Abstract
Ophiorrhiza japonica Bl. is a traditional Chinese materia medica widely used to treat several diseases. Chemical and pharmacological studies on O. japonica have been carried out; however, neither of them has been fully explored. In this study, an array of compounds was isolated from the title plant, including a new anthraquinone, ophiorrhizaquinone A (1), three alkaloids 2–4 and seven other compounds 5–11 with diverse structural types. Additionally, compounds 2, 5, 7, 8, 10 and 11 were isolated from the genus of Ophiorrhiza for the first time. Antioxidant bioassays in vitro using DPPH and ABTS were performed, and the results showed that compound 3 exhibited modest antioxidant activity with IC50 values of 0.0321 mg/mL and 0.0319 mg/mL, respectively. An in silico study of PPARα agonistic activities of compounds 2 and 3 was conducted by molecular docking experiments, revealing that both of them occupied the active site of PPARα via hydrogen bonds and hydrophobic interactions effectively. This study enriched both the phytochemical and pharmacological profiles of O. japonica.
1. Introduction
Ophiorrhiza japonica Bl., belonging to the Rubiaceae family, is widely distributed in Southern China [1]. Its whole plant is commonly referred to as “she gen cao” in traditional Chinese medicine (TCM), and is used to treat bronchitis, rheumatic arthralgia, injuries, irregular menstruation, etc. [2]. Interestingly, previous phytochemical studies of O. japonica revealed that it is composed of mainly alkaloids [3,4,5,6]. Recently, Liu’s group [3] disclosed two monoterpenoid indole alkaloids possessing a novel spirocyclic ring system, ophiorrhines A and B, from the title species. Their continued phytochemical research led to two undescribed alkaloids, ophiorrhines F and G, which are key biogenetic intermediates of ophiorrhines A and B [4]. To the best of our knowledge, there are only two pharmacological studies of the chemical constituents from O. japonica, both of which revealed the potent immunosuppressive activities [3,4]. However, neither phytochemical nor pharmacological profiles of O. japonica have been fully explored. Therefore, more studies on the isolation and identification of novel compounds with potent biological activities are regarded as necessary, as only a few compounds have been reported.
Dozens of research works revealed that the pathogenesis of various diseases, such as cancer, diabetes and neurodegeneration, are relevant to the damages caused by reactive oxygen species (ROS) [7]. Numerous therapeutic strategies were applied for treating ROS-related diseases, of which the antioxidants played important roles in various disciplines due to their abilities to eliminate ROS directly or to balance redox status in vivo [8]. Not surprisingly, the usage of natural antioxidants becomes an attractive topic, owing to the advantages of wide sources, strong activities and good affinities with biological surroundings [8]. It is found that some compounds from the genus Ophiorrhiza exhibited antioxidant activities [6]. However, no antioxidant bioassays were reported for Ophiorrhiza japonica. As reported, the family of peroxisome proliferator-activated receptors (PPARs), which can regulate the ROS levels over a wide range, from inhibition to induction, were the most prominent targets [9]. Their involvements in the regulation of ROS production and degradation underlie the therapeutic effects. Therefore, the PPARα was selected, and the agonistic activity was studied via molecular docking experiments.
Therefore, this study aimed to explore the phytochemical constituents of O. japonica and evaluate its antioxidant activity in vitro and PPARα agonistic activity in silico. To this purpose, the antioxidant potentials of the isolates were evaluated by DPPH (1,1-diphenyl-2-picrylhydrazyl) and ABTS (2,2′-azinobis-3-ethylbenzthiazoline-6-sulfonic acid) assays. Herein, the isolation, structure elucidation and preliminary biological studies in vitro and in silico are described.
2. Results and Discussion
A phytochemical examination of the whole plant of O. japonica led to one new compound 1, along with 10 known compounds 2–11 (Figure 1). Their chemical structures were identified by comparison of their NMR and optical rotation data with those in the literature.
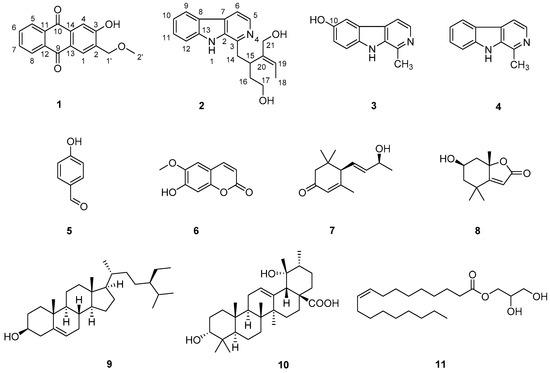
Figure 1.
The chemical structures of compounds 1–11 obtained from O. japonica.
Compound 1 was isolated as a yellow amorphous powder. Its molecular formula was established as C16H12O4 by a ion peak at m/z 267.0660 [M-H]− (calcd. 267.0663 for C16H11O4) in the HREIMS spectrum (Figure S1), indicating 11 degrees of unsaturation. The IR absorption at 3358 and 1658 cm−1 suggested the presence of hydroxyl and carbonyl groups (Figure S8). The 13C NMR, DEPT, and HSQC spectra (Figures S3 and S4) of 1 revealed 16 carbon signals, including one oxygenated methyl (δC 59.1), one oxygenated sp3 methylene (δC 74.0), six sp2 methines (δC 134.2, 133.9, 128.1, 127.3, 127.3, and 114.9), and eight sp2 nonprotonated carbons (six carbons at δC 162.0, 136.5, 133.8, 133.8, 128.2, and 126.5, two ketonic carbonyls at δC 183.0 and 182.3) (Table 1). Six double bonds and two carbonyls accounted for eight of the eleven degrees of unsaturation; the remaining three degrees of unsaturation indicated that a tricyclic ring system belonged to the structure. Its 1H NMR data (Table 1, Figure S2) revealed that compound 1 consist of an ortho-substituted benzene ring (ring A) at δH 8.29 (2H, overlapped) and 7.78 (2H, overlapped), and a 1,2,4,5-tetrasubstituted benzene ring (ring C) at δH 8.02 (1H, s) and 7.72 (1H, s). Herein, a quinone ring (ring B) could be supposed from the left one ring and two ketonic carbonyls. The presence of a quinone ring was further supported by the mutiple absorption maxima between 200 and 400 nm (Figure S9). These spectroscopic features were similar to those of a known compound 3-hydroxy-1-methoxy-2-methoxymethylanthraquinone (12) [10].

Table 1.
1H NMR (δH) and 13C NMR (δC) data of compound 1 in CDCl3.
The only difference was at C-1 position, where the methoxyl functionality in 12 was absent in 1. This replacement was consistent with their 30 mass units’ difference and evidenced by the HMBC correlations from H-1 (δH 8.02) to C-2 (δC 128.2), C-3 (δC 162.0), C-13 (δC 126.5) and C-14 (δC 136.5) (Figure S6). Due to the absence of the methoxyl group, the carbon chemical shift of C-1 (δC 128.1) was prominently shifted upfield (Δδ ≈ +32 ppm) compared to 12. Finally, the structure of 1 was unambiguously elucidated by 1H-1H COSY, HMBC and NOESY experiments (Figure 2, Figures S5–S7). The ortho-substituted benzene ring (ring A) in compound 1 could be deduced from the 1H-1H COSY correlations between the overlapped pairs of H-5/8 (δH 8.29) and H-6/7 (δH 7.78) and the HMBC correlations between the overlapped pairs of H-5/H-8 and C-11/C-12 (δC 133.8). The 1,2,4,5-tetrasubstituted benzene ring (ring C) was indicated by the HMBC correlations from H-1 (δH 8.02) to C-2 (δC 128.2), C-3 (δC 162.0), C-13 (δC 126.5) and C-14 (δC 136.5), and from H-4 (δH 7.72) to C-3 and C-14. The mutual HMBC correlations of H-1′ (δH 4.84)/C-2′ (δC 59.1) and H-2′ (δH 3.54)/C-1′ (δC 74.0) revealed the existence of a methoxymethyl chain. This side chain connected to ring C at C-2, indicating the diagnostic HMBC correlations from H-1′ to C-1 (δC 128.1), C-2 and C-3, which was further supported by the NOESY correlation of H-1′/H-1. A hydroxyl group was suggested at C-3 by the HMBC correlations from OH (δH 8.53) to C-2, C-3 and C-4 (δC 114.9), which was further supported by the NOESY correlation of OH/H-4. The characteristic HMBC correlations of H-5/C-11, H-5/C-10 (δC 183.0), H-8/C-12, H-8/C-9 (δC 182.3), H-4/C-10, H-4/C-14, H-1/C-9 and H-1/C-13 revealed the formation of a quinone ring (ring B). Thus, the structure of 1 was determined as shown in Figure 1.
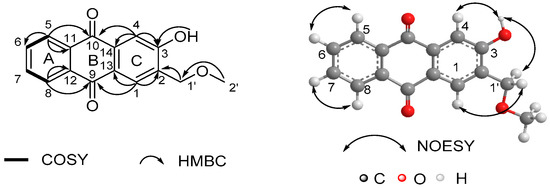
Figure 2.
1H-1H COSY, key HMBC and NOESY correlations of compound 1.
The structures of 10 known compounds 2–11 were established by comparison of their NMR and optical rotation data with those in the literature, such as deppeaninol (2) [11], 6-hydroxyharman (3) [12], harman (4) [13], p-hydroxy benzaldehyde (5) [14], scopoletin (6) [15], 3-oxo-α-ionol (7) [16], loliolide (8) [17], β-sitosterol (9) [18], 3-epi-pomolic acid (10) [10] and 1-monoolein (11) [19], respectively. It may be worth pointing out that compounds 2, 5, 7, 8, 10 and 11 were isolated from the genus of Ophiorrhiza for the first time. Among them, 1 was an anthraquinone, 2–4 were β-carboline-type monoterpenoid indole alkaloids, 5 was a benzene derivative, 6 was a coumarin, 7 and 8 were norsesquiterpenes, 9 was a steroid, 10 was a ursane-type triterpene, and 11 was a lipid. This finding enriched the phytochemical profile of O. japonica.
All the isolated compounds from O. japonica were evaluated for their antioxidant activity by DPPH and ABTS assays. The results revealed that only compounds 2 and 3 exhibited radical scavenging abilities, respectively. Compound 2 exhibited moderate antioxidant activity with IC50 values of 2.70 mg/mL and 2.00 mg/mL, respectively. And compound 3 possessed the best antioxidant activity with IC50 values of 0.0321 mg/mL and 0.0319 mg/mL, while the positive control vitamin C had an IC50 value of 0.0154 mg/mL and 0.0164 mg/mL, respectively. By comparing the structures of the isolated alkaloids 3 and 4, it seemed that the hydroxyl group at C-10 influenced the antioxidant activity, which donate a hydrogen atom to neutralize the free radicals, hence enhancing the oxidation potentials [20]. The comparison of 2 and 4 revealed that the more complex functionality at C-3 could help improve the antioxidant potency.
The in silico study of PPARα agonistic activities of compounds 2 and 3 was conducted preliminarily by the molecular docking experiments, using the highly resolved PPARα crystal structure (PDB: 5HYK with a resolution of 1.83 Å). The molecular docking study revealed that compounds 2 and 3 occupied the same region as the ligand 65W, which was identified as a novel PPARα pan-agonist (also termed AL29-26). Figure 3 displays the binding modes of the isolated compounds with 5HYK.
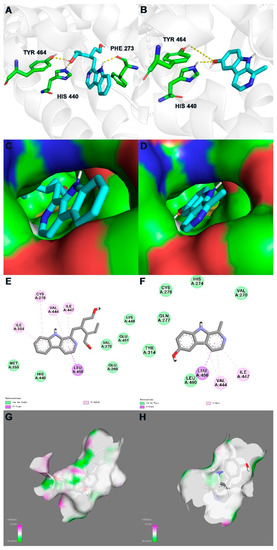
Figure 3.
In silico binding mode of 2 and 3 at PPARα crystal structure 5HYK: first row—the transparent protein surface, in light grey color, and two compounds shown as sticks with atoms colored C cyan, N blue, O red, and H white, are shown to emphasize the clear combination of hydrogen bonds within the target pocket; second row—surfaces of 5HYK with combined compounds; third row—two-dimensional ligand interaction diagrams of two compounds at the PPARα domain; fourth row—receptor surface of H-bond. Left list (A,C,E,G) represents docking results of 2; Right list (B,D,F,H) represents docking results of 3.
For compound 3, its C-10 hydroxy participated in hydrogen bonds with His440 and Tyr464, which were laying in the active site (Figure 3B,D). Discovery Studio Visualizer showed the pyrrole ring and pyridine ring of compound 3 occupied the hydrophobic pocket, which promoted π-σ and π-alkyl interactions with Val444, Ile447 and Leu456 (Figure 3F). For compound 2, except for the same hydrogen bonds and hydrophobic interactions as compound 3, H-1 and the benzene ring also participated in the interaction with Phe273, Cys276 and Ile354 (Figure 3A,C,E). The lower binding affinity of compound 2, compared to that of compound 3 (Table 2), could be deemed to be related to the substituent at C-15 in an environment without an H-bonds donor or acceptor, which may reduce the binding stability with 5HYK (Figure 3G,H).

Table 2.
In silico molecular docking binding affinities of compounds 2 and 3 to PPARα crystal structure (PDB: 5HYK).
3. Materials and Methods
3.1. General Experimental Procedures
NMR spectra were measured on a Bruker DRX-500 or Bruker DRX-600 spectrometer (Bruker Biospin AG, Fällanden, Germany). The LREIMS and HREIMS data were recorded on a Finnigan-MAT-95 mass spectrometer (Finnigan-MAT, San Jose, CA, USA). Commercial silica gel (200–300 and 300–400 mesh, Qingdao Haiyang Chemical Group Co., Ltd., Qingdao, China), Sephadex LH-20 gel (Amersham Biosciences, Amersham, UK) were used for column chromatography, and precoated silica gel plates (GF-254, Yan Tai Zi Fu Chemical Group Co., Yantai, China) were used for analytical TLC. All solvents used for column chromatography were of analytical grade (Shanghai Chemical Reagents Co., Ltd., Shanghai, China)
3.2. Plant Material
O. japonica was collected from the hilly areas of Changning City, Hunan Province, China, and authenticated by Prof. Lei Wu of Central South University of Forestry and Technology (CSUFT). A botanical specimen (WL9962) was deposited at the herbarium of CSUFT.
3.3. Extraction and Isolation
The dried and powdered whole plant materials (1.0 kg) were extracted by maceration with ethanol (3 times, 7 days/time) at room temperature. The ethanol extract was evaporated under reduced pressure to give a dark residue. The ethanol extract was then suspended in water for liquid–liquid extraction and successively extracted with petroleum ether (P), ethyl acetate (E) and chloroform (C) to obtain their corresponding fractions.
The chloroform extract was subjected to silica gel column chromatography (200–300 mesh) by eluting with PE-EA at a ratio of 50:50 to 0:100 and obtained four fractions (Fr. C1–C4). Fr. C3 was purified by silica gel column chromatography (300–400 mesh, dichloromethane (D):methanol (M) 15:1) to yield compound 4 (45.9 mg). Fr. C4 was divided into two sub-fractions (Fr. C4A and C4B) by silica gel column chromatography eluted with D:M (25:1, 15:1). Fr. C4A was further purified by silica gel column chromatography (D:M 22:1) to afford compound 2 (49.7 mg). Similarly, compound 3 (18.7 mg) was obtained from Fr. C4B by chromatographing over silica gel eluted with D:M (10:1).
The ethyl acetate extract was fractioned by silica gel column chromatography, eluted with gradient P-E mixture (75:25 to 0:100) to afford five fractions (Fr. E1–E5). Compound 1 (3.7 mg) was obtained from Fr. E1 by chromatographing over silica gel eluted with 100 % D. Fr. E3 was purified by silica gel column chromatography (D:E 17:1) to yield compound 5 (8.7 mg). Two sub-fractions (Fr. E5A and E5B) were obtained from Fr. E5 by silica gel column chromatography eluted with D:M (10:1 to 0:1). Fr. E5A was further purified by silica gel column chromatography (P:1 1:1) to afford compound 6 (3.0 mg). Compounds 7 (4.1 mg) and 8 (9.7 mg) were obtained from Fr. E5B by chromatographing over Sephadex LH-20 gel eluted with P:D:M (2:1:1).
The petroleum ether extract was separated by silica gel column chromatography, eluted with gradient P-E mixture (100:0 to 0:100), yielding seven fractions (Fr. P1–P7). Fr. P5 was purified by silica gel column chromatography (P:E 8:1) to give compound 9 (48.1 mg). Similarly, compounds 10 (54.0 mg) and 11 (6.3 mg) were afforded from Fr. P5 and Fr. P6 by repeated silica gel column chromatography eluted with D:M (25:1 to 15:1) and D:M (16:1), respectively.
Ophiorrhizaquinone A (1): yellow amorphous powder; UV (MeOH) λmax (log ε): 205 (0.439), 239 (0.303), 243 (0.301), 273 (0.585) nm; IR (KBr) νmax: 3443, 3358, 2922, 2851, 1658, 1632, 1583, 1572, 1344, 1122 cm−1; 1H (CDCl3, 600 MHz) and 13C (CDCl3, 125 MHz) NMR data, see Table 1; HRESIMS m/z 267.0600 [M − H]− (calcd. for C16H11O4, 267.0663).
3.4. In Vitro Antioxidant Assays
The 1,1-diphenyl-2-picrylhydrazyl (DPPH) scavenging capability of the isolates was tested as described in [21]. As standard, vitamin C was used, with methanol as a blank. Similarly, the 2,2′-azino-bis-3-ethylbenzthiazoline-6-sulfonic acid (ABTS) scavenging capability of the isolates was tested as described in [22] using vitamin C for standard calibration. Ethanol was used as a blank. The percentage inhibition was deliberated using the formula below, and the results were expressed as IC50:
where Ac = absorption of the control, and As = absorption of the isolate.
% inhibition = 100 × (Ac − As)/Ac
3.5. Molecular Docking
The cocrystal structure of PPARα (PDB code 5HYK) was obtained from RCSB Protein Data Bank. The water molecules were removed in Autodock, while the binding site of compounds 2 and 3 on the PPARα receptor was consistent with that of 65 W. Moreover, the center of combined area was x = 8.938, y = 34.766, z = −17.821, spacing 0.375 (size x = 13.5; y = 13.5; z = 15.0). Using hydrogens and charges tool in edit, the receptor was prepared for docking. Ligands were sketched in ChemBioDraw program and uploaded to Autodock. Furthermore, ligands were obtained through ‘hydrogens’. The molecular docking module (Autodock Vina) was used for docking, and the simulation with the highest affinity energy which showed hydrogen bond interaction with the active site was analyzed and visualized in Pymol and Discovery Studio Visualizer.
4. Conclusions
In summary, a detailed chemical investigation of O. japonica led to the identification of an array of structurally diverse compounds, including one new anthraquinone (1), three β-carboline-type alkaloids (2–4), one benzene derivative (5), one coumarin (6), two norsesquiterpenes (7 and 8), one steroid (9), one ursane-type triterpene (10) and one lipid (11). All the isolates were evaluated for their antioxidant activities. Among them, compound 3 exhibited the best antioxidant capabilities in the DPPH and ABTS assays, which was similar to that of the positive control vitamin C. Preliminary analysis of the structures of the monoterpenoid indole alkaloids 2–4 revealed the hydroxyl group at C-10 and the substituent at C-3 could be crucial in this compound class for the enhancement of the antioxidant potency. With the assistance of molecular docking analysis, the binding modes of compounds 2 and 3 on PPARα were analyzed, revealing that both of them occupied the active site of PPARα via hydrogen bonds and hydrophobic interactions effectively. The discovery of these compounds of different types as well as their bioactive studies expanded the diversity of phytochemical and pharmacological profiles of O. japonica.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/molecules27165301/s1, Figure S1: HRESIMS spectrum of compound 1; Figure S2: 1H NMR spectrum (600 MHz) of compound 1 in CDCl3; Figure S3: 13C NMR (BB + DEPT) spectrum (125 MHz) of compound 1 in CDCl3; Figure S4: HSQC spectrum (600 MHz) of compound 1 in CDCl3; Figure S5: 1H–1H COSY spectrum (600 MHz) of compound 1 in CDCl3; Figure S6: HMBC spectrum (600 MHz) of compound 1 in CDCl3; Figure S7: NOESY spectrum (600 MHz) of compound 1 in CDCl3; Figure S8: IR spectrum of compound 1; Figure S9: UV spectrum of compound 1.
Author Contributions
Conceptualization, L.-F.L.; methodology, Q.B., Y.J. and M.-J.X.; software, Y.J.; validation, Q.B., Y.J. and M.-J.X.; formal analysis, Q.B., Y.J. and M.-J.X.; investigation, Q.B., Y.J. and M.-J.X.; resources, L.W.; data curation, Q.B., Y.J. and M.-J.X.; writing—original draft preparation, Q.B. and Y.J.; writing—review and editing, L.-F.L.; visualization, Q.B., Y.J. and L.-F.L.; supervision, L.-F.L.; project administration, L.W. and L.-F.L.; funding acquisition, L.W. and L.-F.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (No. 41876194) and the Scientific Research Fund of Hunan Provincial Education Department (18B178).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We thank Y.-W. Guo’s lab at Shanghai Institute of Materia Medica, Chinese Academy of Sciences, for the assistance in measuring MS, IR and NMR spectra.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Flora of China Editorial Committe. Ophiorrhiza japonica Bl. In Flora of China; Science Press: Beijing, China, 1999; Volume 71, p. 163. [Google Scholar]
- Chinese Materia Medica Committee of the State Administration of Traditional Chinese Medicine. Shegencao. In Chinese Materia Medica; Shanghai Science and Technology Press: Shanghai, China, 1999; Volume 18, p. 458. [Google Scholar]
- Feng, T.; Duan, K.-T.; He, S.-J.; Wu, B.; Zheng, Y.-S.; Ai, H.-L.; Li, Z.-H.; He, J.; Zuo, J.-P.; Liu, J.-K. Ophiorrhines A and B, two immunosuppressive monoterpenoid indole alkaloids from Ophiorrhiza japonica. Org. Lett. 2018, 20, 7926–7928. [Google Scholar] [CrossRef]
- Shi, B.-b.; Ai, H.-L.; Duan, K.-T.; Feng, T.; Liu, J.-K. Ophiorrhines F and G, key biogenetic intermediates of ophiorrhine alkaloids from Ophiorrhiza japonica and their immunosuppressant activities. J. Nat. Prod. 2022, 85, 453–457. [Google Scholar] [CrossRef]
- Kitajima, M. Chemical studies on monoterpenoid indole alkaloids from medicinal plant resources Gelsemium and Ophiorrhiza. J. Nat. Med. 2007, 61, 14–23. [Google Scholar] [CrossRef]
- Taher, M.; Shaari, S.S.; Susanti, D.; Arbain, D.; Zakaria, Z.A. Genus Ophiorrhiza: A review of its distribution, traditional uses, phytochemistry, biological activities and propagation. Molecules 2020, 25, 2611. [Google Scholar] [CrossRef] [PubMed]
- Pisoschi, A.M.; Pop, A.; Iordache, F.; Stanca, L.; Predoi, G.; Serban, A.I. Oxidative stress mitigation by antioxidants—An overview on their chemistry and influences on health status. Eur. J. Med. Chem. 2021, 209, 112891. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.-Q. Why natural antioxidants are readily recognized by biological systems? 3D architecture plays a role! Food Chem. 2022, 380, 132143. [Google Scholar] [CrossRef] [PubMed]
- Aleshin, S.; Reiser, G. Role of the peroxisome proliferator-activated receptors (PPAR)-α, β/δ and γ triad in regulation of reactive oxygen species signaling in brain. Biol. Chem. 2013, 394, 1553–1570. [Google Scholar] [CrossRef] [PubMed]
- Chiriboga, X.; Gilardoni, G.; Magnaghi, I.; Vita Finzi, P.; Zanoni, G.; Vidari, G. New anthracene derivatives from Coussarea macrophylla. J. Nat. Prod. 2003, 66, 905–909. [Google Scholar] [CrossRef] [PubMed]
- Kan-Fan, C.; Zuanazzi, J.A.; Quirion, J.-C.; Husson, H.-P.; Henriques, A. Deppeaninol, a new β-carboline alkaloid from Deppea blumenaviensis (Rubiaceae). Nat. Prod. Lett. 1995, 7, 317–321. [Google Scholar] [CrossRef]
- Herath, W.; Mikell, J.R.; Ferreira, D.; Khan, I.A. Microbial metabolites of harman alkaloids. Chem. Pharm. Bull. 2003, 51, 646–648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seki, H.; Hashimoto, A.; Hino, T. The 1H- and 13C-nuclear magnetic resonance spectra of harman. Reinvestigation of the assignments by one- and two-dimensional methods. Chem. Pharm. Bull. 1993, 41, 1169–1172. [Google Scholar] [CrossRef] [Green Version]
- Shubina, L.K.; Makar’eva, T.N.; Denisenko, V.A.; Stonik, V.A. 4-Hydroxybenzaldehyde from the Baikal sponge Lubomirskia baicalensis. Chem. Nat. Compd. 2005, 41, 93–94. [Google Scholar] [CrossRef]
- Li, Y.-H.; Luo, F.; Peng, S.-L.; Liang, J.; Ding, L.-S. A new dihydroisocoumarin from the rhizomes of Notopterygium forbesii. Nat. Prod. Res. 2006, 20, 860–865. [Google Scholar] [CrossRef] [PubMed]
- Yao, D.; Jin, M.; Zhang, C.; Luo, J.; Jiang, Z.; Zheng, M.; Cui, J.; Li, G. Chemical constituents of the leaves of Juglans mandshurica. Chem. Nat. Compd. 2016, 52, 93–95. [Google Scholar] [CrossRef]
- Chen, B.-N.; Yang, G.-E.; Li, J.-K.; Du, H.-J.; Li, Q.-S.; Zhang, Z.-M. Cytotoxic constituents from Viscum coloratum. Chem. Nat. Compd. 2009, 45, 547–549. [Google Scholar] [CrossRef]
- Li, W.-H.; Chang, S.-T.; Chang, S.-C.; Chang, H.-T. Isolation of antibacterial diterpenoids from Cryptomeria japonica bark. Nat. Prod. Res. 2008, 22, 1085–1093. [Google Scholar] [CrossRef]
- Xiao, C.J.; Zhou, X.L.; Han, B.Y.; Tian, X.Y.; Sun, J.Z.; Jiang, B. Chemical constituents from rhizome of Isodon adenantha. Nat. Prod. Res. Dev. 2013, 25, 333–337. [Google Scholar] [CrossRef]
- Muema, F.W.; Liu, Y.; Zhang, Y.; Chen, G.; Guo, M. Flavonoids from Selaginella doederleinii Hieron and their antioxidant and antiproliferative activities. Antioxidants 2022, 11, 1189. [Google Scholar] [CrossRef] [PubMed]
- Abbas, M.W.; Hussain, M.; Akhtar, S.; Ismail, T.; Qamar, M.; Shafiq, Z.; Esatbeyoglu, T. Bioactive compounds, antioxidant, anti-inflammatory, anti-cancer, and toxicity assessment of Tribulus terrestris—In vitro and in vivo studies. Antioxidants 2022, 11, 1160. [Google Scholar] [CrossRef] [PubMed]
- Rhimi, W.; Mohammed, M.A.; Zarea, A.A.; Greco, G.; Tempesta, M.; Otranto, D.; Cafarchia, C. Antifungal, antioxidant and antibiofilm activities of essential oils of Cymbopogon spp. Antibiotics 2022, 11, 829. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).