Binding of Vialinin A and p-Terphenyl Derivatives to Ubiquitin-Specific Protease 4 (USP4): A Molecular Docking Study
Abstract
1. Introduction
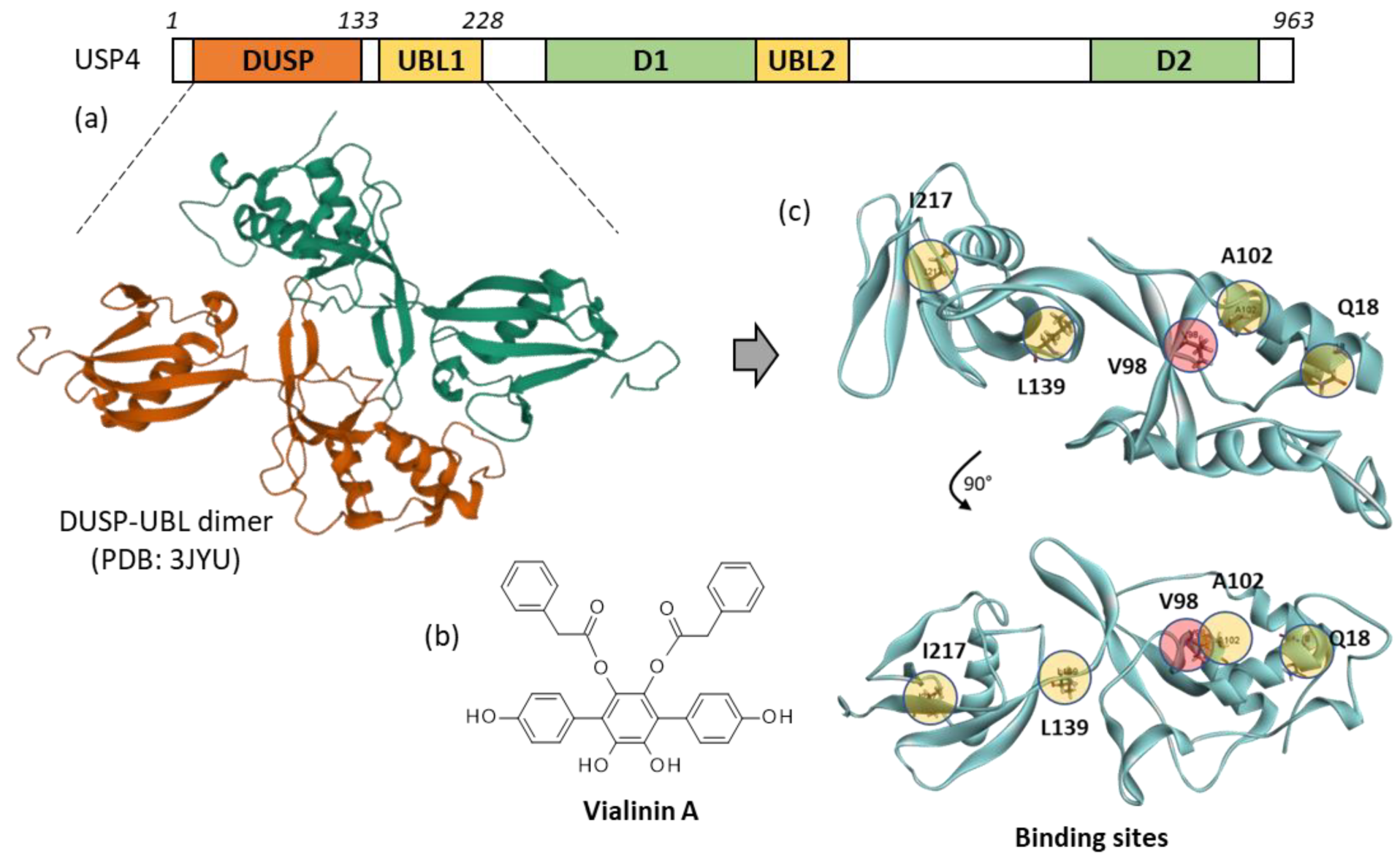
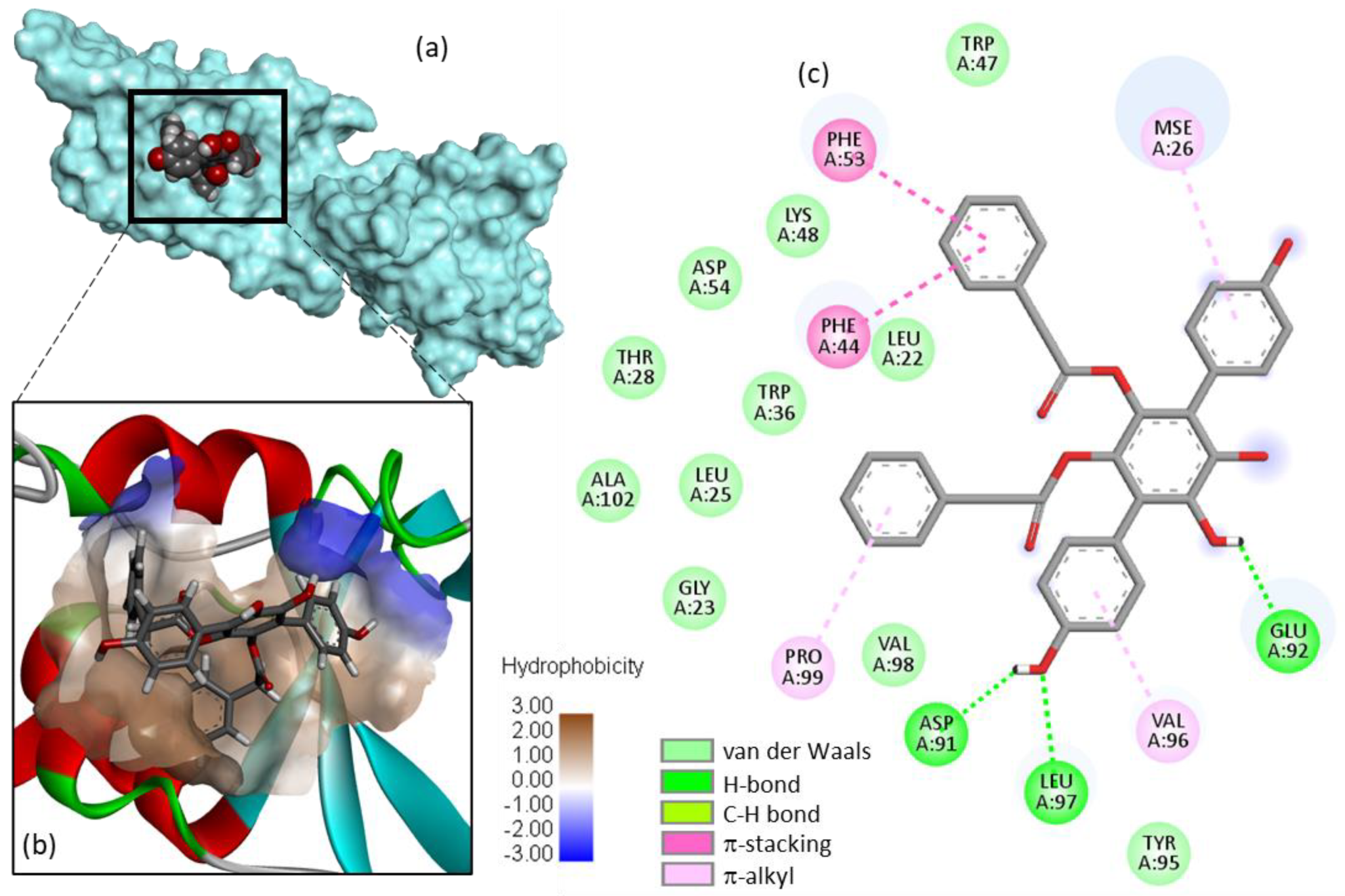
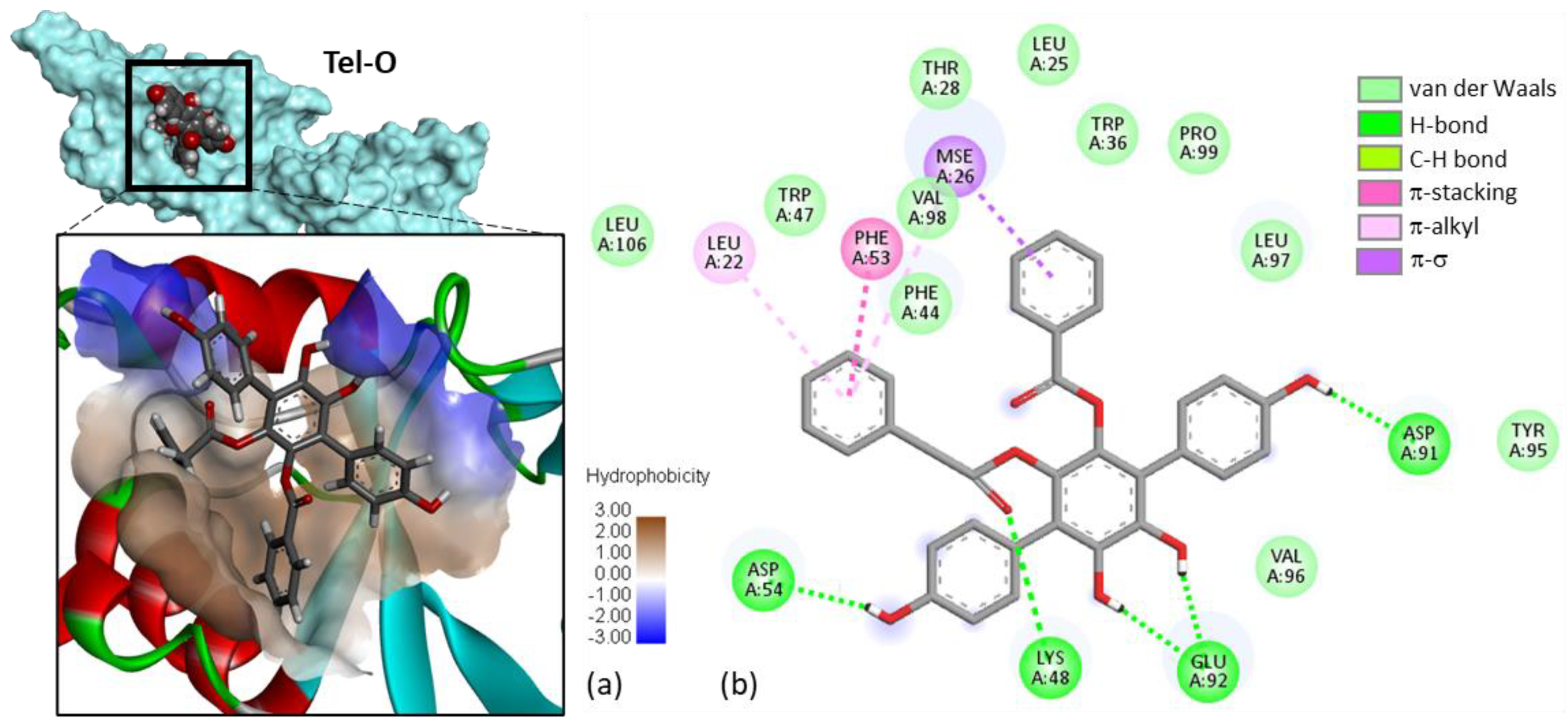
2. Results
3. Discussion and Perspectives
4. Materials and Methods
4.1. Molecular Structures and Software
4.2. In Silico Molecular Docking Procedure
- (1)
- Monte Carlo (MC) conformational search of the ligand using the BOSS (Biochemical and Organic Simulation System) software, freely available to academic users. The structure of each ligand was optimized using a classical MC conformational search procedure, as described in BOSS [52]. A conformational analysis has been performed to define the best starting geometries for each compound. An energy minimization was carried out to identify all minimum-energy conformers, leading to the identification of a unique conformer for the free ligand. Within BOSS, MC simulations were performed in the constant-temperature and constant-pressure ensemble (NPT).
- (2)
- Evaluation of the free energy of hydration for the chosen structure of the ligand. The molecular mechanics/generalized Born surface area (MM/GBSA) procedure was used to evaluate the free energies of hydration (ΔG) [53]. ΔG is directly related to solubility of the compound [54]. MC search and computation of ΔG were performed within BOSS using the xMCGB script according to procedures given in references [53,55]. The best ligand structure is then used in the docking procedure.
- (3)
- Determination of the protein-ligand site of interaction. Potential Mpro-binding sites for the different products were searched using the web server CASTp 3.0 (Computed Atlas of Surface Topography of proteins) and visualized with the molecular modeling system Chimera 1.15 [56,57]. Shape complementarity and geometry considerations favor a docking grid centered in the volume defined by the central amino acid. Within the binding site, the side chains of the specific amino acids were considered fully flexible during docking. With the 3JYU structure, based on shape complementarity criteria, the best USP4 binding site for the ligands has been defined around amino acid residue Val98 (see Results below) and the flexible amino acids are Trp43, Phe44, Trp47, Lys48, Phe53, Glu92, Val96, Val98, Trp103, and Trp109.
- (4)
- Docking procedure using GOLD. In our typical docking process, 100 energetically reasonable poses (according to the ChemPLP scoring function) are retained while searching for the correct binding mode of the ligand. The decision to maintain a trial pose is based on ranked poses, using the PLP fitness scoring function (which is the default in GOLD version 5.3 used here) [58]. Six poses are kept. The empirical potential energy of the interaction ΔE for the ranked complexes was evaluated using the simple expression ΔE(interaction) = E(complex) − [E(protein) + E(ligand)]. Calculations of the final energy are performed on the basis of the SPASIBA spectroscopic force field. The corresponding parameters are derived from vibrational wavenumbers obtained in the infrared and Raman spectra of a large series of compounds including organic molecules, amino-acids, saccharides, nucleic acids and lipids.
- (5)
- Validation using the SPASIBA force field. This last step is considered essential to define the best protein-ligand structure. The spectroscopic SPASIBA (Spectroscopic Potential Algorithm for Simulating Biomolecular conformational Adaptability) force field has been specifically developed to provide refined empirical molecular mechanics force field parameters [59]. It is a spectroscopic molecular-mechanics-derived empirical potential function first devoted to proteins and extended to a large variety of chemical groups (including amino acids, saccharides, and phospholipids). This force field is a very appropriate tool for molecular mechanics which is able to reproduce at the same time structures, energies and vibrational spectra with a much higher precision on the latter than commonly available force fields. SPASIBA introduces the Urey–Bradley F quadratic force constant related to the 1–3 nonbonded distance in a bond angle, solves the redundancy problem among internal coordinates, and adds specific parameters to correctly fit spectra [60,61]. SPASIBA (integrated into CHARMM) [62] has been shown to be excellent for reproducing crystal phase infrared data. The same procedure was used to establish molecular models for the various natural products listed in Table 1, including the reference product Vi-A.
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Hou, X.; Zhu, F.; Ni, Y.; Chen, T.; Du, J.; Liu, X.; Han, Y.; Liu, Y.; Du, W.; Li, Y.; et al. USP4 is pathogenic in allergic airway inflammation by inhibiting regulatory T cell response. Life Sci. 2021, 281, 119720. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Zhang, D.; Zhao, K.; Wang, Y.; Pei, L.; Fu, Q.; Ma, X. Spotlight on USP4: Structure, Function, and Regulation. Front. Cell. Dev. Biol. 2021, 9, 595159. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhou, L.; Lu, J.; Jiang, B.; Liu, C.; Guo, J. USP4 function and multifaceted roles in cancer: A possible and potential therapeutic target. Cancer Cell Int. 2020, 20, 298. [Google Scholar] [CrossRef] [PubMed]
- Lin, R.; Nie, J.; Ren, J.; Liang, R.; Li, D.; Wang, P.; Gao, C.; Zhuo, C.; Yang, C.; Li, B. USP4 interacts and positively regulates IRF8 function via K48-linked deubiquitination in regulatory T cells. FEBS Lett. 2017, 591, 1677–1686. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Xu, P.; Ge, S.; Zhang, C.; Zheng, X.; Xu, J.; Liu, Z.; Li, B.; Ge, S. Ubiquitin specific peptidase 4 stabilizes interferon regulatory factor protein and promotes its function to facilitate interleukin-4 expression in T helper type 2 cells. Int. J. Mol. Med. 2017, 40, 979–986. [Google Scholar] [CrossRef]
- Kwon, S.K.; Kim, E.H.; Baek, K.H. RNPS1 is modulated by ubiquitin-specific protease 4. FEBS Lett. 2017, 591, 369–381. [Google Scholar] [CrossRef]
- Milojevic, T.; Reiterer, V.; Stefan, E.; Korkhov, V.M.; Dorostkar, M.M.; Ducza, E.; Ogris, E.; Boehm, S.; Freissmuth, M.; Nanoff, C. The ubiquitin-specific protease Usp4 regulates the cell surface level of the A2A receptor. Mol. Pharmacol. 2006, 69, 1083–1094. [Google Scholar] [CrossRef]
- Das, T.; Kim, E.E.; Song, E.J. Phosphorylation of USP15 and USP4 Regulates Localization and Spliceosomal Deubiquitination. J. Mol. Biol. 2019, 431, 3900–3912. [Google Scholar] [CrossRef]
- Das, T.; Lee, E.Y.; You, H.J.; Kim, E.E.; Song, E.J. USP15 and USP4 facilitate lung cancer cell proliferation by regulating the alternative splicing of SRSF1. Cell Death Discov. 2022, 8, 24. [Google Scholar] [CrossRef]
- Sarri, N.; Wang, K.; Tsioumpekou, M.; Castillejo-López, C.; Lennartsson, J.; Heldin, C.H.; Papadopoulos, N. Deubiquitinating enzymes USP4 and USP17 finetune the trafficking of PDGFRβ and affect PDGF-BB-induced STAT3 signalling. Cell. Mol. Life Sci. 2022, 79, 85. [Google Scholar] [CrossRef]
- Li, F.; Hu, Q.; He, T.; Xu, J.; Yi, Y.; Xie, S.; Ding, L.; Fu, M.; Guo, R.; Xiao, Z.J.; et al. The Deubiquitinase USP4 Stabilizes Twist1 Protein to Promote Lung Cancer Cell Stemness. Cancers 2020, 12, 1582. [Google Scholar] [CrossRef] [PubMed]
- Murali, S.K.; Aroankins, T.S.; Moeller, H.B.; Fenton, R.A. The Deubiquitylase USP4 Interacts with the Water Channel AQP2 to Modulate Its Apical Membrane Accumulation and Cellular Abundance. Cells 2019, 8, 265. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Wang, Y.; Wang, X.; Lin, L.; Wang, P.; Sun, J.; Jiang, L. The Effects of the Transforming Growth Factor-β1 (TGF-β1) Signaling Pathway on Cell Proliferation and Cell Migration are Mediated by Ubiquitin Specific Protease 4 (USP4) in Hypertrophic Scar Tissue and Primary Fibroblast Cultures. Med. Sci. Monit. 2020, 26, 920736. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Xing, M.; Wang, X.; Cao, W.; Wang, H. MiR-148a suppresses invasion and induces apoptosis of breast cancer cells by regulating USP4 and BIM expression. Int. J. Clin. Exp. Pathol. 2017, 10, 8361–8368. [Google Scholar] [PubMed]
- Liang, Y.; Song, X.; Li, Y.; Ma, T.; Su, P.; Guo, R.; Chen, B.; Zhang, H.; Sang, Y.; Liu, Y.; et al. Targeting the circBMPR2/miR-553/USP4 Axis as a Potent Therapeutic Approach for Breast Cancer. Mol. Ther. Nucleic Acids. 2019, 17, 347–361. [Google Scholar] [CrossRef]
- Geng, N.; Li, Y.; Zhang, W.; Wang, F.; Wang, X.; Jin, Z.; Xing, Y.; Li, D.; Zhang, H.; Li, Y.; et al. A PAK5-DNPEP-USP4 axis dictates breast cancer growth and metastasis. Int. J. Cancer. 2020, 146, 1139–1151. [Google Scholar] [CrossRef]
- Nguyen, H.H.; Kim, T.; Nguyen, T.; Hahn, M.J.; Yun, S.I.; Kim, K.K. A Selective Inhibitor of Ubiquitin-Specific Protease 4 Suppresses Colorectal Cancer Progression by Regulating β-Catenin Signaling. Cell. Physiol. Biochem. 2019, 53, 157–171. [Google Scholar]
- Lai, C.Y.; Yeh, D.W.; Lu, C.H.; Liu, Y.L.; Chuang, Y.C.; Ruan, J.W.; Kao, C.Y.; Huang, L.R.; Chuang, T.H. Epigenetic Silencing of Ubiquitin Specific Protease 4 by Snail1 Contributes to Macrophage-Dependent Inflammation and Therapeutic Resistance in Lung Cancer. Cancers 2020, 12, 148. [Google Scholar] [CrossRef]
- Zhou, Y.; Liang, P.; Ji, W.; Yu, Z.; Chen, H.; Jiang, L. Ubiquitin-specific protease 4 promotes glioblastoma multiforme via activating ERK pathway. Onco Targets Ther. 2019, 12, 1825–1839. [Google Scholar] [CrossRef]
- Qin, N.; Han, F.; Li, L.; Ge, Y.; Lin, W.; Wang, J.; Wu, L.; Zhao, G.; Deng, Y.; Zhang, J. Deubiquitinating enzyme 4 facilitates chemoresistance in glioblastoma by inhibiting P53 activity. Oncol Lett. 2019, 17, 958–964. [Google Scholar] [CrossRef]
- Ward, J.A.; McLellan, L.; Stockley, M.; Gibson, K.R.; Whitlock, G.A.; Knights, C.; Harrigan, J.A.; Jacq, X.; Tate, E.W. Quantitative Chemical Proteomic Profiling of Ubiquitin Specific Proteases in Intact Cancer Cells. ACS Chem. Biol. 2016, 11, 3268–3272. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Koshino, H.; Esumi, Y.; Takahashi, S.; Yoshikawa, K.; Abe, N. Vialinin A, a novel 2,2-diphenyl-1-picrylhydrazyl (DPPH) radical scavenger from an edible mushroom in China. Biosci. Biotechnol. Biochem. 2005, 69, 2326–2332. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Onose, J.; Xie, C.; Ye, Y.Q.; Sugaya, K.; Takahashi, S.; Koshino, H.; Yasunaga, K.; Abe, N.; Yoshikawa, K. Vialinin A, a novel potent inhibitor of TNF-alpha production from RBL-2H3 cells. Biol. Pharm. Bull. 2008, 31, 831–833. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Okada, K.; Ye, Y.Q.; Taniguchi, K.; Yoshida, A.; Akiyama, T.; Yoshioka, Y.; Onose, J.; Koshino, H.; Takahashi, S.; Yajima, A.; et al. Vialinin A is a ubiquitin-specific peptidase inhibitor. Bioorg. Med. Chem. Lett. 2013, 23, 4328–4331. [Google Scholar] [CrossRef] [PubMed]
- Yoshioka, Y.; Ye, Y.Q.; Okada, K.; Taniguchi, K.; Yoshida, A.; Sugaya, K.; Onose, J.; Koshino, H.; Takahashi, S.; Yajima, A.; et al. Ubiquitin-specific peptidase 5, a target molecule of vialinin A; is a key molecule of TNF-α production in RBL-2H3 cells. PLoS ONE 2013, 8, e80931. [Google Scholar] [CrossRef]
- Ye, Y.Q.; Negishi, C.; Hongo, Y.; Koshino, H.; Onose, J.; Abe, N.; Takahashi, S. Structural elucidation and synthesis of vialinin C, a new inhibitor of TNF-alpha production. Bioorg. Med. Chem. 2014, 22, 2442–2446. [Google Scholar] [CrossRef]
- Ye, Y.Q.; Koshino, H.; Onose, J.; Yoshikawa, K.; Abe, N.; Takahashi, S. Studies on natural p-terphenyls: Total syntheses of vialinin A and terrestrin B. Biosci. Biotechnol. Biochem. 2010, 74, 140–146. [Google Scholar] [CrossRef]
- Ye, Y.Q.; Koshino, H.; Onose, J.; Yoshikawa, K.; Abe, N.; Takahashi, S. Expeditious synthesis of vialinin B, an extremely potent inhibitor of TNF-alpha production. Org. Lett. 2009, 11, 5074–5077. [Google Scholar] [CrossRef]
- Fujiwara, K.; Sato, T.; Sano, Y.; Norikura, T.; Katoono, R.; Suzuki, T.; Matsue, H. Total synthesis of thelephantin O, vialinin A/terrestrin A, and terrestrins B-D. J. Org. Chem. 2012, 77, 5161–5166. [Google Scholar] [CrossRef]
- Onose, J.; Yoshioka, Y.; Ye, Y.Q.; Sugaya, K.; Yajima, A.; Taniguchi, K.; Okada, K.; Yajima, S.; Takahashi, S.; Koshino, H.; et al. Inhibitory effects of vialinin A and its analog on tumor necrosis factor-alpha release and production from RBL-2H3 cells. Cell Immunol. 2012, 279, 140–144. [Google Scholar] [CrossRef]
- Xu, J.; Chen, D.; Jin, L.; Chen, Z.; Tu, Y.; Huang, X.; Xue, F.; Xu, J.; Chen, M.; Wang, X.; et al. Ubiquitously specific protease 4 inhibitor-Vialinin A attenuates inflammation and fibrosis in S100-induced hepatitis mice through Rheb/mTOR signalling. J. Cell. Mol. Med. 2021, 25, 1140–1150. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Na, S.; Pan, S.; Long, S.; Xin, Y.; Jiang, Q.; Lai, Z.; Yan, J.; Cao, Z. Inhibition of USP4 attenuates pathological scarring by downregulation of the TGF-β/Smad signaling pathway. Mol. Med. Rep. 2019, 20, 1429–1435. [Google Scholar] [CrossRef] [PubMed]
- Zhou, G.; Zhu, T.; Che, Q.; Zhang, G.; Li, D. Structural diversity and biological activity of natural p-terphenyls. Mar. Life Sci. Technol. 2022, 4, 62–73. [Google Scholar] [CrossRef]
- Bailly, C. Anti-inflammatory and anticancer p-terphenyl derivatives from fungi of the genus Thelephora. Bioorg. Med. Chem. 2022, 70, 116935. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Koshino, H.; Esumi, Y.; Onose, J.; Yoshikawa, K.; Abe, N. Vialinin B, a novel potent inhibitor of TNF-alpha production, isolated from an edible mushroom, Thelephora vialis. Bioorg. Med. Chem. Lett. 2006, 16, 5424–5426. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Li, X.B.; Lou, H.X. Structural and biological diversity of natural p-terphenyls. J. Asian Nat. Prod. Res. 2018, 20, 1–13. [Google Scholar] [CrossRef]
- Wang, S.M.; Han, J.J.; Ma, K.; Jin, T.; Bao, L.; Pei, Y.F.; Liu, H.W. New α-glucosidase inhibitors with p-terphenyl skeleton from the mushroom Hydnellum concrescens. Fitoterapia 2014, 98, 149–155. [Google Scholar] [CrossRef]
- Hu, L.; Liu, J.K. p-Terphenyls from the basidiomycete Thelephora aurantiotincta. Z. Naturforsch. C J Biosci. 2003, 58, 452–454. [Google Scholar]
- Clerici, M.; Luna-Vargas, M.P.; Faesen, A.C.; Sixma, T.K. The DUSP-Ubl domain of USP4 enhances its catalytic efficiency by promoting ubiquitin exchange. Nat. Commun. 2014, 5, 5399. [Google Scholar] [CrossRef]
- Quang, D.N.; Hashimoto, T.; Nukada, M.; Yamamoto, I.; Hitaka, Y.; Tanaka, M.; Asakawa, Y. Thelephantins A, B and C: Three benzoyl p-terphenyl derivatives from the inedible mushroom Thelephora aurantiotincta. Phytochemistry 2003, 62, 109–113. [Google Scholar] [CrossRef]
- Ngoc Quang, D.; Hashimoto, T.; Hitaka, Y.; Tanaka, M.; Nukada, M.; Yamamoto, I.; Asakawa, Y. Thelephantins D-H: Five p-terphenyl derivatives from the inedible mushroom Thelephora aurantiotincta. Phytochemistry 2003, 63, 919–924. [Google Scholar] [CrossRef]
- Norikura, T.; Fujiwara, K.; Narita, T.; Yamaguchi, S.; Morinaga, Y.; Iwai, K.; Matsue, H. Anticancer activities of thelephantin O and vialinin A isolated from Thelephora aurantiotincta. J. Agric. Food Chem. 2011, 59, 6974–6979. [Google Scholar] [CrossRef] [PubMed]
- Norikura, T.; Fujiwara, K.; Yanai, T.; Sano, Y.; Sato, T.; Tsunoda, T.; Kushibe, K.; Todate, A.; Morinaga, Y.; Iwai, K.; et al. p-terphenyl derivatives from the mushroom Thelephora aurantiotincta suppress the proliferation of human hepatocellular carcinoma cells via iron chelation. J. Agric. Food Chem. 2013, 61, 1258–1264. [Google Scholar] [CrossRef] [PubMed]
- Gregoire-Mitha, S.; Gray, D.A. What deubiquitinating enzymes, oncogenes, and tumor suppressors actually do: Are current assumptions supported by patient outcomes? Bioessays 2021, 43, 2000269. [Google Scholar] [CrossRef]
- Hwang, S.J.; Lee, H.W.; Kim, H.R.; Lee, H.; Shin, C.H.; Yun, S.I.; Lee, D.H.; Kim, D.H.; Kim, K.K.; Joo, K.M.; et al. Ubiquitin-specific protease 4 controls metastatic potential through β-catenin stabilization in brain metastatic lung adenocarcinoma. Sci. Rep. 2016, 6, 21596. [Google Scholar] [CrossRef]
- Ye, Y.Q.; Onose, J.; Abe, N.; Koshino, H.; Takahashi, S. Design and synthesis of a vialinin A analog with a potent inhibitory activity of TNF-α production and its transformation into a couple of bioprobes. Bioorg. Med. Chem. Lett. 2012, 22, 2385–2387. [Google Scholar] [CrossRef]
- Sonowal, H.; Shukla, K.; Kota, S.; Saxena, A.; Ramana, K.V. Vialinin A, an Edible Mushroom-Derived p-Terphenyl Antioxidant, Prevents VEGF-Induced Neovascularization In Vitro and In Vivo. Oxid. Med. Cell. Longev. 2018, 2018, 1052102. [Google Scholar] [CrossRef]
- Zhang, M.; Chang, W.; Shi, H.; Li, Y.; Zheng, S.; Li, W.; Lou, H. Floricolin C elicits intracellular reactive oxygen species accumulation and disrupts mitochondria to exert fungicidal action. FEMS Yeast Res. 2018, 18, 1–6. [Google Scholar] [CrossRef]
- Lu, Z.; Xiao, P.; Zhou, Y.; Li, Z.; Yu, X.; Sun, J.; Shen, Y.; Zhao, B. Identification of HN252 as a potent inhibitor of protein phosphatase PPM1B. J. Cell. Mol. Med. 2020, 24, 13463–13471. [Google Scholar] [CrossRef]
- Chen, W.; Zhang, J.; Qi, X.; Zhao, K.; Pang, X.; Lin, X.; Liao, S.; Yang, B.; Zhou, X.; Liu, S.; et al. p-Terphenyls as Anti-HSV-1/2 Agents from a Deep-Sea-Derived Penicillium sp. J. Nat. Prod. 2021, 84, 2822–2831. [Google Scholar] [CrossRef]
- Spiliotopoulos, A.; Blokpoel Ferreras, L.; Densham, R.M.; Caulton, S.G.; Maddison, B.C.; Morris, J.R.; Dixon, J.E.; Gough, K.C.; Dreveny, I. Discovery of peptide ligands targeting a specific ubiquitin-like domain-binding site in the deubiquitinase USP11. J. Biol. Chem. 2019, 294, 424–436. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, W.L.; Tirado-Rives, J. Monte Carlo versus Molecular Dynamics for conformational sampling. J. Phys. Chem. 1996, 100, 14508–14513. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Tirado-Rives, J. Molecular modeling of organic and biomolecular systems using BOSS and MCPRO. J. Comput. Chem. 2005, 26, 1689–1700. [Google Scholar] [CrossRef] [PubMed]
- Zafar, A.; Reynisson, J. Hydration free energy as a molecular descriptor in drug design: A feasibility study. Mol. Inform. 2016, 35, 207–214. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Ulmschneider, J.P.; Tirado-Rives, J. Free energies of hydration from a generalized Born model and an ALL-atom force field. J. Phys. Chem. B 2004, 108, 16264–16270. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Tian, W.; Chen, C.; Lei, X.; Zhao, J.; Liang, J. CASTp 3.0: Computed atlas of surface topography of proteins. Nucleic Acids Res. 2018, 46, W363–W367. [Google Scholar] [CrossRef]
- Jones, G.; Willett, P.; Glen, R.C.; Leach, A.R.; Taylor, R. Development and validation of a genetic algorithm for flexible docking. J. Mol. Biol. 1997, 267, 727–748. [Google Scholar] [CrossRef]
- Meziane-Tani, M.; Lagant, P.; Semmoud, A.; Vergoten, G. The SPASIBA force field for chondroitin sulfate: Vibrational analysis of D-glucuronic and N-acetyl-D-galactosamine 4-sulfate sodium salts. J. Phys. Chem. A 2006, 110, 11359–11370. [Google Scholar] [CrossRef]
- Vergoten, G.; Mazur, I.; Lagant, P.; Michalski, J.C.; Zanetta, J.P. The SPASIBA force field as an essential tool for studying the structure and dynamics of saccharides. Biochimie 2003, 85, 65–73. [Google Scholar] [CrossRef]
- Lagant, P.; Nolde, D.; Stote, R.; Vergoten, G.; Karplus, M. Increasing Normal Modes Analysis Accuracy: The SPASIBA Spectroscopic Force Field Introduced into the CHARMM Program. J. Phys. Chem. A 2004, 108, 4019–4029. [Google Scholar] [CrossRef]
- Homans, S.W. A molecular mechanical force field for the conformational analysis of oligosaccharides: Comparison of theoretical and crystal structures of Man alpha 1-3Man beta 1-4GlcNAc. Biochemistry 1990, 29, 9110–9118. [Google Scholar] [CrossRef] [PubMed]

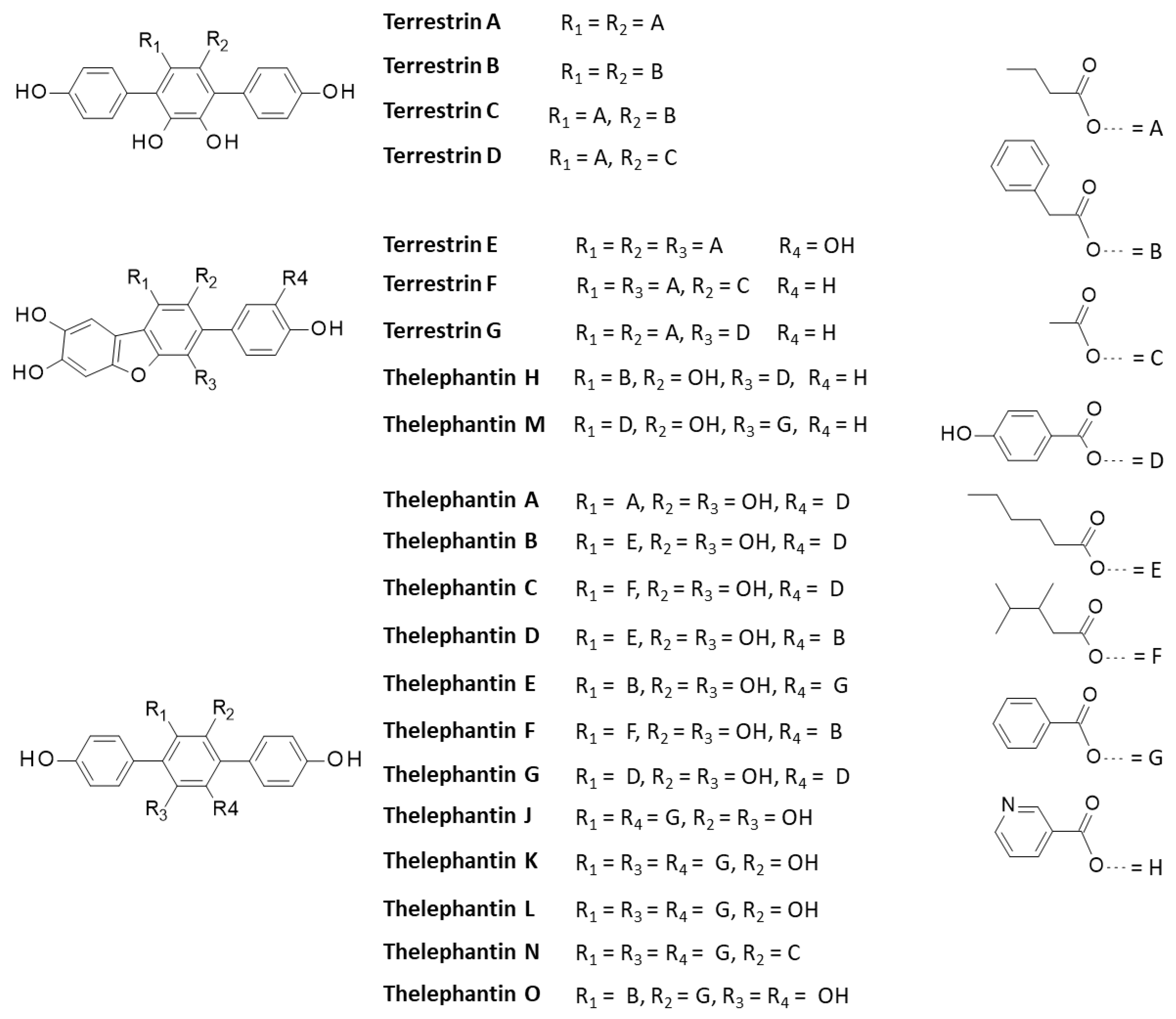
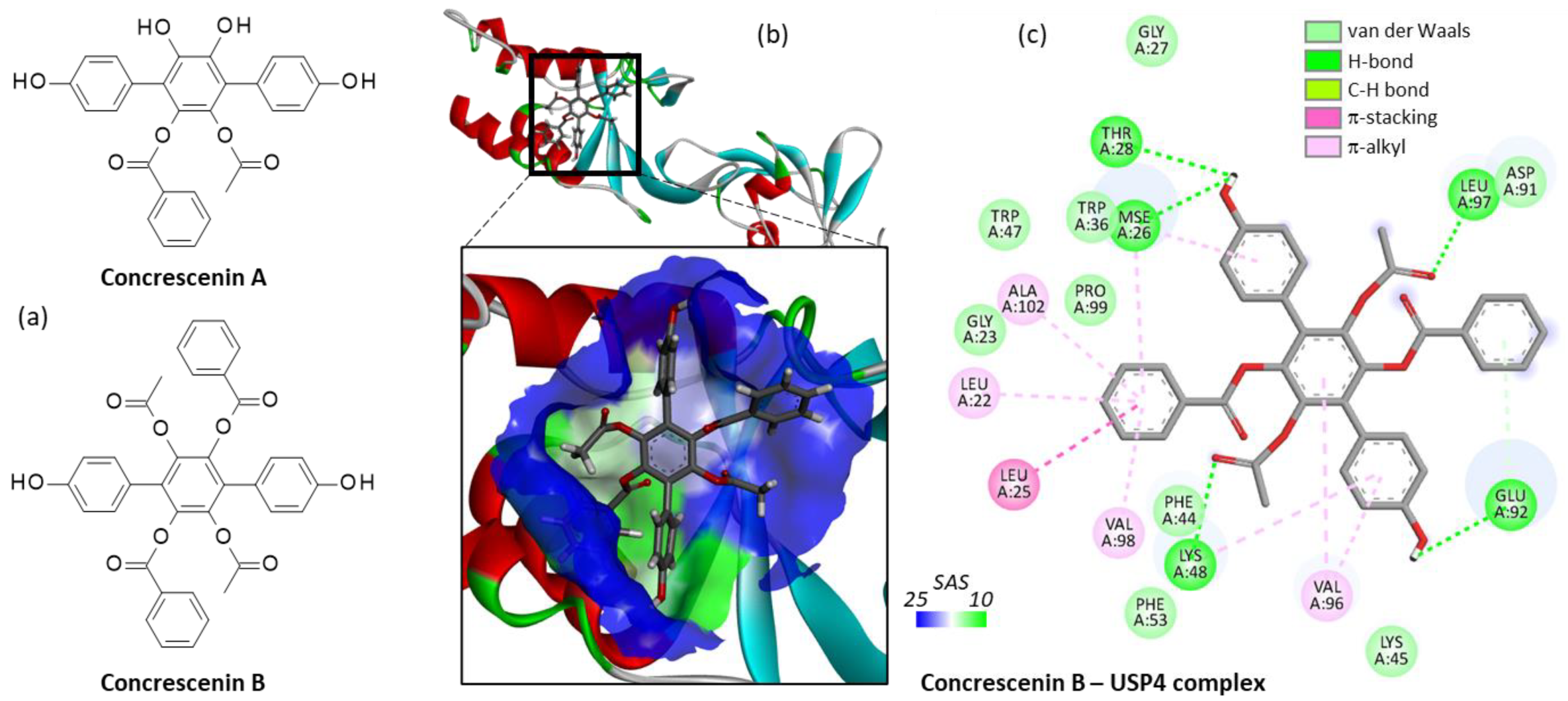
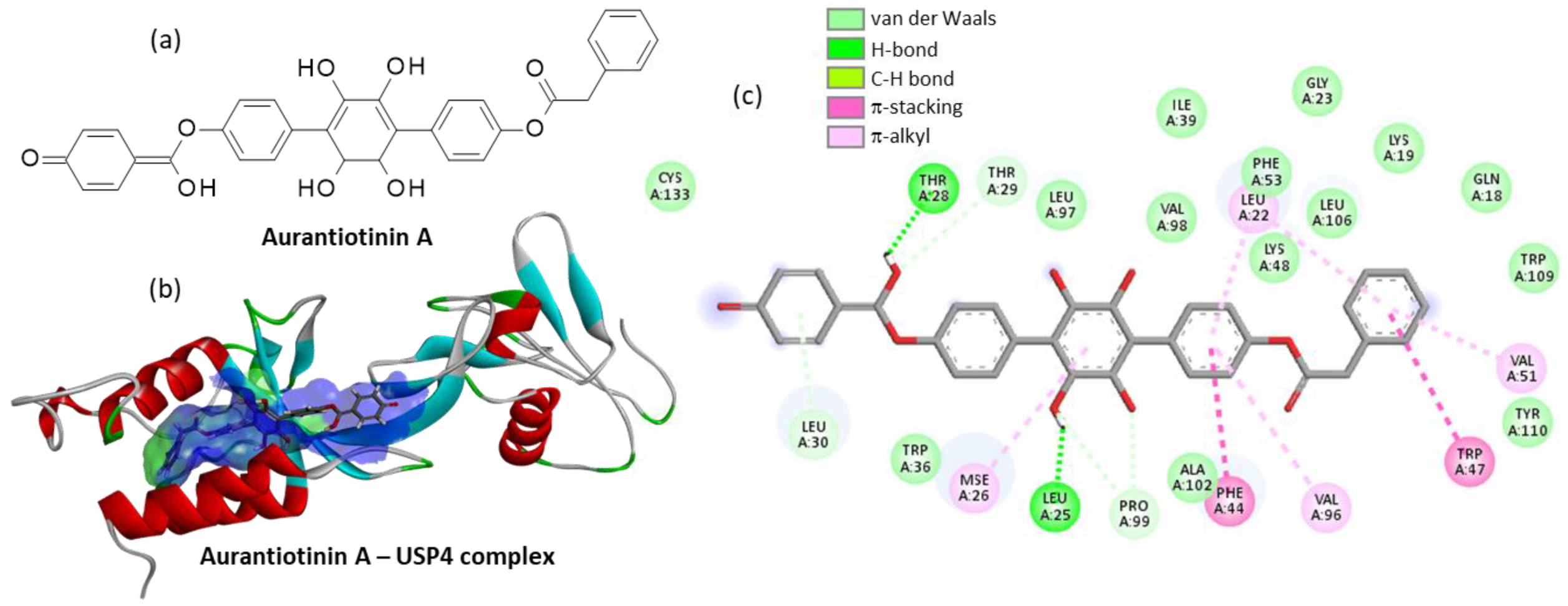
| Compound | CID#a | ΔE |
|---|---|---|
| Atromentin | 99148 | −74.93 |
| Aurantiotinin A | (*) | −118.40 |
| Concrescenin A | 139584566 | −76.79 |
| Concrescenin B | 139587861 | −109.22 |
| DMT | (*) | −64.45 |
| Terferol | 167947 | −79.50 |
| Terphenyllin | 100437 | −75.24 |
| Terrestrin B | 60202116 | −87.55 |
| Terrestrin C | 101743853 | −88.55 |
| Terrestrin D | 60201958 | −89.62 |
| Terrestrin E | 101743854 | −62.05 |
| Terrestrin F | 101743855 | −102.70 |
| Terrestrin G | 101743856 | −103.65 |
| Thelephantin A | 5321927 | −79.97 |
| Thelephantin B | 5321928 | −92.13 |
| Thelephantin C | 5321929 | −76.47 |
| Thelephantin D | 101260622 | −96.31 |
| Thelephantin E | 139583228 | −78.31 |
| Thelephantin F | 10126023 | −103.24 |
| Thelephantin G | 101245355 | −87.40 |
| Thelephantin H | 101260624 | −102.10 |
| Thelephantin I | 21577381 | −82.80 |
| Thelephantin J | 21577382 | −72.04 |
| Thelephantin K | 21577383 | −84.50 |
| Thelephantin L | 21577384 | −70.92 |
| Thelephantin M | (*) | −104.40 |
| Thelephantin N | 21577386 | −92.40 |
| Thelephantin O | 53375718 | −105.70 |
| Thelephorin A | 10325564 | −98.47 |
| Vialinin A | 11563133 | −108.06 |
| Vialinin B | 16049791 | −116.76 |
| Vialinin C | 86279343 | −106.26 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bailly, C.; Vergoten, G. Binding of Vialinin A and p-Terphenyl Derivatives to Ubiquitin-Specific Protease 4 (USP4): A Molecular Docking Study. Molecules 2022, 27, 5909. https://doi.org/10.3390/molecules27185909
Bailly C, Vergoten G. Binding of Vialinin A and p-Terphenyl Derivatives to Ubiquitin-Specific Protease 4 (USP4): A Molecular Docking Study. Molecules. 2022; 27(18):5909. https://doi.org/10.3390/molecules27185909
Chicago/Turabian StyleBailly, Christian, and Gérard Vergoten. 2022. "Binding of Vialinin A and p-Terphenyl Derivatives to Ubiquitin-Specific Protease 4 (USP4): A Molecular Docking Study" Molecules 27, no. 18: 5909. https://doi.org/10.3390/molecules27185909
APA StyleBailly, C., & Vergoten, G. (2022). Binding of Vialinin A and p-Terphenyl Derivatives to Ubiquitin-Specific Protease 4 (USP4): A Molecular Docking Study. Molecules, 27(18), 5909. https://doi.org/10.3390/molecules27185909






