Which Extraction Solvents and Methods Are More Effective in Terms of Chemical Composition and Biological Activity of Alcea fasciculiflora from Turkey?
Abstract
:1. Introduction
2. Results and Discussion
2.1. Chemical Profiling
2.2. Hydroxybenzoic, Hydroxycinnamic Acids and Derivatives
2.3. Hexaric Acid Derivatives
2.4. Coumarins
2.5. Flavonoids
2.6. Cinnamic Acid Amides
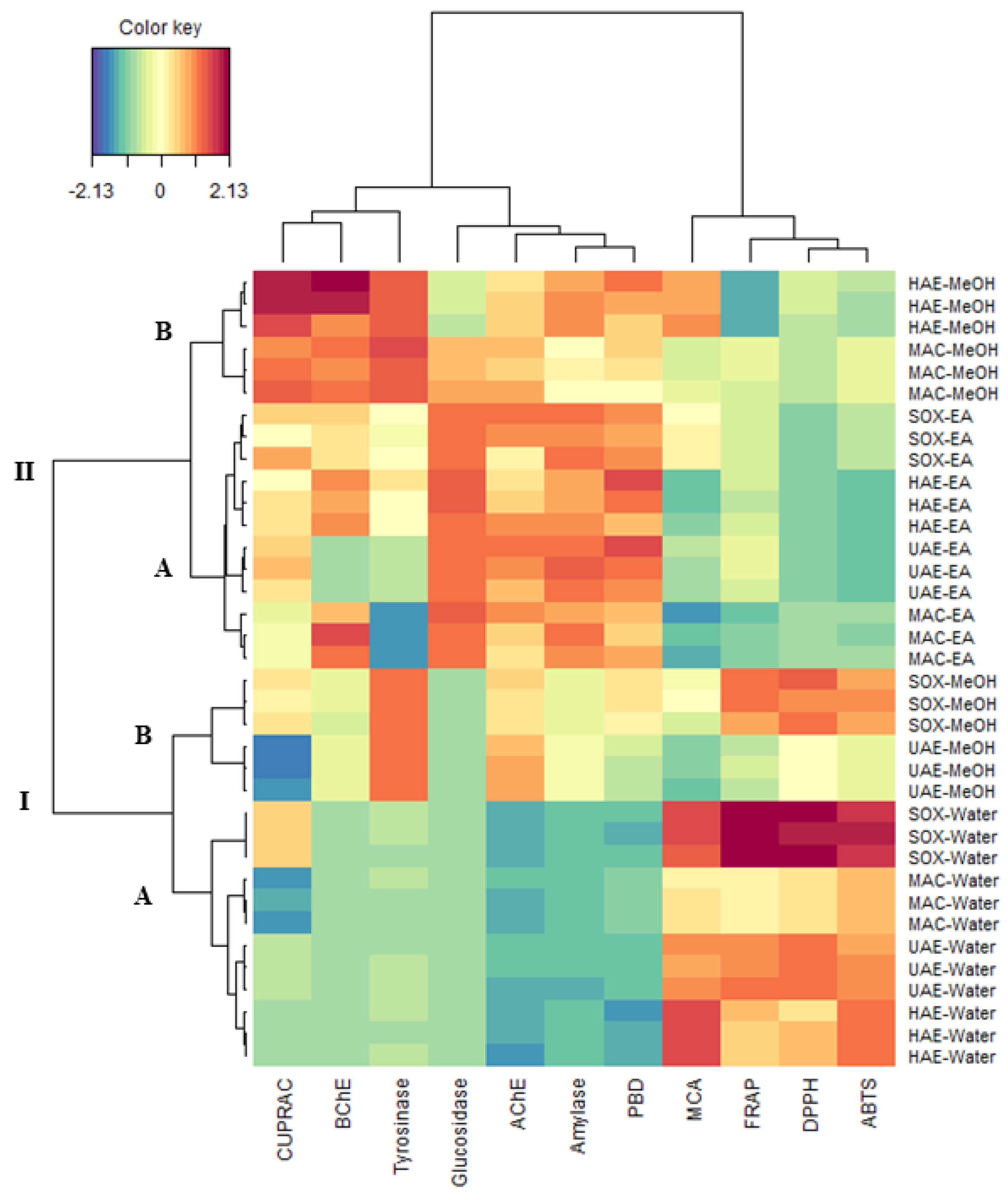
2.7. Chemometrics Analysis
2.8. Antioxidant Properties
2.9. Enzyme Inhibition Effects
2.10. Data Mining
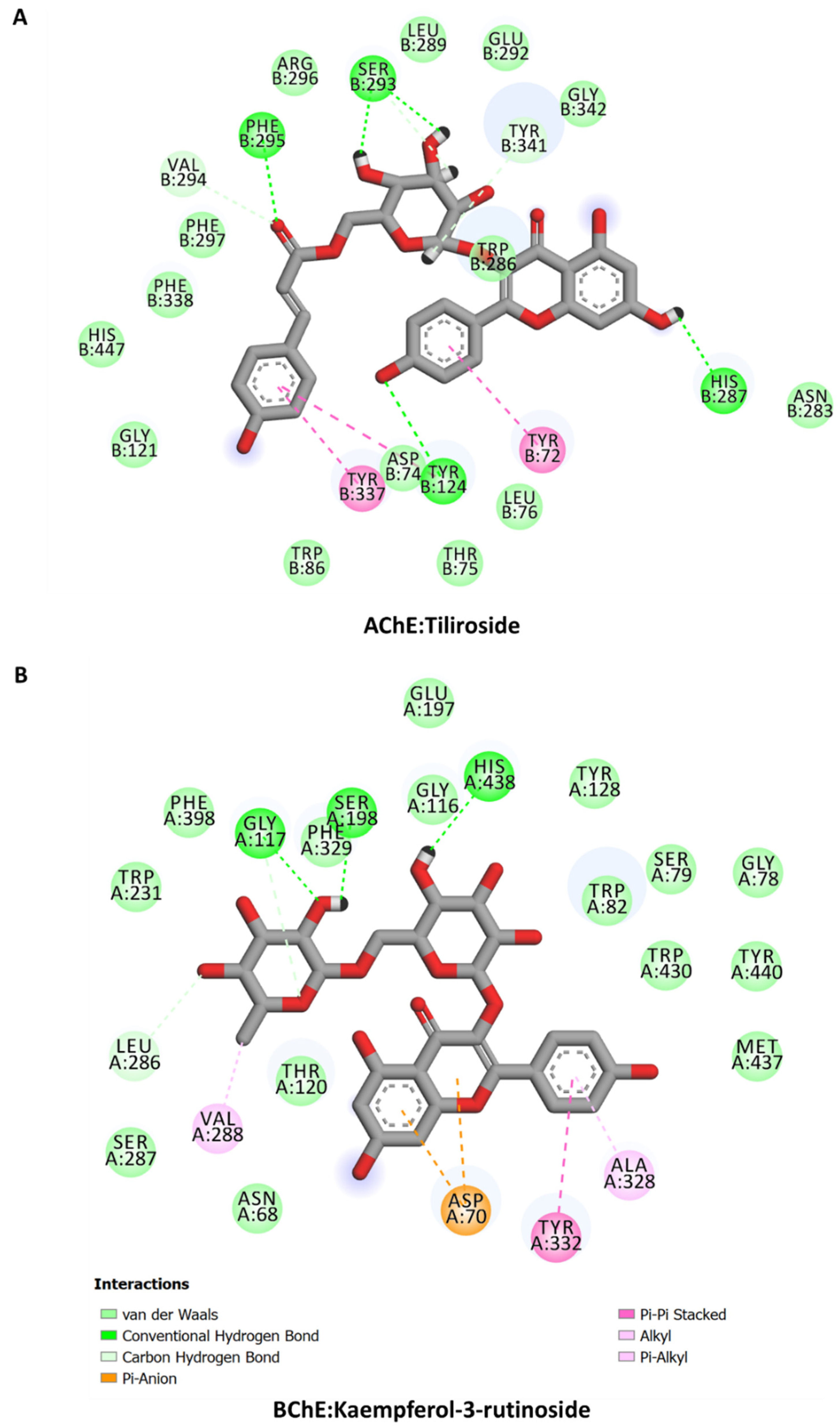
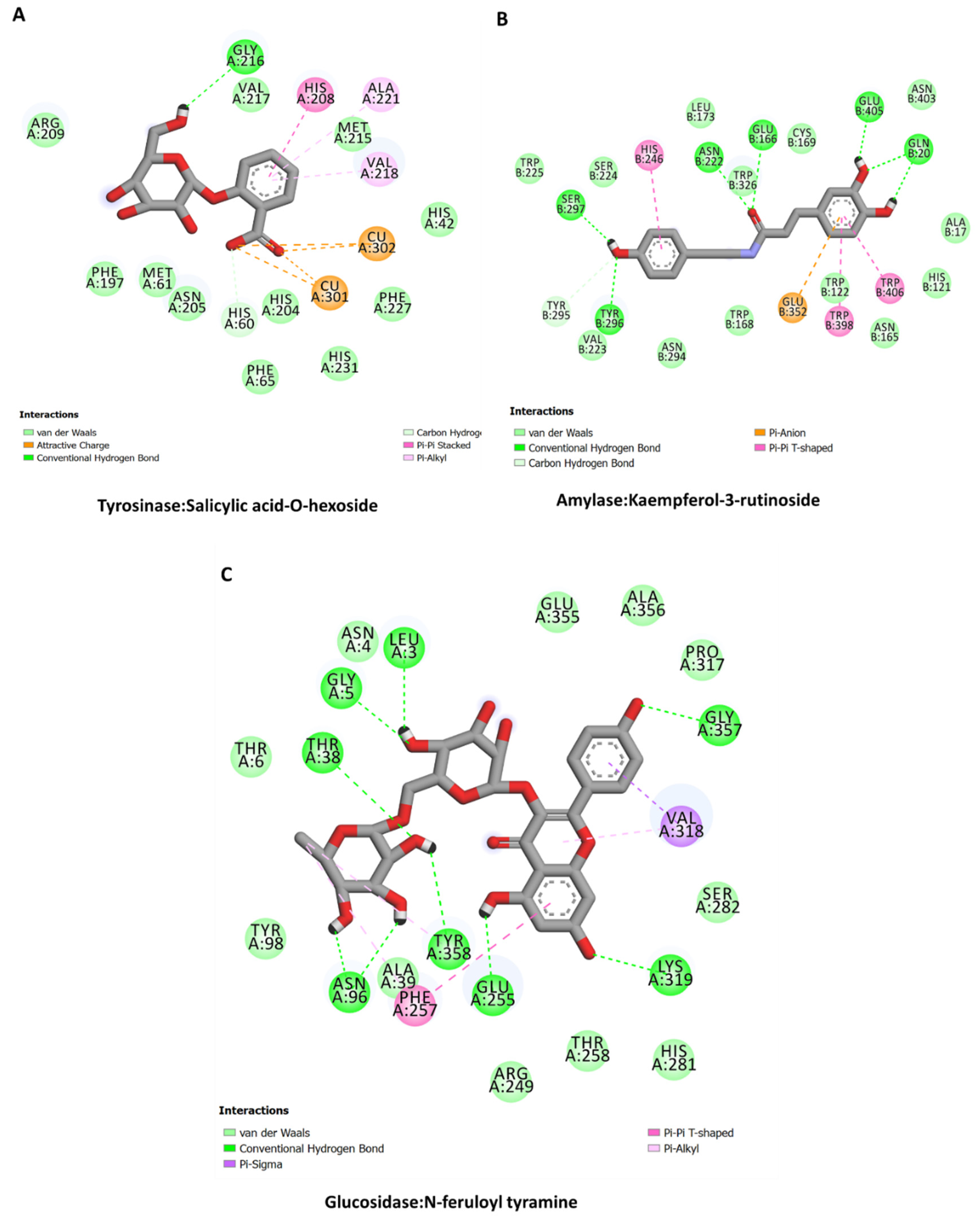
2.11. Molecular Docking
3. Materials and Methods
3.1. Plant Materials and Preparation of Extracts
3.2. Total Phenolic and Flavonoid Content
3.3. Ultra-High-Performance Liquid Chromatography Coupled with Hybrid Quadrupol-Orbitrap High Resolution Mass Spectrometry (UHPLC-HRMS)
3.4. Antioxidant Assays
3.5. Enzyme Inhibitory Assays
3.6. Molecular Modeling
3.7. Data Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Eshete, M.A.; Molla, E.L. Cultural significance of medicinal plants in healing human ailments among Guji semi-pastoralist people, Suro Barguda District, Ethiopia. J. Ethnobiol. Ethnomed. 2021, 17, 1–18. [Google Scholar] [CrossRef]
- Nadiroğlu, M.; Behçet, L.; Çakılcıoğlu, U. An ethnobotanical survey of medicinal plants in Karlıova (Bingöl-Turkey). Indian J. Tradit. Knowl. 2019, 18, 76–87. [Google Scholar]
- Azab, A. Alcea: Traditional medicine, current research and future opportunities. Eur. Chem. Bull. 2016, 5, 505–514. [Google Scholar]
- Ertas, A.; Boga, M.; Gazioglu, I.; Yesil, Y.; Hasimi, N.; Ozaslan, C.; Yilmaz, H.; Kaplan, M. Fatty acid, essential oil and phenolic compositions of alcea pallida and alcea apterocarpa with antioxidant, anticholinesterase and antimicrobial activities. Chiang Mai J. Sci. 2016, 43, 1143–1153. [Google Scholar]
- Pakravan, M. Alcea ilamica, A New Species from Iran. Rostaniha 2003, 4, 93–97. [Google Scholar]
- Uzunhisarcikli, M.E.; Vural, M. The taxonomic revision of Alcea and Althaea (Malvaceae) in Turkey. Turk. J. Bot. 2012, 36, 603–636. [Google Scholar] [CrossRef]
- Rezaeipour, N.; Jafari, F.; Rezaeizadeh, H.; Nasseri, M.; Kamalinejad, M.; Ghobadi, A.; Shamsipour, M.; Zargaran, A.; Ameri, A. Efficacy of a persian medicine herbal compound (Alcea digitata alef and Malva sylvestris l.) on prevention of radiation induced acute mucositis in patients with head and neck cancer: A pilot study. Int. J. Cancer Manag. 2017, 10, e8642. [Google Scholar] [CrossRef] [Green Version]
- Ahmadi, M.; Rad, A.K.; Rajaei, Z.; Mohammadian, N.; Tabasi, N.S. Alcea rosea root extract as a preventive and curative agent in ethylene glycol-induced urolithiasis in rats. Indian J. Pharmacol. 2012, 44, 304. [Google Scholar]
- Bussmann, R.; Glenn, A. Medicinal plants used in Peru for the treatment of respiratory disorders. Rev. Peru. Biol. 2010, 17, 331–346. [Google Scholar]
- Hanif, M.; Mehmood, M.H.; Ishrat, G.; Virji, S.N.; Malik, A.; Ahmed, M.; Gilani, A.-H. Pharmacological basis for the medicinal use of Alcea rosea in airways disorders and chemical characterization of its fixed oils through GC-MS. Pak. J. Pharm. Sci. 2019, 32, 2347–2355. [Google Scholar] [PubMed]
- Göç, F.; Erel, E.; Sarı, A. Plants used in Traditional Treatment for Boils in Turkey. Int. J. Tradit. Complement. Med. Res. 2021, 2, 49–61. [Google Scholar]
- Bajes, H.R.; Oran, S.A.; Bustanji, Y.K. Chemical composition and antiproliferative and antioxidant activities of methanolic extract of Alcea setosa A. Malvaceae. Res. J. Pharm. Technol. 2021, 14, 6447–6454. [Google Scholar] [CrossRef]
- Abdel-Salam, N.A.; Ghazy, N.M.; Sallam, S.M.; Radwan, M.M.; Wanas, A.S.; ElSohly, M.A.; El-Demellawy, M.A.; Abdel-Rahman, N.M.; Piacente, S.; Shenouda, M.L. Flavonoids of Alcea rosea L. and their immune stimulant, antioxidant and cytotoxic activities on hepatocellular carcinoma HepG-2 cell line. Nat. Prod. Res. 2018, 32, 702–706. [Google Scholar] [CrossRef]
- Alhage, J.; Elbitar, H. In vitro screening for antioxidant and antimicrobial properties of three lebanese medicinal plants crude extracts. Pharmacogn. Res. 2019, 11, 127–133. [Google Scholar]
- Anlas, C.; Ustuner, O.; Alkan, F.U.; Bakirel, T.; Aydogan, M.N.; Erel, S.B. Comparative study on the antioxidant activities and phenolic contents of different extracts of Achillea nobilis subsp. sipylea and Alcea apterocarpa (Fenzl) Boiss, endemic plants in Turkey. Fresenius Environ. Bull 2017, 26, 1423–1430. [Google Scholar]
- Azadeh, Z.; Saeidi, K.; Lorigooini, Z.; Kiani, M.; Maggi, F. Organ-oriented phytochemical profiling and radical scavenging activity of Alcea spp.(Malvaceae) from Iran. SN Appl. Sci. 2020, 2, 1–9. [Google Scholar] [CrossRef]
- Kartal, D.İ.; Neşe, E.; Öztürk, F. Van İlinin Çaldıran İlçesinde Yetişen Bazı Tıbbi Bitkilerin Toplam Fenolik ve Flavonoid Madde Miktarlarının Belirlenmesi. Kahramanmaraş Sütçü İmam Üniversitesi Tarım Ve Doğa Derg. 2020, 23, 611–616. [Google Scholar]
- Taşkin, T.; Kahvecİoğlu, D.; Türkoğlu, E.A.; Dogan, A.; Müslüm, K. In vitro Biological Activities of Different Extracts from Alcea dissecta. Clin. Exp. Health Sci. 2022, 12, 53–60. [Google Scholar] [CrossRef]
- Zakizadeh, M.; Nabavi, S.; Nabavi, S.; Ebrahimzadeh, M. In vitro antioxidant activity of flower, seed and leaves of Alcea hyrcana Grossh. Eur. Rev. Med. Pharmacol. Sci. 2011, 15, 406–412. [Google Scholar] [PubMed]
- Kłeczek, N.; Michalak, B.; Malarz, J.; Kiss, A.K.; Stojakowska, A. Carpesium divaricatum Sieb. & Zucc. revisited: Newly identified constituents from aerial parts of the plant and their possible contribution to the biological activity of the Plant. Molecules 2019, 24, 1614. [Google Scholar]
- Zhou, L.; Li, J.; Yan, C. Simultaneous determination of three flavonoids and one coumarin by LC–MS/MS: Application to a comparative pharmacokinetic study in normal and arthritic rats after oral administration of Daphne genkwa extract. Biomed. Chromatogr. 2018, 32, e4233. [Google Scholar] [CrossRef] [PubMed]
- Voynikov, Y.; Zheleva-Dimitrova, D.; Gevrenova, R.; Lozanov, V.; Zaharieva, M.M.; Tsvetkova, I.; Najdenski, H.; Yagi, S.; Almoulah, N.F.; Momekov, G. Hydroxycinnamic acid amide profile of Solanum schimperianum Hochst by UPLC-HRMS. Int. J. Mass Spectrom. 2016, 408, 42–50. [Google Scholar] [CrossRef]
- Šamec, D.; Maretić, M.; Lugarić, I.; Mešić, A.; Salopek-Sondi, B.; Duralija, B. Assessment of the differences in the physical, chemical and phytochemical properties of four strawberry cultivars using principal component analysis. Food Chem. 2016, 194, 828–834. [Google Scholar] [CrossRef]
- Mohammad Salamatullah, A.; Hayat, K.; Mabood Husain, F.; Asif Ahmed, M.; Arzoo, S.; Musaad Althbiti, M.; Alzahrani, A.; Al-Zaied, B.A.M.; Kahlil Alyahya, H.; Albader, N. Effects of different solvents extractions on total polyphenol content, HPLC analysis, antioxidant capacity, and antimicrobial properties of peppers (red, yellow, and green (Capsicum annum L.)). Evid. Based Complement. Altern. Med. 2022, 2022, 7372101. [Google Scholar] [CrossRef]
- Lourenço, S.C.; Moldão-Martins, M.; Alves, V.D. Antioxidants of natural plant origins: From sources to food industry applications. Molecules 2019, 24, 4132. [Google Scholar] [CrossRef] [Green Version]
- Abeyrathne, E.D.N.S.; Nam, K.; Huang, X.; Ahn, D.U. Plant-and Animal-Based Antioxidants’ Structure, Efficacy, Mechanisms, and Applications: A Review. Antioxidants 2022, 11, 1025. [Google Scholar] [CrossRef] [PubMed]
- Stepankova, S.; Komers, K. Cholinesterases and cholinesterase inhibitors. Curr. Enzym. Inhib. 2008, 4, 160–171. [Google Scholar] [CrossRef]
- Ahmed, F.; Ghalib, R.M.; Sasikala, P.; Ahmed, K.M. Cholinesterase inhibitors from botanicals. Pharmacogn. Rev. 2013, 7, 121. [Google Scholar] [CrossRef] [Green Version]
- Barbosa Filho, J.M.; Medeiros, K.C.P.; Diniz, M.d.F.F.; Batista, L.M.; Athayde-Filho, P.F.; Silva, M.S.; da Cunha, E.V.; Almeida, J.R.; Quintans-Júnior, L.J. Natural products inhibitors of the enzyme acetylcholinesterase. Rev. Bras. De Farmacogn. 2006, 16, 258–285. [Google Scholar] [CrossRef] [Green Version]
- Mohammadi, R.; Zarei, M.; Ghobadi, S. Investigation and Determination of Acetylcholinesterase Inhibition by Methanol Extract of the Aerial Parts of Alcea kurdica (Schlecht.) Alef and Astragaluse glumaceus Bioss. J. Med. Plants 2016, 15, 54–62. [Google Scholar]
- Zolghadri, S.; Bahrami, A.; Hassan Khan, M.T.; Munoz-Munoz, J.; Garcia-Molina, F.; Garcia-Canovas, F.; Saboury, A.A. A comprehensive review on tyrosinase inhibitors. J. Enzym. Inhib. Med. Chem. 2019, 34, 279–309. [Google Scholar] [CrossRef] [Green Version]
- Souza, P.M.; Elias, S.T.; Simeoni, L.A.; de Paula, J.E.; Gomes, S.M.; Guerra, E.N.S.; Fonseca, Y.M.; Silva, E.C.; Silveira, D.m.; Magalhaes, P.O. Plants from Brazilian Cerrado with potent tyrosinase inhibitory activity. PLoS ONE 2012, 7, e48589. [Google Scholar] [CrossRef] [Green Version]
- Sim, Y.Y.; Tan, C.P.; Cheong, L.Z.; Nyam, K.L. Hibiscus cannabinus L. leaf and seed in cosmetic formulation: An integrated approach as antioxidant and melanogenesis inhibitor. Sustain. Mater. Technol. 2022, 33, e00457. [Google Scholar] [CrossRef]
- Mohan, S.; Prabhakaran, V.-S.; Narayanaswamy, R. In Silico Analysis of Cissus rotundifolia Constituents as Human Neutrophil Elastase (HNE), Matrix Metalloproteinases (MMP 2 and MMP 9), and Tyrosinase Inhibitors. Appl. Biochem. Biotechnol. 2022, 194, 232–245. [Google Scholar] [CrossRef]
- Güven, Z.B.; Dogan, Z.; Saracoglu, I.; Picot, L.; Nagatsu, A.; Basaran, A.A. Food plant with antioxidant, tyrosinase inhibitory and antimelanoma activity: Prunus mahaleb L. Food Biosci. 2022, 48, 101804. [Google Scholar] [CrossRef]
- Namjoyan, F.; Jahangiri, A.; Azemi, M.E.; Arkian, E.; Mousavi, H. Inhibitory effects of Physalis alkekengi L., Alcea rosea L., Bunium persicum B. Fedtsch. and Marrubium vulgare L. on Mushroom Tyrosinase. Jundishapur J. Nat. Pharm. Prod. 2015, 10, e23356. [Google Scholar] [CrossRef] [Green Version]
- Gong, L.; Feng, D.; Wang, T.; Ren, Y.; Liu, Y.; Wang, J. Inhibitors of α-amylase and α-glucosidase: Potential linkage for whole cereal foods on prevention of hyperglycemia. Food Sci. Nutr. 2020, 8, 6320–6337. [Google Scholar] [CrossRef]
- Zhao, Y.; Wang, M.; Zhang, J.; Xiong, C.; Huang, G. The mechanism of delaying starch digestion by luteolin. Food Funct. 2021, 12, 11862–11871. [Google Scholar] [CrossRef] [PubMed]
- Tadera, K.; Minami, Y.; Takamatsu, K.; Matsuoka, T. Inhibition of α-glucosidase and α-amylase by flavonoids. J. Nutr. Sci. Vitaminol. 2006, 52, 149–153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zengin, G.; Aktumsek, A. Investigation of antioxidant potentials of solvent extracts from different anatomical parts of Asphodeline anatolica E. Tuzlaci: An endemic plant to Turkey. Afr. J. Tradit. Complement. Altern. Med. 2014, 11, 481–488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uysal, S.; Zengin, G.; Locatelli, M.; Bahadori, M.B.; Mocan, A.; Bellagamba, G.; De Luca, E.; Mollica, A.; Aktumsek, A. Cytotoxic and enzyme inhibitory potential of two Potentilla species (P. speciosa L. and P. reptans Willd.) and their chemical composition. Front. Pharmacol. 2017, 8, 290. [Google Scholar] [CrossRef]
- Grochowski, D.M.; Uysal, S.; Aktumsek, A.; Granica, S.; Zengin, G.; Ceylan, R.; Locatelli, M.; Tomczyk, M. In vitro enzyme inhibitory properties, antioxidant activities, and phytochemical profile of Potentilla thuringiaca. Phytochem. Lett. 2017, 20, 365–372. [Google Scholar] [CrossRef]
- Gerlits, O.; Ho, K.-Y.; Cheng, X.; Blumenthal, D.; Taylor, P.; Kovalevsky, A.; Radić, Z. A new crystal form of human acetylcholinesterase for exploratory room-temperature crystallography studies. Chem. Biol. Interact. 2019, 309, 108698. [Google Scholar] [CrossRef] [PubMed]
- Rosenberry, T.; Brazzolotto, X.; Macdonald, I.; Wandhammer, M.; Trovaslet-Leroy, M.; Darvesh, S.; Nachon, F. Comparison of the Binding of Reversible Inhibitors to Human Butyrylcholinesterase and Acetylcholinesterase: A Crystallographic, Kinetic and Calorimetric Study. Molecules 2017, 22, 2098. [Google Scholar] [CrossRef] [Green Version]
- Ielo, L.; Deri, B.; Germanò, M.P.; Vittorio, S.; Mirabile, S.; Gitto, R.; Rapisarda, A.; Ronsisvalle, S.; Floris, S.; Pazy, Y.; et al. Exploiting the 1-(4-fluorobenzyl)piperazine fragment for the development of novel tyrosinase inhibitors as anti-melanogenic agents: Design, synthesis, structural insights and biological profile. Eur. J. Med. Chem. 2019, 178, 380–389. [Google Scholar] [CrossRef] [PubMed]
- Božić, N.; Rozeboom, H.J.; Lončar, N.; Slavić, M.Š.; Janssen, D.B.; Vujčić, Z. Characterization of the starch surface binding site on Bacillus paralicheniformis α-amylase. Int. J. Biol. Macromol. 2020, 165, 1529–1539. [Google Scholar] [CrossRef] [PubMed]
- Karade, S.S.; Hill, M.L.; Kiappes, J.L.; Manne, R.; Aakula, B.; Zitzmann, N.; Warfield, K.L.; Treston, A.M.; Mariuzza, R.A. N-Substituted Valiolamine Derivatives as Potent Inhibitors of Endoplasmic Reticulum α-Glucosidases I and II with Antiviral Activity. J. Med. Chem. 2021, 64, 18010–18024. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
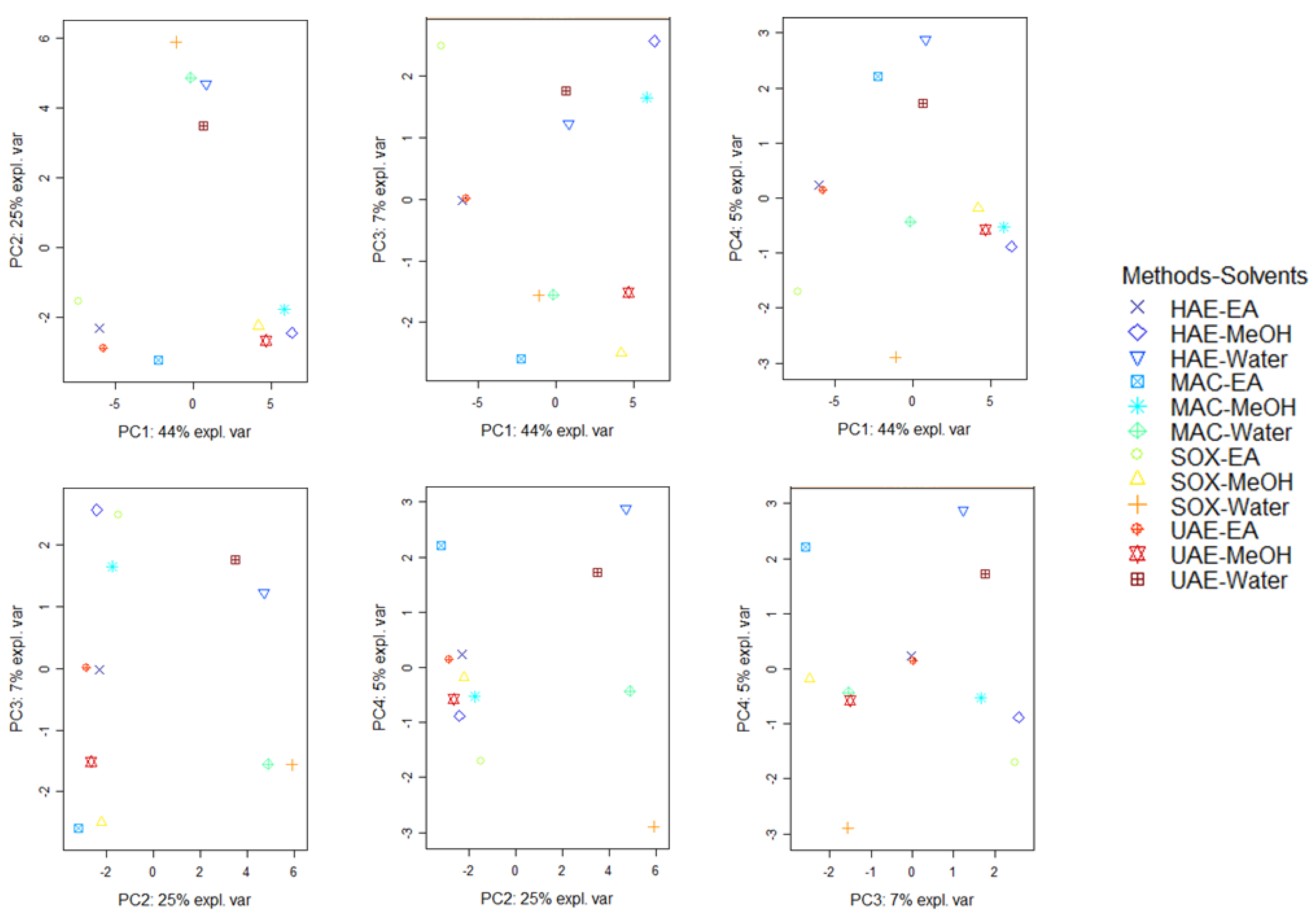
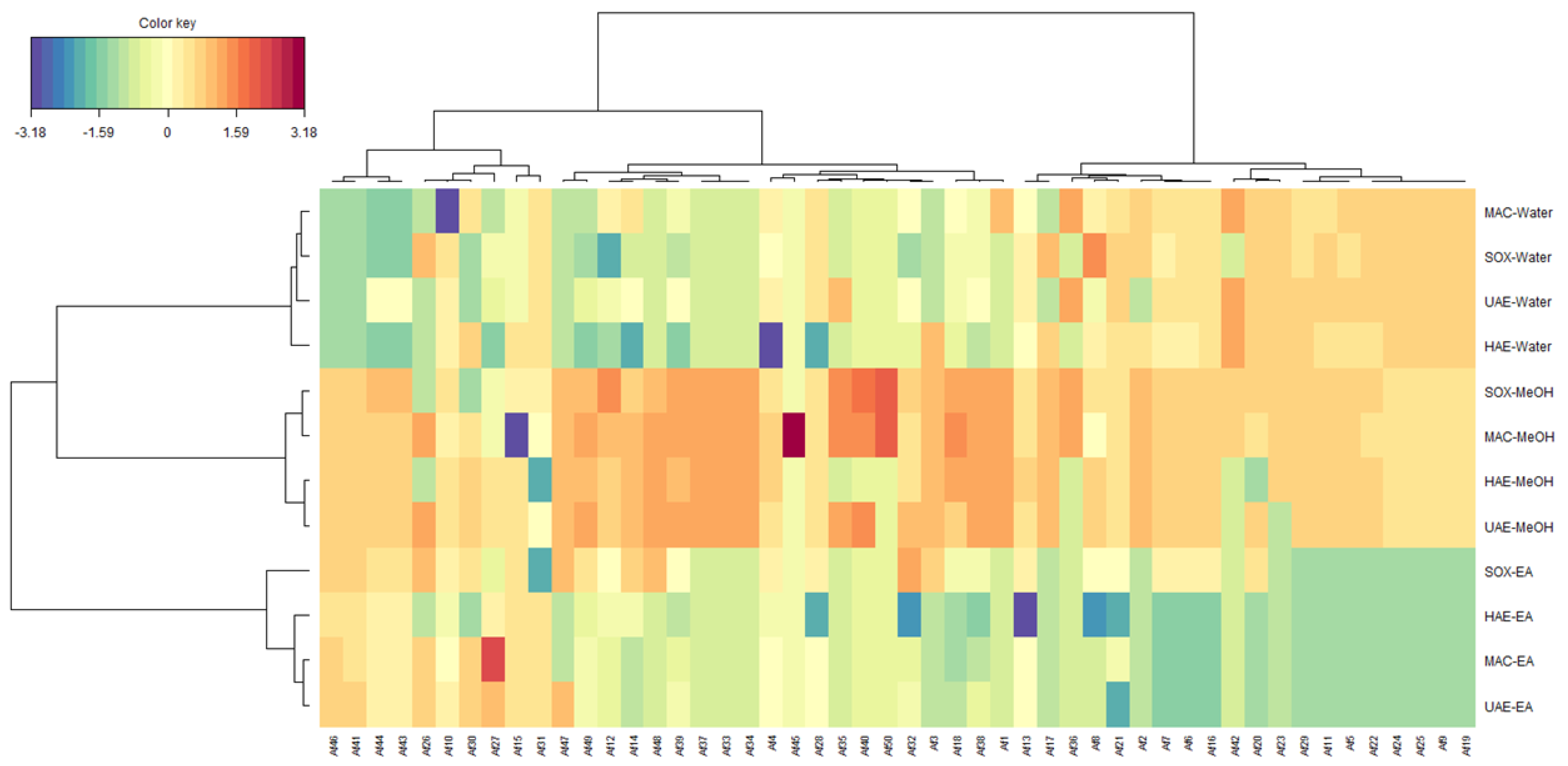
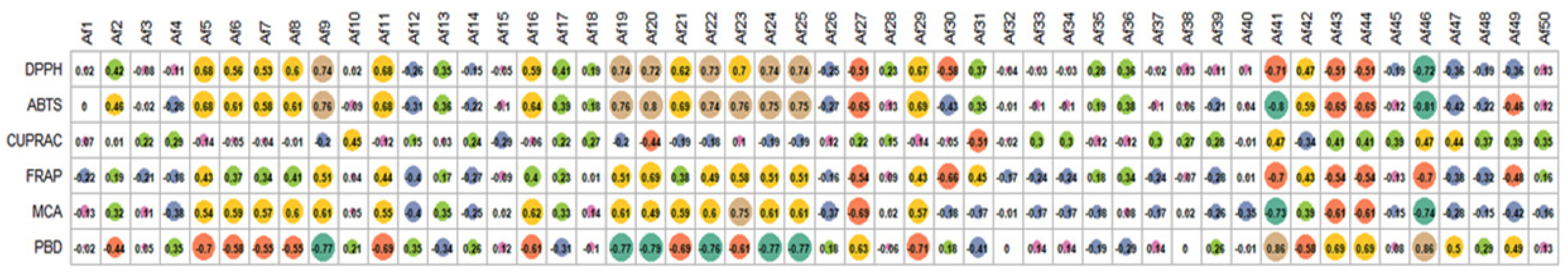
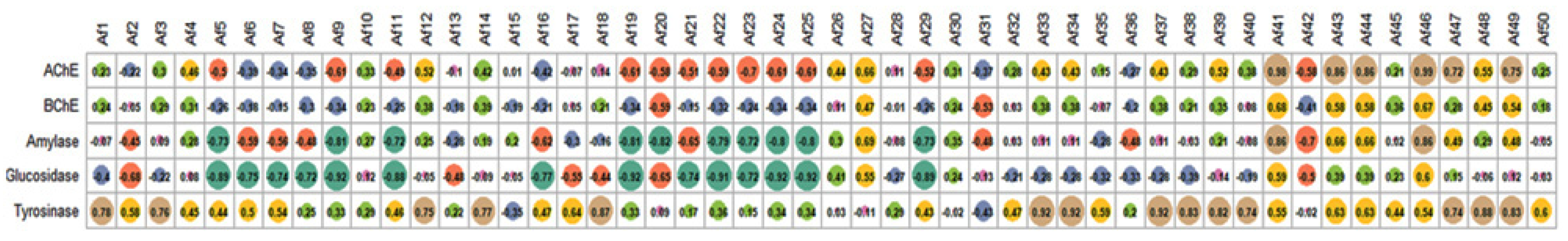

| Extraction Methods | Solvents | TPC (mg GAE/g) | TFC (mg RE/g) |
|---|---|---|---|
| MAC | EA | 20.53 ± 0.12 b | 1.99 ± 0.26 gh |
| MeOH | 15.34 ± 0.62 d | 3.26 ± 0.28 fgh | |
| Water | 18.75 ± 0.14 c | 4.43 ± 0.10 ef | |
| SOX | EA | 24.87 ± 1.13 a | 2.13 ± 0.05 gh |
| MeOH | 19.58 ± 0.23 bc | 25.26 ± 0.28 a | |
| Water | 23.70 ± 0.03 a | 6.02 ± 0.18 de | |
| HAE | EA | 20.61 ± 0.45 b | 1.68 ± 0.23 h |
| MeOH | 24.66 ± 0.26 a | 20.98 ± 1.61 b | |
| Water | 19.86 ± 0.02 bc | 3.55 ± 0.64 fg | |
| UAE | EA | 20.51 ± 0.29 b | 2.44 ± 0.48 gh |
| MeOH | 14.25 ± 0.19 d | 18.47 ± 0.35 c | |
| Water | 20.04 ± 0.21 b | 6.37 ± 0.14 d |
| No. (Af) | Identified/Tentatively Annotated Compound | Molecular Formula | Exact Mass [M-H]− | Fragmentation Pattern in (−) ESI-MS/MS | tR (Min) | Δ ppm | Distribution |
|---|---|---|---|---|---|---|---|
| Hydroxybenzoic, hydroxycinnamic acids and derivatives | |||||||
| 1. | gallic acid-O-hexoside | C13H16O10 | 331.0674 | 331.0674 (100), 313.0565 (6.63), 169.0130 (4.40), 16830054 (38.44), 125.0231 (35.01), 97.0281 (1.99) | 1.25 | 2.417 | 2,3,5,8,11 |
| 2. | protocatechuic acid-O-hexoside | C13H16O9 | 315.0725 | 315.0725 (100), 153.0183 (26.65), 152.0103 (58.78), 123.0075 (2.44), 108.0202 (75.18) | 1.68 | 1.126 | 2,3,5,6,8,9,11 |
| 3. | vanillic acid a | C8H8O4 | 167.0350 | 167.0336 (17.07), 152.0103 (100), 123.0071 (20.77), 108.0203 (48.42), 95.0122 (4.33) | 1.79 | −7.196 | 2,4,5,8,9,11 |
| 4. | vanillic alcochol-O-hexoside | C13H16O9 | 315.0729 | 315.0729 (59.95), 153.0545 (100), 135.0435 (0.25), 123.0437 (53.52), 109.0280 (49.09) | 2.15 | 2.395 | 1,2,3,5,6,7,8,10,11,12 |
| 5. | p-hydroxyphenylacetic acid-O-hexoside | C14H18O8 | 313.0938 | 313.0932 (100), 151.0389 (19.28), 123.0441 (4.41), 93.0329 (28.23) | 2.16 | 2.936 | 2,3,5,6,8,9,11,12 |
| 6. | syringic acid 4-O-hexoside | C15H20O10 | 359.0992 | 359.0989 (7.49), 197.0449 (100), 182.0213 (19.70), 153.0547 (16.59), 123.0074 (33.00) | 2.27 | 2.172 | 2,3,4,5,6,8,9,11,12 |
| 7. | caffeic acid-O-hexoside | C15H18O9 | 341.0881 | 341.0881 (4.51), 179.0341 (100), 135.0439 (58.82), 109.2430 (0.70) | 2.39 | 0.952 | 2,3,4,5,6,8,9,11,12 |
| 8. | protocatechuic acid a | C7H6O4 | 153.0183 | 153.0182 (16.31), 109.0280 (100), 91.0173 (1.33), 81.0331 (1.51) | 2.03 | −6.548 | 1,2,3,4,5,6,7,8,9,10,11,12 |
| 9. | vanillic alcohol | C8H10O3 | 153.0547 | 153.0545 (15.63), 137.0236 (0.46), 123.0437 (100), 109.0279 (1.08), 95.0487 (1.82) | 2.14 | −6.648 | 2,3,4,5,6,8,9,11,12 |
| 10. | ferulic acid-hexoside | C16H20O9 | 355.1040 | 355.1040 (1.0), 193.0498 (100), 178.0262 (9.5), 149.0596 (21.5), 134.0360 (57.7) | 2.96 | 1.618 | 1,2,4,5,6,7,8,9,10,11,12 |
| 11. | salicylic acid-O-hexoside | C13H16O8 | 299.0774 | 299.0774 (100), 271.0774 (0.88), 137.0232 (20.46), | 3.00 | 0.399 | 1,2,3,4,5,6,8,9,11,12 |
| 12. | caffeic acid a | C9H8O4 | 179.0341 | 179.0341 (18.86), 135.0438 (100), 117.0336 (0.59), 107.0489 (1.37), 91.0538 (0.55) | 3.54 | −4.982 | 1,2,3,4,5,6,7,8,9,10,11,12 |
| 13. | ferulic acid-hexoside isomer | C16H20O9 | 355.1038 | 355.1038 (9.3), 193.0499 (100), 178.0262 (16.2), 149.0596 (12.0), 134.0360 (36.8) | 3.76 | 0.971 | 1,3,4,5,6,8,9,10,11,12 |
| 14. | m-coumaric acid a | C9H8O3 | 163.0390 | 163.0390 (7.97), 119.0488 (100), 93.0330 (0.99) | 4.55 | −6.608 | 1,2,3,4,5,6,7,8,9,10,11,12 |
| 15. | shikimic acid | C7H10O5 | 173.0455 | 173.0808 (46.78), 155.0697 (1.81), 129.0907 (5.60), 111.0800 (100) | 4.93 | −6.657 | 1,2,3,5,7,8,9,10,11,12 |
| 16. | ferulic acid a | C10H10O4 | 193.0501 | 193.0499 (28.83), 178.0262 (83.59), 149.0596 (29.31), 134.0360 (100) | 5.14 | −2.860 | 2,3,4,5,6,8,9,11,12 |
| 17. | o-coumaric acid a | C9H8O3 | 163.0390 | 163.0390 (8.41), 135.0438 (0.16), 119.0487 (100), 93.0330 (1.27) | 5.55 | −6.547 | 2,5,6,8,9,11 |
| 18. | salicylic acid a | C7H6O3 | 137.0233 | 137.0231 (10.65), 108.0203 (0.51), 93.0330 (100) | 6.29 | −8.519 | 1,2,3,4,5,6,7,8,9,10,11,12 |
| Hexaric acids | |||||||
| 19. | p-coumaroylhexaric acid | C15H16O10 | 355.0674 | 355.0674 (6.2), 209.0298 (22.9), 191.0190 (40.2), 163.0389 (3.6), 147.0286 (16.2), 129.0181 (9.5), 119.0488 (2.3), 111.0073 (5.1), 85.0279 (100) | 2.42 | 1.042 | 2,3,5,6,8,9,11,12 |
| 20. | p-coumaroylhexaric acid isomer | C15H16O10 | 355.0677 | 355.0677 (6.6), 209.0296 (23.6), 191.0189 (40.5), 163.0391 (3.0), 147.0287 (16.0), 129.0179 (10.0), 119.0488 (2.0), 111.0074 (4.5), 85.0279 (100) | 2.42 | 1.747 | 2,3,5,6,9,11,12 |
| 21. | p-coumaroylhexaric acid isomer | C15H16O10 | 355.0677 | 355.0677 (5.1), 209.0297 (25.1), 191.0190 (40.7), 163.0392 (3.8), 147.0287 (16.6), 129.0177 (7.7), 119.0488 (1.6), 111.0074 (4.1), 85.0279 (100) | 3.15 | −1.182 | 1,2,3,4,5,6,8,9,11,12 |
| 22. | feruloylhexaric acid | C16H18O11 | 385.0784 | 385.0784 (10.1), 209.0297 (9.9), 193.0500 (3.2), 191.0189 (51.0), 147.0284 (17.2), 134.0360 (2.9), 129.0180 (10.7), 111.0072 (3.3), 85.0279 (100) | 2.96 | 1.988 | 1,2,3,5,6,8,9,11,12 |
| 23. | feruloylhexaric acid isomer | C16H18O11 | 385.0774 | 385.0774 (7.1), 209.0294 (9.5), 193.0501 (2.3), 191.0190 (54.1), 147.0287 (18.9), 134.0362 (3.2), 129.0180 (10.5), 111.0074 (3.6), 85.0279 (100) | 3.18 | −0.635 | 2,3,5,6,8,9,12 |
| 24. | feruloylhexaric acid isomer | C16H18O11 | 385.0771 | 385.0771 (12.9), 209.0296 (12.5), 193.0500 (2.9), 191.0190 (50.6), 149.0232 (12.5), 134.0359 (2.8), 129.0180 (11.4), 85.0279 (100) | 3.38 | −1.386 | 2,3,5,6,8,9,11,12 |
| 25. | feruloylhexaric acid isomer | C16H18O11 | 385.0778 | 385.0778 (8.6), 209.0296 (10.9), 193.0500 (1.9), 191.0189 (57.6), 147.0287 (15.2), 134.0361 (4.6), 111.0073 (4.8), 85.0279 (100) | 3.61 | 0.482 | 2,3,5,6,8,9,11,12 |
| Coumarins | |||||||
| 26. | aesculin | C15H16O9 | 339.0729 | 339.0724 (23.13), 177.0183 (100), 149.0233 (1.33), 133.0281 (10.44), 105.0331 (3.72), 89.0381 (2.24) | 2.69 | 2.137 | 1,2,4,6,10,11 |
| 27. | aesculetin | C9H6O4 | 177.0186 | 177.0184 (100), 149.0233 (3.62), 133.0281 (21.03), 121.0281 (0.94), 105.0331 (10.06), 89.0381 (8.06) | 3.45 | −4.191 | 1,2,3,4,5,6,7,8,9,10,11,12 |
| 28. | fraxin | C16H16O10 | 369.0832 | 369.0830 (15.63), 207.0292 (100), 192.0056 (49.59), 164.0103 (1.53), 123.0073 (1.42) | 3.52 | 1.192 | 1,2,3,5,6,8,10,11,12 |
| 29. | umbelliferone | C9H6O3 | 161.0234 | 161.0233 (77.03), 133.0281 (100), 115.0178 (1.47), 105.0332 (2.02), 89.0379 (1.09) | 3.78 | −6.442 | 2,3,5,6,8,9,11,12 |
| 30. | fraxetin | C10H8O5 | 207.0294 | 207.0292 (28.75), 192.0055 (100), 164.0103 (3.60), 123.0075 (2.94) | 4.22 | −2.544 | 1,2,3,4,8,9,10,11 |
| 31. | coumarin | C9H6O2 | 145.0284 | 145.0283 (35.47), 117.0332 (100), 102.0545 (5.52), 89.0381 (1.73) | 4.52 | −7.604 | 1,2,3,5,6,7,9,10,11,12 |
| 32. | scopoletin | C10H8O4 | 191.0343 | 191.0342 (19.51), 176.0105 (100), 148.0154 (15.95), 104.0253 (18.79) | 5.05 | −3.465 | 1,2,3,4,5,6,7,8,9,10,11,12 |
| Flavonoids | |||||||
| 33. | myricetin-3-O-rutinoside | C27H30O17 | 625.1418 | 625.1418 (100), 317.0289 (18.42), 316.0226 (81.70), 287.0200 (16.61), 271.0251 (25.56), 178.9976 (4.46), 157.0022 (4.21), 137.0231 (0.78), 107.0127 (0.67) | 4.48 | 1.324 | 2,5,8,11 |
| 34. | myricetin-3-O-hexoside | C21H20O13 | 479.0839 | 479.0835 (100), 317.0297 (18.03), 316.0228 (89.81), 287.0201 (15.13), 271.0248 (21.32), 178.9977 (1.63), 151.0018 (4.89) | 4.58 | 1.683 | 2,5,8,11 |
| 35. | isorhamnetin 3-O-rutinoside a | C26H28O15 | 623.1618 | 623.1625 (100), 315.0492 (8.44), 314.0436 (70.10), 300.0261 (8.99), 299.0200 (54.91), 271.0251 (26.91), 255.0299 (3.40), 243.0297 (5.02), 227.0347 (3.24), 151.0024 (0.25) | 5.13 | 0.758 | 2,5,11,12 |
| 36. | patuletin-3-O-glucoside | C22H22O13 | 493.0997 | 493.0995 (100), 331.0416 (15.90), 330.0384 (48.73), 316.0223 (22.3), 315.0151 (33.33), 287.0205 (12.67), 271.0253 (4.11), 259.0246 (2.94), 243.0296 (3.30), 178.9979 (0.79), 139.0024 (2.11), 136.9866 (0.72), 151.0008 (0.45), 121.0275 (0.58) | 5.26 | 1.959 | 2,3,5,12 |
| 37. | isoquercitrin a | C21H20O12 | 463.0888 | 463.0888 (100), 301.0351 (41.40), 300.0277 (77.04), 271.0249 (36.72), 255.0298 (16.35), 227.0347 (2.25), 151.0023 (5.9), 121.0248 (1.25), 107.0124 (2.58) | 5.29 | 1.254 | 2,5,6,8,11 |
| 38. | kaempferol-3-O-rutinoside a | C27H30O15 | 593.1522 | 593.1518 (100), 285.0404 (99.85), 284.0327 (57.50), 255.0298 (42.81), 227.0347 (28.43), 151.0024 (2.61), 135.0078 (1.02), 107.0121 (2.54) | 5.64 | −0.107 | 1,2,3,4,5,7,8,9,10,11,12 |
| 39. | kaempferol-3-O-glucoside a | C21H20O11 | 447.0941 | 447.0938 (100), 285.0399 (23.82), 284.0329 (58.22), 255.0250 (36.72), 227.0347 (2.25), 151.0025 (2.12), 107.0125 (0.79) | 5.85 | −0.145 | 1,2,3,4,5,6,7,8,9,10,11,12 |
| 40. | quercetin-O-hexuronide | C21H18O13 | 477.0677 | 477.0677 (100), 301.0356 (100), 300.0273 (2.57), 227.0343 (2.11), 151.0032 (0.52), 107.0126 (0.34) | 6.85 | 0.600 | 2,5,11 |
| 41. | luteolin a | C15H10O6 | 285.0410 | 285.0406 (100), 151.0024 (4.29), 149.0229 (0.29), 133.0282 (22.21), 121.0281 (0.80), 107.0124 (3.33) | 7.57 | 2.065 | 1,2,4,5,7,8,10,11 |
| 42. | quercetin a | C15H10O7 | 301.0356 | 301.0356 (100), 273.0399 (0.97), 257.0470 (5.93), 178.9976 (22.06), 151.0025 (43.82), 149.0239 (1.38) 121.0281 (14.69), 107.0122 (12.18) | 7.63 | 0.578 | 2,3,5,9,12 |
| 43. | tiliroside a | C30H26O13 | 593.1301 | 593.1307 (100), 447.0959 (1.89), 285.0404 (79.06), 284.0327 (56.62), 255.0298 (38.90), 227.0345 (28.83), 151.0025 (4.18), 107.0123 (2.48), 135.0073 (1.59) | 7.68 | 0.010 | 1,2,4,5,7,8,10,11,12 |
| 44. | kaempferol-O-p-coumaroyl-O-hexoside | C30H26O13 | 593.1308 | 593.1307 (100), 447.0925 (2.51), 285.0404 (76.82), 284.0328 (61.03), 255.0298 (41.36), 227.0345 (28.72), 151.0021 (3.15), 107.0124 (2.27), 135.0077 (1.58) | 7.94 | 1.241 | 1,2,4,5,7,8,10,11,12 |
| 45. | naringenin | C15H12O5 | 271.0615 | 271.0615 (100), 151.0025 (68.67), 119.0488 (55.13), 107.0123 (18.70) | 8.56 | 1.082 | 2 |
| 46. | apigenin a | C15H10O5 | 269.0456 | 269.0456 (100), 225.0549 (1.28), 151.0024 (4.59), 149.0232 (5.01), 117.0331 (17.57), 107.0123 (5.21) | 8.63 | 0.273 | 1,2,4,5,7,8,10,11 |
| 47. | kaempferol a | C15H10O6 | 285.0409 | 285.0405 (100), 227.0348 (0.48), 151.0026 (1.69), 117.0331 (0.78), 107.0123 (0.82)135.0074 (0.4), 107.0124 (1.4) | 8.80 | 0.136 | 2,4,5,8,10,11 |
| Hydroxycinnamic acid amides | |||||||
| No. | Identified/tentatively annotated compound | Molecular formula | Exact mass [M + H]+ | Fragmentation pattern in (+) ESI-MS/MS | tR (min) | Δ ppm | Distribution |
| 48. | N-coumaroyl tyramine | C17H17 O3N | 284.1287 | 284.1275 (24.8), 147.0439 (100), 139.0908 (0.7), 121.0649 (38.0), 119.0493 (15.8), 103.0546 (6.5) | 6.95 | −2.076 | 2,4,5,8,11 |
| 49. | N-feruloyl tyramine | C18H19O4N | 314.1377 | 314.1380 (46.4), 177.0545 (100), 149.0596 (5.0), 145.0283 (30.9), 121.0649 (43.6), 103.0546 (7.9) | 7.22 | −2.179 | 1,2,3,4,5,6,7,8,9,10,11,12 |
| 50. | N-feruloyl-3-methoxytyramine | C19H21O5N | 344.1489 | 344.1487 (8.5), 177.0544 (100), 149.0596 (4.5), 145.0282 (29.9), 117.0336 (17.4) | 7.51 | −0.298 | 2,5 |
| Extraction Methods | Solvents | DPPH (mg TE/g) | ABTS (mg TE/g) | CUPRAC (mg TE/g) | FRAP (mg TE/g) | MCA (mg EDTAE/g) | PBD (mmol TE/g) |
|---|---|---|---|---|---|---|---|
| MAC | EA | 4.86 ± 0.51 gh | 21.40 ± 1.79 g | 56.11 ± 0.21 d | 19.53 ± 0.70 f | 18.28 ± 1.57 g | 1.88 ± 0.08 abc |
| MeOH | 7.26 ± 0.09 f | 31.26 ± 0.44 e | 72.89 ± 1.50 b | 24.35 ± 0.77 e | 26.78 ± 0.74 de | 1.67 ± 0.11 c | |
| Water | 17.65 ± 0.28 d | 54.18 ± 1.03 d | 42.26 ± 0.30 f | 28.21 ± 0.26 d | 33.54 ± 1.04 c | 1.07 ± 0.03 de | |
| SOX | EA | 3.37 ± 0.20 h | 24.84 ± 0.37 f | 64.13 ± 4.74 c | 23.75 ± 0.46 e | 32.54 ± 0.77 c | 1.98 ± 0.05 a |
| MeOH | 26.75 ± 1.40 b | 58.19 ± 1.71 c | 62.26 ± 1.12 c | 35.20 ± 1.03 b | 29.09 ± 2.39 d | 1.68 ± 0.03 bc | |
| Water | 35.33 ± 0.71 a | 76.27 ± 1.13 a | 64.13 ± 0.10 c | 42.58 ± 0.07 a | 45.67 ± 0.17 a | 0.96 ± 0.02 e | |
| HAE | EA | 2.63 ± 0.16 h | 14.45 ± 0.76 h | 61.00 ± 1.87 cd | 23.06 ± 0.62 e | 19.97 ± 0.97 g | 2.07 ± 0.18 a |
| MeOH | 7.15 ± 0.92 fg | 23.21 ± 0.80 fg | 78.32 ± 2.26 a | 17.51 ± 0.29 g | 40.05 ± 1.12 b | 1.93 ± 0.13 ab | |
| Water | 20.54 ± 1.87 c | 64.02 ± 0.85 b | 49.12 ± 0.33 e | 31.52 ± 0.63 c | 46.71 ± 0.20 a | 0.90 ± 0.03 e | |
| UAE | EA | 2.66 ± 0.07 h | 13.46 ± 0.65 h | 64.22 ± 1.58 c | 24.37 ± 0.93 e | 23.56 ± 0.51 ef | 2.12 ± 0.12 a |
| MeOH | 13.49 ± 0.15 e | 33.15 ± 0.45 e | 40.38 ± 1.25 f | 22.77 ± 0.72 e | 20.74 ± 1.25 fg | 1.25 ± 0.05 d | |
| Water | 26.80 ± 0.61 b | 58.70 ± 1.05 c | 50.55 ± 0.36 e | 35.09 ± 0.80 b | 40.76 ± 0.25 b | 1.03 ± 0.01 de |
| Extraction Methods | Solvents | AChE (mg GALAE/g) | BChE (mg GALAE/g) | Tyrosinase (mg KAE/g) | Amylase (mmol ACAE/g) | Glucosidase (mmol ACAE |
|---|---|---|---|---|---|---|
| MAC | EA | 2.44 ± 0.42 a | 4.61 ± 1.08 ab | 1.79 ± 0.16 e | 1.03 ± 0.06 ab | 0.73 ± 0.06 a |
| MeOH | 2.45 ± 0.19 a | 4.63 ± 0.23 ab | 58.93 ± 0.62 a | 0.65 ± 0.03 c | 0.56 ± 0.02 b | |
| Water | 0.33 ± 0.03 b | na | 16.15 ± 1.21 d | 0.14 ± 0.01 e | na | |
| SOX | EA | 2.58 ± 0.61 a | 3.04 ± 0.16 c | 30.26 ± 1.59 c | 1.07 ± 0.03 ab | 0.74 ± 0.01 a |
| MeOH | 2.17 ± 0.05 a | 1.17 ± 0.19 d | 54.44 ± 0.15 b | 0.50 ± 0.01 d | na | |
| Water | 0.21 ± 0.01 b | na | 18.12 ± 1.66 d | 0.12 ± 0.01 e | na | |
| HAE | EA | 2.33 ± 0.36 a | 4.09 ± 0.20 bc | 33.36 ± 3.20 c | 0.99 ± 0.03 b | 0.76 ± 0.04 a |
| MeOH | 2.23 ± 0.06 a | 5.80 ± 1.22 a | 57.23 ± 0.82 ab | 1.02 ± 0.04 ab | 0.11 ± 0.03 c | |
| Water | 0.17 ± 0.02 b | na | 17.28 ± 1.04 d | 0.14 ± 0.01 e | na | |
| UAE | EA | 2.80 ± 0.27 a | na | 17.85 ± 1.08 d | 1.11 ± 0.05 a | 0.73 ± 0.02 a |
| MeOH | 2.58 ± 0.13 a | 1.23 ± 0.11 d | 53.43 ± 0.67 b | 0.55 ± 0.02 d | na | |
| Water | 0.39 ± 0.09 b | na | 17.73 ± 1.65 d | 0.11 ± 0.01 e | na |
| S/N | Compound | AChE | BChE | Tyrosinase | Amylase | Glucosidase |
|---|---|---|---|---|---|---|
| Kcal/moL | ||||||
| 11 | Salicylic acid-O-hexoside | −9.43 | −10.53 | −9.48 | −6.96 | −10.05 |
| 16 | Ferulic acid | −6.25 | −8.31 | −8.80 | −3.76 | −6.76 |
| 18 | Salicylic acid | −5.57 | −7.70 | −8.60 | −3.08 | −4.41 |
| 31 | Coumarin | −7.90 | −4.65 | −2.69 | −3.62 | −6.32 |
| 38 | Kaempferol-3-rutinoside | −11.42 | −11.92 | −7.50 | −9.95 | −10.65 |
| 39 | Kaempferol-3-glucoside | −10.22 | −9.60 | −5.61 | −7.57 | −8.50 |
| 43 | Tiliroside | −13.91 | −10.63 | −5.64 | −6.99 | −4.55 |
| 44 | Kaempferol-O-p-coumaroyl-O-hexoside | −11.35 | −7.34 | −5.72 | −3.51 | −5.66 |
| 46 | Apigenin | −9.98 | −7.34 | −4.29 | −6.22 | −7.06 |
| 49 | N-feruloyl tyramine | −10.53 | −9.56 | −5.94 | −7.67 | −10.25 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ozturk, R.B.; Zengin, G.; Sinan, K.I.; Montesano, D.; Zheleva-Dimitrova, D.; Gevrenova, R.; Uba, A.I.; Çakılcıoğlu, U.; Kaplan, A.; Jugreet, S.; et al. Which Extraction Solvents and Methods Are More Effective in Terms of Chemical Composition and Biological Activity of Alcea fasciculiflora from Turkey? Molecules 2022, 27, 5011. https://doi.org/10.3390/molecules27155011
Ozturk RB, Zengin G, Sinan KI, Montesano D, Zheleva-Dimitrova D, Gevrenova R, Uba AI, Çakılcıoğlu U, Kaplan A, Jugreet S, et al. Which Extraction Solvents and Methods Are More Effective in Terms of Chemical Composition and Biological Activity of Alcea fasciculiflora from Turkey? Molecules. 2022; 27(15):5011. https://doi.org/10.3390/molecules27155011
Chicago/Turabian StyleOzturk, Refiye Beyza, Gokhan Zengin, Kouadio Ibrahime Sinan, Domenico Montesano, Dimitrina Zheleva-Dimitrova, Reneta Gevrenova, Abdullahi Ibrahim Uba, Uğur Çakılcıoğlu, Alevcan Kaplan, Sharmeen Jugreet, and et al. 2022. "Which Extraction Solvents and Methods Are More Effective in Terms of Chemical Composition and Biological Activity of Alcea fasciculiflora from Turkey?" Molecules 27, no. 15: 5011. https://doi.org/10.3390/molecules27155011
APA StyleOzturk, R. B., Zengin, G., Sinan, K. I., Montesano, D., Zheleva-Dimitrova, D., Gevrenova, R., Uba, A. I., Çakılcıoğlu, U., Kaplan, A., Jugreet, S., Dall’Acqua, S., & Mahomoodally, M. F. (2022). Which Extraction Solvents and Methods Are More Effective in Terms of Chemical Composition and Biological Activity of Alcea fasciculiflora from Turkey? Molecules, 27(15), 5011. https://doi.org/10.3390/molecules27155011












