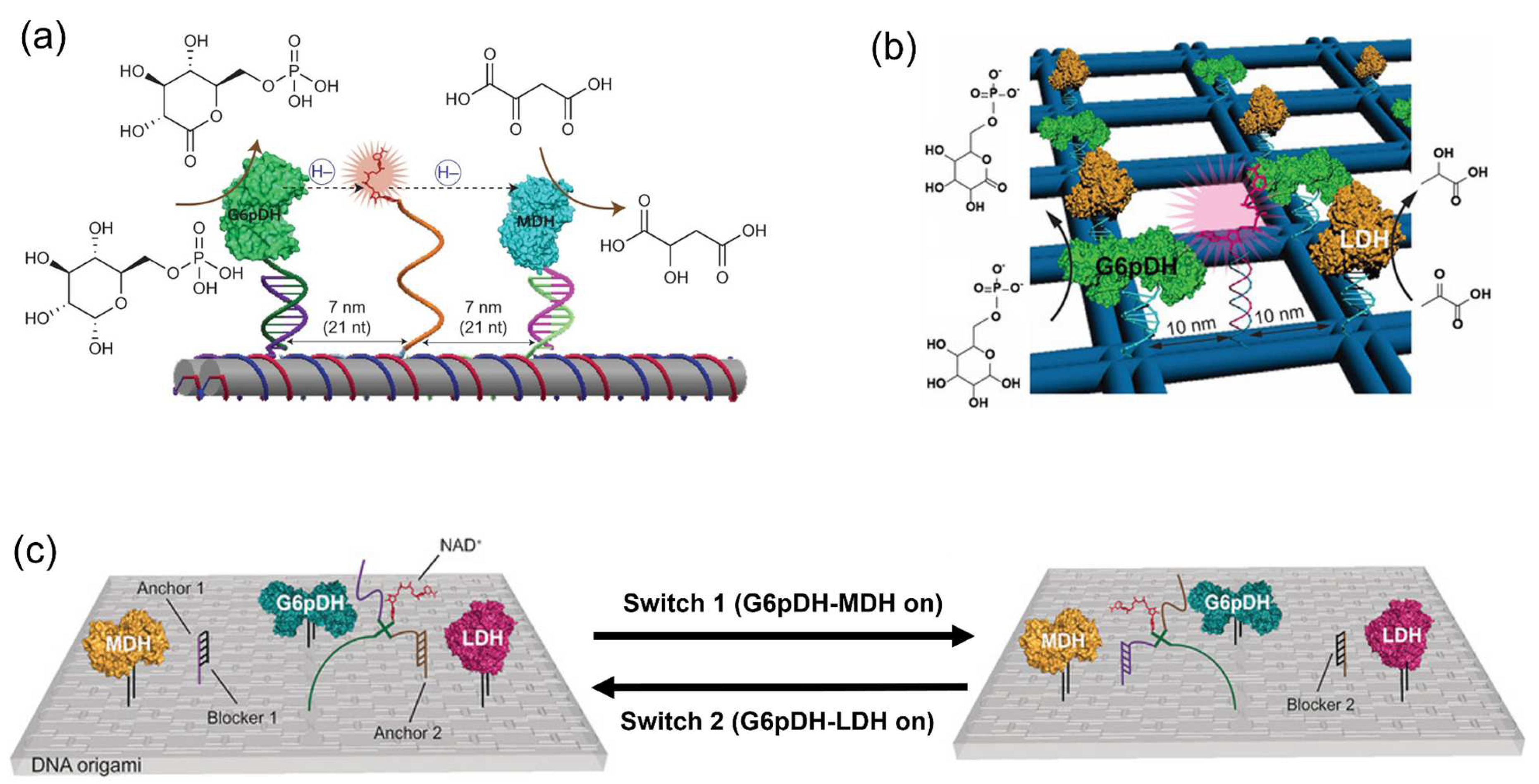
4.3. Proximity Effect: Controversy Remains

4.4. Enzyme Kinetics of Cascade Reactions
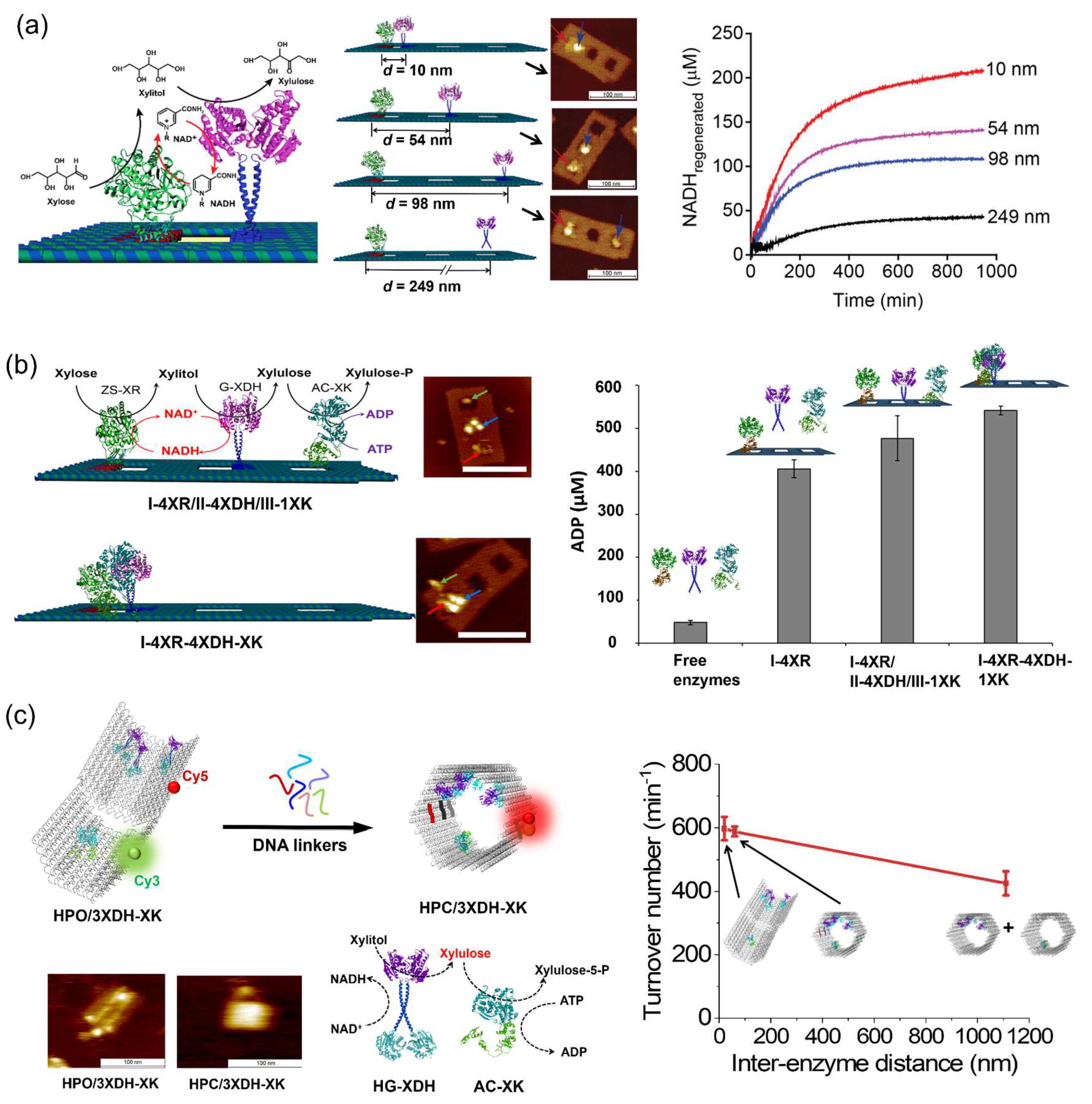
4.5. Spatial Arrangement of Enzymes
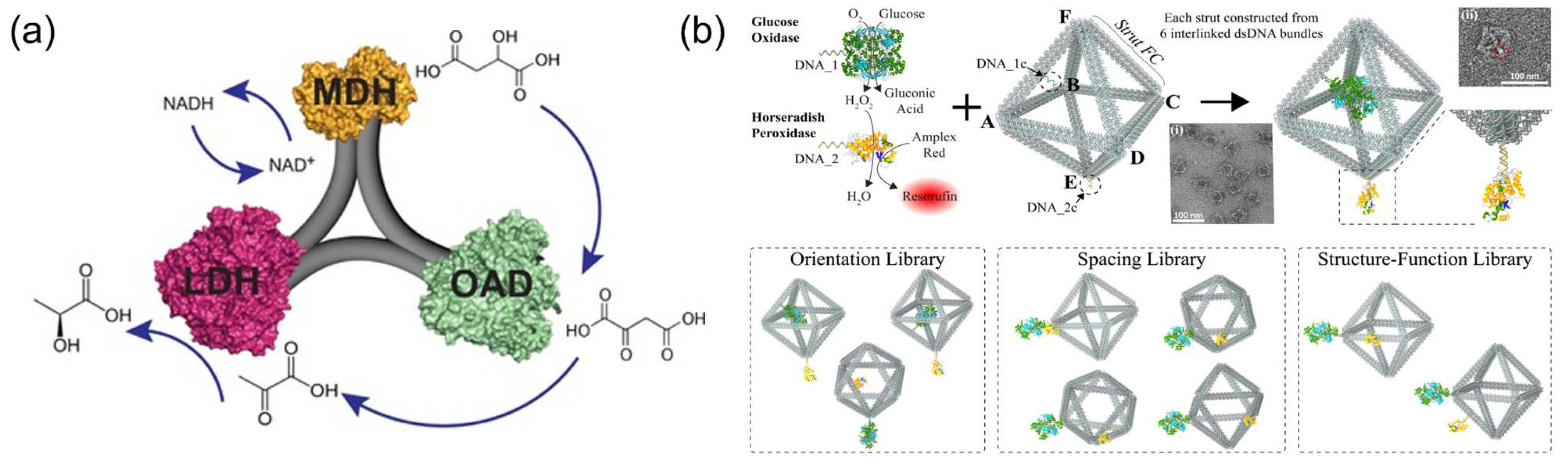
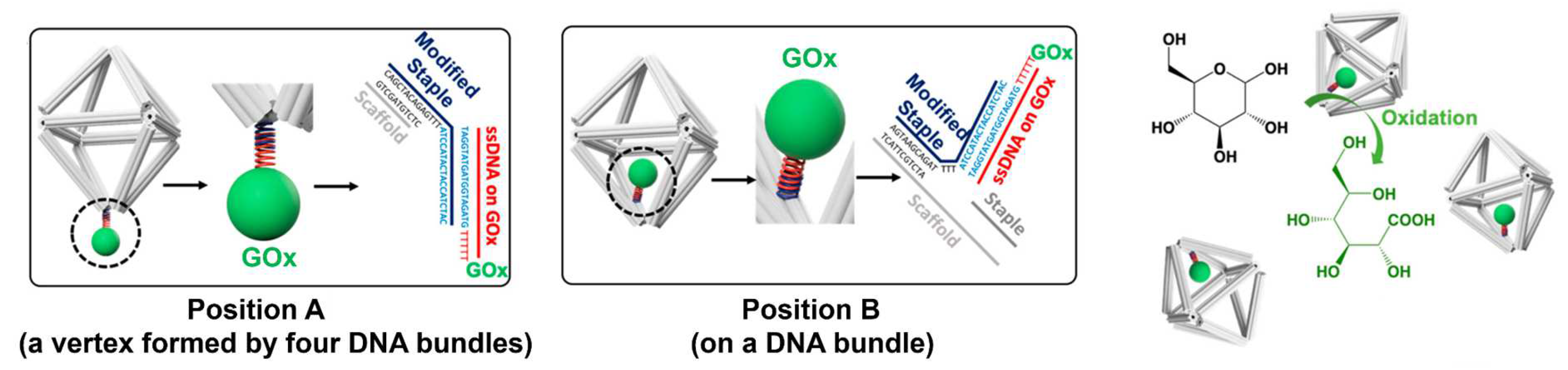
4.6. Effect of 3D Confinement
5. Conclusions and Future Perspectives

Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| RuBisCO | Ribulose 1,5-bisphosphate carboxylase/oxygenase |
| 2D | two-dimensional |
| 3D | three-dimensional |
| MDH–CS | malate dehydrogenase–citrate synthase |
| DNPB | de novo purine biosynthesis |
| GFP | green fluorescent protein |
| OFP | orange fluorescent protein |
| Hsp90 | heat shock protein 90 kDa |
| CA | carbonic anhydrase |
| PRPP | phosphoribosyl pyrophosphate |
| GMP | guanosine 5′-monophosphate |
| AMP | adenosine 5′-monophosphate |
| FGAMS | formylglycinamidine ribonucleotide synthase |
| GART | glycinamide ribonucleotide transformylase |
| PPAT | phosphoribosyl pyrophosphate amidotransferase |
| ADSL | adenylosuccinate lyase |
| PAICS | phosphoribosylaminoimidazole succinocarboxamide synthetase |
| IMPDH | inosine monophosphate dehydrogenase |
| ADSS | adenylosuccinate synthetase |
| GOx | glucose oxidase |
| HRP | horseradish peroxidase |
| OmpF | outer membrane protein F |
| CMP-Neu5Ac | CMP-N-acetylneuraminic acid |
| NAL | N-Acetylneuraminate lyase |
| CSS | CMP-sialic acid synthetase |
| AGE | N-Acyl-D-glucosamine 2-epimerase |
| EutM | ethanolamine utilization bacterial microcompartment |
| ADH | alcohol dehydrogenase |
| AmDH | amine dehydrogenase |
| CH | chlorohexane |
| AP | p-Aminophenol |
| P | phenol |
| HBA | p-hydroxybenzoic acid |
| OPD | o-phenylenediamine |
| TMB | 3,3′,5,5′-tetramethylbenzidine |
| ABTS | 2,2′-azino-bis3-ethylbenzothiazoline-6-sulphonic acid |
| G6pDH | glucose-6-phosphate dehydrogenase |
| MDH | malic dehydrogenase |
| LDH | lactic dehydrogenase |
| XR | xylose reductase |
| XDH | xylitol dehydrogenase |
| XK | xylulose kinase |
| ZS-XR | modular adaptor ZF-SNAP fused XR |
| G-XDH | adaptor GCN4 fused XDH |
| HG-XDH | modular adaptor Halo-GCN4 fused XDH |
| AC-XK | modular adaptor AZ-CLIP fused XK |
| ZS-CA | modular adaptor ZF-SNAP fused CA |
| sXR | DNA-scaffolded ZS-XR |
| sXDH | DNA-scaffolded HG-XDH |
| HP | DNA hexagonal prism |
| HPO | open state of HP |
| HPC | closed state of HPC |
| p-NPA | p-nitrophenyl acetate |
| p-NPB | p-nitrophenyl butyrate |
| p-NPV | p-nitrophenyl valerate |
| OAD | oxaloacetate decarboxylase |
References
- Hossain, G.S.; Nadarajan, S.P.; Zhang, L.; Ng, T.K.; Foo, J.L.; Ling, H.; Choi, W.J.; Chang, M.W. Rewriting the metabolic blueprint: Advances in pathway diversification in microorganisms. Front. Microbiol. 2018, 9, 155. [Google Scholar] [CrossRef] [PubMed]
- Chen, A.H.; Silver, P.A. Designing biological compartmentalization. Trends Cell Biol. 2012, 22, 662–670. [Google Scholar] [CrossRef] [PubMed]
- Agapakis, C.M.; Boyle, P.M.; Silver, P.A. Natural strategies for the spatial optimization of metabolism in synthetic biology. Nat. Chem. Biol. 2012, 8, 527–535. [Google Scholar] [CrossRef] [PubMed]
- Bonacci, W.; Teng, P.K.; Afonso, B.; Niederholtmeyer, H.; Grob, P.; Silver, P.A.; Savage, D.F. Modularity of a carbon-fixing protein organelle. Proc. Natl. Acad. Sci. USA 2012, 109, 478–483. [Google Scholar] [CrossRef]
- Šrejber, M.; Navrátilová, V.; Paloncýová, M.; Bazgier, V.; Berka, K.; Anzenbacher, P.; Otyepka, M. Membrane-attached mammalian cytochromes P450: An overview of the membrane’s effects on structure, drug binding, and interactions with redox partners. J. Inorg. Biochem. 2018, 183, 117–136. [Google Scholar] [CrossRef]
- Kastritis, P.L.; Gavin, A.C. Enzymatic complexes across scales. Essays Biochem. 2018, 62, 501–514. [Google Scholar] [CrossRef]
- Zhang, Y.H.P. Substrate channeling and enzyme complexes for biotechnological applications. Biotechnol. Adv. 2011, 29, 715–725. [Google Scholar] [CrossRef]
- Seo, M.J.; Schmidt-Dannert, C. Organizing multi-enzyme systems into programmable materials for biocatalysis. Catalysts 2021, 11, 409. [Google Scholar] [CrossRef]
- Rabe, K.S.; Müller, J.; Skoupi, M.; Niemeyer, C.M. Cascades in compartments: En route to machine-assisted biotechnology. Angew. Chem. Int. Ed. 2017, 56, 13574–13589. [Google Scholar] [CrossRef]
- Küchler, A.; Yoshimoto, M.; Luginbühl, S.; Mavelli, F.; Walde, P. Enzymatic reactions in confined environments. Nat. Nanotechnol. 2016, 11, 409–420. [Google Scholar] [CrossRef]
- Vázquez-González, M.; Wang, C.; Willner, I. Biocatalytic cascades operating on macromolecular scaffolds and in confined environments. Nat. Catal. 2020, 3, 256–273. [Google Scholar] [CrossRef]
- Dubey, N.C.; Tripathi, B.P. Nature inspired multienzyme immobilization: Strategies and concepts. ACS Appl. Bio Mater. 2021, 4, 1077–1114. [Google Scholar] [CrossRef]
- Madsen, M.; Gothelf, K.V. Chemistries for DNA nanotechnology. Chem. Rev. 2019, 119, 6384–6458. [Google Scholar] [CrossRef]
- Rothemund, P.W. Folding DNA to create nanoscale shapes and patterns. Nature 2006, 440, 297–302. [Google Scholar] [CrossRef]
- Douglas, S.M.; Dietz, H.; Liedl, T.; Högberg, B.; Graf, F.; Shih, W.M. Self-assembly of DNA into nanoscale three-dimensional shapes. Nature 2009, 459, 414–418. [Google Scholar] [CrossRef]
- Hong, F.; Zhang, F.; Liu, Y.; Yan, H. DNA origami: Scaffolds for creating higher order structures. Chem. Rev. 2017, 117, 12584–12640. [Google Scholar] [CrossRef]
- Fu, J.; Wang, Z.; Liang, X.H.; Oh, S.W.; Iago-McRae, E.S.; Zhang, T. DNA-scaffolded proximity assembly and confinement of multienzyme reactions. Top. Curr. Chem. 2020, 378, 38. [Google Scholar] [CrossRef]
- Du, P.; Xu, S.; Xu, Z.K.; Wang, Z.G. Bioinspired Self-Assembling Materials for Modulating Enzyme Functions. Adv. Funct. Mater. 2021, 31, 2104819. [Google Scholar] [CrossRef]
- Jaekel, A.; Stegemann, P.; Saccà, B. Manipulating enzymes properties with DNA nanostructures. Molecules 2019, 24, 3694. [Google Scholar] [CrossRef]
- Zhang, Y.; Ge, J.; Liu, Z. Enhanced activity of immobilized or chemically modified enzymes. ACS Catal. 2015, 5, 4503–4513. [Google Scholar] [CrossRef]
- Linko, V.; Nummelin, S.; Aarnos, L.; Tapio, K.; Toppari, J.J.; Kostiainen, M.A. DNA-based enzyme reactors and systems. Nanomaterials 2016, 6, 139. [Google Scholar] [CrossRef]
- Kong, G.; Xiong, M.; Liu, L.; Hu, L.; Meng, H.M.; Ke, G.; Zhang, X.B.; Tan, W. DNA origami-based protein networks: From basic construction to emerging applications. Chem. Soc. Rev. 2021, 50, 1846–1873. [Google Scholar] [CrossRef]
- Ellis, G.A.; Klein, W.P.; Lasarte-Aragones, G.; Thakur, M.; Walper, S.A.; Medintz, I.L. Artificial multienzyme scaffolds: Pursuing in vitro substrate channeling with an overview of current progress. ACS Catal. 2019, 9, 10812–10869. [Google Scholar] [CrossRef]
- Fu, J.; Oh, S.W.; Monckton, K.; Arbuckle-Keil, G.; Ke, Y.; Zhang, T. Biomimetic compartments scaffolded by nucleic acid nanostructures. Small 2019, 15, 1900256. [Google Scholar] [CrossRef]
- Schmitt, D.L.; An, S. Spatial organization of metabolic enzyme complexes in cells. Biochemistry 2017, 56, 3184–3196. [Google Scholar] [CrossRef]
- Wheeldon, I.; Minteer, S.D.; Banta, S.; Barton, S.C.; Atanassov, P.; Sigman, M. Substrate channelling as an approach to cascade reactions. Nat. Chem. 2016, 8, 299–309. [Google Scholar] [CrossRef]
- Maria-Solano, M.A.; Iglesias-Fernández, J.; Osuna, S. Deciphering the allosterically driven conformational ensemble in tryptophan synthase evolution. J. Am. Chem. Soc. 2019, 141, 13049–13056. [Google Scholar] [CrossRef]
- Miles, E.W.; Rhee, S.; Davies, D.R. The molecular basis of substrate channeling. J. Biol. Chem. 1999, 274, 12193–12196. [Google Scholar] [CrossRef]
- Wu, F.; Minteer, S. Krebs cycle metabolon: Structural evidence of substrate channeling revealed by cross-linking and mass spectrometry. Angew. Chem. Int. Ed. 2015, 54, 1851–1854. [Google Scholar] [CrossRef]
- Sweetlove, L.J.; Fernie, A.R. The role of dynamic enzyme assemblies and substrate channelling in metabolic regulation. Nat. Commun. 2018, 9, 2136. [Google Scholar] [CrossRef] [PubMed]
- Pedley, A.M.; Benkovic, S.J. A new view into the regulation of purine metabolism: The purinosome. Trends Biochem Sci. 2017, 42, 141–154. [Google Scholar] [CrossRef] [PubMed]
- Kotopka, B.J.; Smolke, C.D. Production of the cyanogenic glycoside dhurrin in yeast. Metab. Eng. Commun. 2019, 9, e00092. [Google Scholar] [CrossRef] [PubMed]
- Araiza-Olivera, D.; Chiquete-Felix, N.; Rosas-Lemus, M.; Sampedro, J.G.; Peña, A.; Mujica, A.; Uribe-Carvajal, S. A glycolytic metabolon in S accharomyces cerevisiae is stabilized by F-actin. FEBS J. 2013, 280, 3887–3905. [Google Scholar] [CrossRef] [PubMed]
- An, S.; Kumar, R.; Sheets, E.D.; Benkovic, S.J. Reversible compartmentalization of de novo purine biosynthetic complexes in living cells. Science 2008, 320, 103–106. [Google Scholar] [CrossRef]
- Pedley, A.M.; Pareek, V.; Benkovic, S.J. The Purinosome: A Case Study for a Mammalian Metabolon. Annu. Rev. Biochem. 2022, 91, 89–106. [Google Scholar] [CrossRef]
- Pareek, V.; Tian, H.; Winograd, N.; Benkovic, S.J. Metabolomics and mass spectrometry imaging reveal channeled de novo purine synthesis in cells. Science 2020, 368, 283–290. [Google Scholar] [CrossRef]
- Pedley, A.M.; Boylan, J.P.; Chan, C.Y.; Kennedy, E.L.; Kyoung, M.; Benkovic, S.J. Purine biosynthetic enzymes assemble into liquid-like condensates dependent on the activity of chaperone protein HSP90. J. Biol. Chem. 2022, 298, 101845. [Google Scholar] [CrossRef]
- Wang, H.; Yan, X.; Aigner, H.; Bracher, A.; Nguyen, N.D.; Hee, W.Y.; Long, B.M.; Price, G.D.; Hartl, F.U.; Hayer-Hartl, M. Rubisco condensate formation by CcmM in β-carboxysome biogenesis. Nature 2019, 566, 131–135. [Google Scholar] [CrossRef]
- Liu, L.N. Distribution and dynamics of electron transport complexes in cyanobacterial thylakoid membranes. Biochim. Biophys. Acta Bioenerg. 2016, 1857, 256–265. [Google Scholar] [CrossRef]
- Liang, W.; Xu, H.; Carraro, F.; Maddigan, N.K.; Li, Q.; Bell, S.G.; Huang, D.M.; Tarzia, A.; Solomon, M.B.; Amenitsch, H.; et al. Enhanced activity of enzymes encapsulated in hydrophilic metal–organic frameworks. J. Am. Chem. Soc. 2019, 141, 2348–2355. [Google Scholar] [CrossRef]
- Yue, L.; Wang, S.; Wulf, V.; Willner, I. Stiffness-switchable DNA-based constitutional dynamic network hydrogels for self-healing and matrix-guided controlled chemical processes. Nat. Commun. 2019, 10, 4774. [Google Scholar] [CrossRef]
- Lin, P.; Zhang, Y.; Ren, H.; Wang, Y.; Wang, S.; Fang, B. Assembly of graphene oxide-formate dehydrogenase composites by nickel-coordination with enhanced stability and reusability. Eng. Life Sci. 2018, 18, 326–333. [Google Scholar] [CrossRef]
- Walde, P.; Ichikawa, S. Enzymes inside lipid vesicles: Preparation, reactivity and applications. Biomol. Eng. 2001, 18, 143–177. [Google Scholar] [CrossRef]
- Klermund, L.; Poschenrieder, S.T.; Castiglione, K. Biocatalysis in polymersomes: Improving multienzyme cascades with incompatible reaction steps by compartmentalization. ACS Catal. 2017, 7, 3900–3904. [Google Scholar] [CrossRef]
- Zhang, G.; Quin, M.B.; Schmidt-Dannert, C. Self-assembling protein scaffold system for easy in vitro coimmobilization of biocatalytic cascade enzymes. ACS Catal. 2018, 8, 5611–5620. [Google Scholar] [CrossRef]
- Rajendran, A.; Nakata, E.; Nakano, S.; Morii, T. Nucleic-Acid-Templated Enzyme Cascades. ChemBioChem 2017, 18, 696–716. [Google Scholar] [CrossRef]
- Yoshimoto, M.; Wang, S.; Fukunaga, K.; Fournier, D.; Walde, P.; Kuboi, R.; Nakao, K. Novel immobilized liposomal glucose oxidase system using the channel protein OmpF and catalase. Biotechnol. Bioeng. 2005, 90, 231–238. [Google Scholar] [CrossRef]
- Frey, R.; Mantri, S.; Rocca, M.; Hilvert, D. Bottom-up construction of a primordial carboxysome mimic. J. Am. Chem. Soc. 2016, 138, 10072–10075. [Google Scholar] [CrossRef]
- Tapio, K.; Bald, I. The potential of DNA origami to build multifunctional materials. Multifunct. Mater. 2020, 3, 032001. [Google Scholar] [CrossRef]
- Wang, P.; Meyer, T.A.; Pan, V.; Dutta, P.K.; Ke, Y. The beauty and utility of DNA origami. Chem 2017, 2, 359–382. [Google Scholar] [CrossRef]
- Castro, C.E.; Kilchherr, F.; Kim, D.N.; Shiao, E.L.; Wauer, T.; Wortmann, P.; Bathe, M.; Dietz, H. A primer to scaffolded DNA origami. Nat. Methods. 2011, 8, 221–229. [Google Scholar] [CrossRef]
- Douglas, S.M.; Marblestone, A.H.; Teerapittayanon, S.; Vazquez, A.; Church, G.M.; Shih, W.M. Rapid prototyping of 3D DNA-origami shapes with caDNAno. Nucleic Acids Res. 2009, 37, 5001–5006. [Google Scholar] [CrossRef]
- Andersen, E.S.; Dong, M.; Nielsen, M.M.; Jahn, K.; Subramani, R.; Mamdouh, W.; Golas, M.M.; Sander, B.; Stark, H.; Oliveira, C.L.P.; et al. Self-assembly of a nanoscale DNA box with a controllable lid. Nature 2009, 459, 73–76. [Google Scholar] [CrossRef]
- Han, D.; Pal, S.; Nangreave, J.; Deng, Z.; Liu, Y.; Yan, H. DNA origami with complex curvatures in three-dimensional space. Science 2011, 332, 342–346. [Google Scholar] [CrossRef]
- Veneziano, R.; Ratanalert, S.; Zhang, K.; Zhang, F.; Yan, H.; Chiu, W.; Bathe, M. Designer nanoscale DNA assemblies programmed from the top down. Science 2016, 352, 1534. [Google Scholar] [CrossRef]
- DeLuca, M.; Shi, Z.; Castro, C.E.; Arya, G. Dynamic DNA nanotechnology: Toward functional nanoscale devices. Nanoscale Horiz. 2020, 5, 182–201. [Google Scholar] [CrossRef]
- Zhang, D.Y.; Seelig, G. Dynamic DNA nanotechnology using strand-displacement reactions. Nat. Chem. 2011, 3, 103–113. [Google Scholar] [CrossRef]
- Rangel, A.E.; Hariri, A.A.; Eisenstein, M.; Soh, H.T. Engineering aptamer switches for multifunctional stimulus-responsive nanosystems. Adv. Mater. 2020, 32, 2003704. [Google Scholar] [CrossRef]
- Turek, V.A.; Chikkaraddy, R.; Cormier, S.; Stockham, B.; Ding, T.; Keyser, U.F.; Baumberg, J.J. Thermo-Responsive Actuation of a DNA Origami Flexor. Adv. Funct. Mater. 2018, 28, 1706410. [Google Scholar] [CrossRef]
- Ijäs, H.; Hakaste, I.; Shen, B.; Kostiainen, M.A.; Linko, V. Reconfigurable DNA origami nanocapsule for pH-controlled encapsulation and display of cargo. ACS Nano 2019, 13, 5959–5967. [Google Scholar] [CrossRef]
- Marras, A.E.; Shi, Z.; Lindell III, M.G.; Patton, R.A.; Huang, C.M.; Zhou, L.; Su, H.J.; Arya, G.; Castro, C.E. Cation-activated avidity for rapid reconfiguration of DNA nanodevices. ACS Nano 2018, 12, 9484–9494. [Google Scholar] [CrossRef] [PubMed]
- Kopperger, E.; List, J.; Madhira, S.; Rothfischer, F.; Lamb, D.C.; Simmel, F.C. A self-assembled nanoscale robotic arm controlled by electric fields. Science 2018, 359, 296–301. [Google Scholar] [CrossRef] [PubMed]
- Ijäs, H.; Nummelin, S.; Shen, B.; Kostiainen, M.A.; Linko, V. Dynamic DNA origami devices: From strand-displacement reactions to external-stimuli responsive systems. Int. J. Mol. Sci. 2018, 19, 2114. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekaran, A.R.; Anderson, N.; Kizer, M.; Halvorsen, K.; Wang, X. Beyond the fold: Emerging biological applications of DNA origami. ChemBioChem 2016, 17, 1081–1089. [Google Scholar] [CrossRef]
- McCluskey, J.B.; Clark, D.S.; Glover, D.J. Functional Applications of Nucleic Acid–Protein Hybrid Nanostructures. Trends Biotechnol. 2020, 38, 976–989. [Google Scholar] [CrossRef]
- Jiao, Y.; Shang, Y.; Li, N.; Ding, B. DNA-based enzymatic systems and their applications. Iscience 2022, 25, 104018. [Google Scholar] [CrossRef]
- Jiang, Q.; Liu, S.; Liu, J.; Wang, Z.G.; Ding, B. Rationally designed DNA-origami nanomaterials for drug delivery in vivo. Adv. Mater. 2019, 31, 1804785. [Google Scholar] [CrossRef]
- Tan, Y.Q.; Xue, B.; Yew, W.S. Genetically Encodable Scaffolds for Optimizing Enzyme Function. Molecules 2021, 26, 1389. [Google Scholar] [CrossRef]
- Glettenberg, M.; Niemeyer, C.M. Tuning of peroxidase activity by covalently tethered DNA oligonucleotides. Bioconjugate Chem. 2009, 20, 969–975. [Google Scholar] [CrossRef]
- Rudiuk, S.; Venancio-Marques, A.; Baigl, D. Enhancement and Modulation of Enzymatic Activity through Higher-Order Structural Changes of Giant DNA–Protein Multibranch Conjugates. Angew. Chem. Int. Ed. 2012, 51, 12694–12698. [Google Scholar] [CrossRef]
- Timm, C.; Niemeyer, C.M. Assembly and purification of enzyme-functionalized DNA origami structures. Angew. Chem. Int. Ed. 2015, 54, 6745–6750. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Kim, K.R.; Ahn, D.R.; Lee, J.E.; Yang, E.G.; Kim, S.Y. Reversible regulation of enzyme activity by pH-responsive encapsulation in DNA nanocages. ACS Nano 2017, 11, 9352–9359. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.L.; Wheeldon, I. Kinetic enhancements in DNA–enzyme nanostructures mimic the sabatier principle. ACS Catal. 2013, 3, 560–564. [Google Scholar] [CrossRef]
- Kosinski, R.; Perez, J.M.; Schöneweiß, E.C.; Ruiz-Blanco, Y.B.; Ponzo, I.; Bravo-Rodriguez, K.; Erkelenz, M.; Schlücker, S.; Uhlenbrock, G.; Sanchez-Garcia, E.; et al. The role of DNA nanostructures in the catalytic properties of an allosterically regulated protease. Sci. Adv. 2022, 8, eabk0425. [Google Scholar] [CrossRef]
- Zhao, Z.; Fu, J.; Dhakal, S.; Johnson-Buck, A.; Liu, M.; Zhang, T.; Woodbury, N.W.; Liu, Y.; Walter, N.G.; Yan, H. Nanocaged enzymes with enhanced catalytic activity and increased stability against protease digestion. Nat. Commun. 2016, 7, 10619. [Google Scholar] [CrossRef]
- Zhang, Y.; Tsitkov, S.; Hess, H. Proximity does not contribute to activity enhancement in the glucose oxidase–horseradish peroxidase cascade. Nat. Commun. 2016, 7, 13982. [Google Scholar] [CrossRef]
- Xiong, Y.; Huang, J.; Wang, S.T.; Zafar, S.; Gang, O. Local environment affects the activity of enzymes on a 3D molecular scaffold. ACS Nano 2020, 14, 14646–14654. [Google Scholar] [CrossRef]
- Lin, P.; Dinh, H.; Morita, Y.; Zhang, Z.; Nakata, E.; Kinoshita, M.; Morii, T. Evaluation of the role of the DNA surface for enhancing the activity of scaffolded enzymes. Chem. Commun. 2021, 57, 3925–3928. [Google Scholar] [CrossRef]
- Nakata, E.; Dinh, H.; Ngo, T.A.; Saimura, M.; Morii, T. A modular zinc finger adaptor accelerates the covalent linkage of proteins at specific locations on DNA nanoscaffolds. Chem. Commun. 2015, 51, 1016–1019. [Google Scholar] [CrossRef]
- Ngo, T.A.; Nakata, E.; Saimura, M.; Morii, T. Spatially organized enzymes drive cofactor-coupled cascade reactions. J. Am. Chem. Soc. 2016, 138, 3012–3021. [Google Scholar] [CrossRef]
- Nguyen, T.M.; Nakata, E.; Saimura, M.; Dinh, H.; Morii, T. Design of modular protein tags for orthogonal covalent bond formation at specific DNA sequences. J. Am. Chem. Soc. 2017, 139, 8487–8496. [Google Scholar] [CrossRef]
- Ngo, T.A.; Dinh, H.; Nguyen, T.M.; Liew, F.F.; Nakata, E.; Morii, T. Protein adaptors assemble functional proteins on DNA scaffolds. Chem. Commun. 2019, 55, 12428–12446. [Google Scholar] [CrossRef]
- Nguyen, T.M.; Nakata, E.; Zhang, Z.; Saimura, M.; Dinh, H.; Morii, T. Rational design of a DNA sequence-specific modular protein tag by tuning the alkylation kinetics. Chem. Sci. 2019, 10, 9315–9325. [Google Scholar] [CrossRef]
- Zhang, Z.; Nakata, E.; Dinh, H.; Saimura, M.; Rajendran, A.; Matsuda, K.; Morii, T. Tuning the Reactivity of a Substrate for SNAP-Tag Expands Its Application for Recognition-Driven DNA-Protein Conjugation. Chem. Eur. J. 2021, 27, 18118–18128. [Google Scholar] [CrossRef]
- Dinh, H.; Nakata, E.; Mutsuda-Zapater, K.; Saimura, M.; Kinoshita, M.; Morii, T. Enhanced enzymatic activity exerted by a packed assembly of a single type of enzyme. Chem. Sci. 2020, 11, 9088–9100. [Google Scholar] [CrossRef]
- Zhang, Y.; Hess, H. Toward rational design of high-efficiency enzyme cascades. ACS Catal. 2017, 7, 6018–6027. [Google Scholar] [CrossRef]
- Abdallah, W.; Hong, X.; Banta, S.; Wheeldon, I. Microenvironmental effects can masquerade as substrate channelling in cascade biocatalysis. Curr. Opin. Biotechnol. 2022, 73, 233–239. [Google Scholar] [CrossRef]
- Klein, W.P.; Thomsen, R.P.; Turner, K.B.; Walper, S.A.; Vranish, J.; Kjems, J.; Ancona, M.G.; Medintz, I.L. Enhanced catalysis from multienzyme cascades assembled on a DNA origami triangle. ACS Nano 2019, 13, 13677–13689. [Google Scholar] [CrossRef]
- Lim, S.; Kim, J.; Kim, Y.; Xu, D.; Clark, D.S. CRISPR/Cas-directed programmable assembly of multi-enzyme complexes. Chem. Commun. 2020, 56, 4950–4953. [Google Scholar] [CrossRef]
- Fu, J.; Yang, Y.R.; Johnson-Buck, A.; Liu, M.; Liu, Y.; Walter, N.G.; Woodbury, N.W.; Yan, H. Multi-enzyme complexes on DNA scaffolds capable of substrate channelling with an artificial swinging arm. Nat. Nanotechnol. 2014, 9, 531–536. [Google Scholar] [CrossRef]
- Yang, Y.R.; Fu, J.; Wootten, S.; Qi, X.; Liu, M.; Yan, H.; Liu, Y. 2D enzyme cascade network with efficient substrate channeling by swinging arms. ChemBioChem 2018, 19, 212–216. [Google Scholar] [CrossRef]
- Ke, G.; Liu, M.; Jiang, S.; Qi, X.; Yang, Y.R.; Wootten, S.; Zhang, F.; Zhu, Z.; Liu, Y.; Yang, C.J.; et al. Directional regulation of enzyme pathways through the control of substrate channeling on a DNA origami scaffold. Angew. Chem. Int. Ed. 2016, 55, 7483–7486. [Google Scholar] [CrossRef]
- Chen, Y.; Ke, G.; Ma, Y.; Zhu, Z.; Liu, M.; Liu, Y.; Yan, H.; Yang, C.J. A synthetic light-driven substrate channeling system for precise regulation of enzyme cascade activity based on DNA origami. J. Am. Chem. Soc. 2018, 140, 8990–8996. [Google Scholar] [CrossRef]
- Wilner, O.I.; Weizmann, Y.; Gill, R.; Lioubashevski, O.; Freeman, R.; Willner, I. Enzyme cascades activated on topologically programmed DNA scaffolds. Nat. Nanotechnol. 2009, 4, 249–254. [Google Scholar] [CrossRef]
- Fu, J.; Liu, M.; Liu, Y.; Woodbury, N.W.; Yan, H. Interenzyme substrate diffusion for an enzyme cascade organized on spatially addressable DNA nanostructures. J. Am. Chem. Soc. 2012, 134, 5516–5519. [Google Scholar] [CrossRef]
- Xin, L.; Zhou, C.; Yang, Z.; Liu, D. Regulation of an enzyme cascade reaction by a DNA machine. Small 2013, 9, 3088–3091. [Google Scholar] [CrossRef]
- Idan, O.; Hess, H. Origins of activity enhancement in enzyme cascades on scaffolds. ACS Nano 2013, 7, 8658–8665. [Google Scholar] [CrossRef]
- Kuzmak, A.; Carmali, S.; von Lieres, E.; Russell, A.J.; Kondrat, S. Can enzyme proximity accelerate cascade reactions? Sci. Rep. 2019, 9, 1–7. [Google Scholar] [CrossRef]
- Cao, Y.; Li, X.; Xiong, J.; Wang, L.; Yan, L.T.; Ge, J. Investigating the origin of high efficiency in confined multienzyme catalysis. Nanoscale 2019, 11, 22108–22117. [Google Scholar] [CrossRef]
- Dinh, H.; Nakata, E.; Lin, P.; Saimura, M.; Ashida, H.; Morii, T. Reaction of ribulose biphosphate carboxylase/oxygenase assembled on a DNA scaffold. Bioorg. Med. Chem. 2019, 27, 115120. [Google Scholar] [CrossRef]
- Ngo, T.A.; Nakata, E.; Saimura, M.; Kodaki, T.; Morii, T. A protein adaptor to locate a functional protein dimer on molecular switchboard. Methods 2014, 67, 142–150. [Google Scholar] [CrossRef] [PubMed]
- Lin, P.; Dinh, H.; Nakata, E.; Morii, T. Conditional dependence of enzyme cascade reaction efficiency on the inter-enzyme distance. Chem. Commun. 2021, 57, 11197–11200. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Fu, J.; Qi, X.; Wootten, S.; Woodbury, N.W.; Liu, Y.; Yan, H. A Three-Enzyme Pathway with an Optimised Geometric Arrangement to Facilitate Substrate Transfer. ChemBioChem 2016, 17, 1097–1101. [Google Scholar] [CrossRef] [PubMed]
- Kahn, J.S.; Xiong, Y.; Huang, J.; Gang, O. Cascaded Enzyme Reactions over a Three-Dimensional, Wireframe DNA Origami Scaffold. JACS Au 2022, 2, 357–366. [Google Scholar] [CrossRef]
- Chado, G.R.; Stoykovich, M.P.; Kaar, J.L. Role of dimension and spatial arrangement on the activity of biocatalytic cascade reactions on scaffolds. ACS Catal. 2016, 6, 5161–5169. [Google Scholar] [CrossRef]
- Rossetti, M.; Bertucci, A.; Patiño, T.; Baranda, L.; Porchetta, A. Programming DNA-based systems through effective molarity enforced by biomolecular confinement. Chem. Eur. J. 2020, 26, 9826–9834. [Google Scholar] [CrossRef]
- Fu, Y.; Zeng, D.; Chao, J.; Jin, Y.; Zhang, Z.; Liu, H.; Li, D.; Ma, H.; Huang, Q.; Gothelf, K.V.; et al. Single-step rapid assembly of DNA origami nanostructures for addressable nanoscale bioreactors. J. Am. Chem. Soc. 2013, 135, 696–702. [Google Scholar] [CrossRef]
- Linko, V.; Eerikäinen, M.; Kostiainen, M.A. A modular DNA origami-based enzyme cascade nanoreactor. Chem. Commun. 2015, 51, 5351–5354. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, P.; Yang, H.; Nakata, E.; Morii, T. Mechanistic Aspects for the Modulation of Enzyme Reactions on the DNA Scaffold. Molecules 2022, 27, 6309. https://doi.org/10.3390/molecules27196309
Lin P, Yang H, Nakata E, Morii T. Mechanistic Aspects for the Modulation of Enzyme Reactions on the DNA Scaffold. Molecules. 2022; 27(19):6309. https://doi.org/10.3390/molecules27196309
Chicago/Turabian StyleLin, Peng, Hui Yang, Eiji Nakata, and Takashi Morii. 2022. "Mechanistic Aspects for the Modulation of Enzyme Reactions on the DNA Scaffold" Molecules 27, no. 19: 6309. https://doi.org/10.3390/molecules27196309
APA StyleLin, P., Yang, H., Nakata, E., & Morii, T. (2022). Mechanistic Aspects for the Modulation of Enzyme Reactions on the DNA Scaffold. Molecules, 27(19), 6309. https://doi.org/10.3390/molecules27196309








